Abstract
Bacterial endosymbionts, in genera Wolbachia and Cardinium, infect various arthropods and some nematode groups. Manipulating these microbial symbionts presents a promising biocontrol strategy for managing disease-causing parasites. However, the diversity of Wolbachia and Cardinium in nematodes remains unclear. This study employed a genome skimming strategy to uncover their occurrence in plant-parasitic nematodes, analyzing 52 populations of 12 species. A metagenome analysis revealed varying endosymbiont genome content, leading to the categorization of strong, weak, and no evidence for endosymbiont genomes. Strong evidence for Wolbachia was found in five populations, and for Cardinium in one population, suggesting a limited occurrence. Strong Wolbachia evidence was noted in Pratylenchus penetrans and Radopholus similis from North/South America and Africa. Heterodera glycines from North America showed strong Cardinium evidence. Weak genomic evidence for Wolbachia was observed in Globodera pallida, Meloidogyne incognita, Rotylenchus reniformis, Pratylechus coffeae, Pratylenchus neglectus, and Pratylenchus thornei; for Cardinium was found in G. pallida, R. reniformis and P. neglectus; 27/52 populations exhibited no endosymbiont evidence. Wolbachia and Cardinium presence varied within nematode species, suggesting non-obligate mutualism. Wolbachia and Cardinium genomes differed among nematode species, indicating potential species-specific functionality. This study advances knowledge of plant-parasitic nematode–bacteria symbiosis, providing insights for downstream eco-friendly biocontrol strategies.
1. Introduction
Advancements in sequencing technologies have revolutionized the field of applied microbiology, enabling researchers to explore complex microbial communities with unprecedented depth and precision. Among these cutting-edge approaches, genome skimming has emerged as a rapid and efficient tool for analyzing genomes of organisms and their associated microorganisms [1,2]. One of the key applications of genome skimming in applied microbiology lies in the investigation of microbial symbioses. Symbiotic relationships between microbes and their host organisms play a pivotal role in shaping various ecological processes and can have profound implications for human health, agriculture, and the environment. With the ability to efficiently and cost-effectively analyze the genetic material from diverse samples, genome skimming has enabled researchers to uncover previously unknown associations between microbes and their hosts [1]. Managing disease-causing parasites through the manipulation of their microbial partners offers a promising avenue for biological control (‘biocontrol’), potentially reducing the use of chemical controls. Plant-parasitic nematodes are ubiquitous in agricultural soils, and losses caused by plant-parasitic nematodes are estimated to be over USD 100 billion globally [3,4]. Over 4100 plant-parasitic nematode species have been described, and they are considered one of the most important agricultural pests [5]. The ‘top ten’ damaging plant-parasitic nematodes include the root knot nematode (Meloidogyne), cyst nematodes (Globodera and Heterodera), root-lesion nematode (Pratylenchus), burrowing nematode (Radopholus), bulb and stem nematode (Ditylenchus), pine-wilt nematode (Bursaphelenchus), reniform nematode (Rotylenchulus), dagger nematode (Xiphinema), potato rosary nematode (Nacobbus), and foliar nematode (Aphelenchoides) [6]. The most common plant-parasitic nematode control strategy is the use of fumigant and non-fumigant nematicides. Although nematicides offer effective strategies that are widely implemented around the world, these compounds can also be toxic and cause damage to human health and the environment [7,8]. These limitations of nematicide-based management have motivated a search for alternative methods for nematode management such as cultural and biological control.
Managing plant-parasitic nematodes through the manipulation of their microbial symbionts is an appealing biocontrol strategy to combat plant-parasitic nematodes. The development of symbiont-dependent biocontrol strategies depends on the knowledge of functional relationships between specific symbionts and their hosts. Simply put, if the microbial symbionts have parasitic effects that reduce the fitness of target host, then the biocontrol strategy centers on symbiont spread. If the symbionts are beneficial to hosts, then symbiont demise is the biocontrol goal.
Bacterial endosymbiosis in plant-parasitic nematodes has received limited research attention, with the exception of the bacterial genus Pasteuria [9]. Pasteuria is an obligate bacterial parasite of plant-parasitic nematodes that has shown promise as a biocontrol agent [10,11]. Pasteuria endospores persist in the soil until a suitable host nematode comes into physical contact with the bacterium. Once the Pasteuria spores adhere to the nematode cuticle, they germinate and invade the body of the infected nematode; when an infected host dies, new spores are released to the environment. Therefore, Pasteuria are transmitted horizontally between hosts. Pasteuria bacteria act in a species-specific manner, and most economically important plant-parasitic nematode species are affected by some member of this group of bacteria [12]. For example, Pas. penetrans infects Meloidogyne sp., Pas. thornei infects Pratylenchus sp., and Pas. nishizawae infects Heterodera and Globodera [13,14].
Other important bacterial endosymbionts reported in plant-parasitic nematodes so far include Wolbachia, Cardinium, and Xiphinematobacter [15]. Unlike Pasteuria, these bacteria are vertically transmitted through the maternal germline of host nematodes. These three endosymbionts infect some of the most agriculturally important plant-parasitic nematodes. Xiphinematobacter is a species-specific endosymbiont that occurs in at least 27 species of Xiphinema spp. globally [15,16]. Xiphinematobacter is hypothesized to be an obligate mutualist with a possible role associated with plant-parasitic nematode nutrition [17]. Cardinium has been reported in three species of Heterodera (H. glycines, H. avenae, and H. goettingiana), G. rostochiensis, and Pratylenchus penetrans [18,19,20]. In P. penetrans, Cardinium is reported to co-occur with Wolbachia [1,20,21]. The role of Cardinium in plant-parasitic nematodes remains unclear, though it is speculated that Cardinium endosymbionts are mild parasites or commensal symbionts [18,19]. To date, Wolbachia has been reported in two species of Radopholus (R. similis and R. arabocoffeae) and in P. penetrans [21,22]. A previous study suggests the potential association of Wolbachia infection with host reproductive manipulation in P. penetrans [21].
Although knowledge on Wolbachia and Cardinium distribution, diversity, and functional impacts in plant-parasitic nematodes is limited, more is known about these endosymbionts in arthropods. Cardinium occurs in the arthropod orders Hymenoptera, Hemiptera, Diptera, and Arachnida, where it occurs in 6–7% of species [15]. Cardinium is not known to occur in other nematode groups outside those plant-parasitic nematode genera where it has been characterized [15]. Wolbachia is widespread and well-studied in arthropods, and is known to occur in some filarial animal-parasitic nematodes [23]. It is estimated that more than 65% of the arthropod species carry Wolbachia (e.g., Hymenoptera, Lepidoptera, Heteroptera, Crustacea, Collembola, Coleoptera, and Arachnida) [24].
Both Cardinium and Wolbachia are associated with various modes of reproductive manipulation in their hosts. Cytoplasmic incompatibility (CI) is the most common reproductive abnormality caused by Wolbachia and Cardinium endosymbionts. CI is a form of conditional sterility in which no viable offspring are produced when infected males mate with uninfected females. These phenotypes can be exploited for environment-friendly biocontrol methods for agricultural pests. In fact, Wolbachia has been the target for many biological control measures in arthropods and filarial nematodes. For example, Wolbachia has been used in male-release programs to control mosquitoes where it acts as a reproductive manipulator [25]. In contrast, Wolbachia is an essential mutualist in filarial nematode species, and simple antibiotic treatments targeting the endosymbiont are used in filarial nematode control schemes [26]. Development of endosymbiont-based biocontrol in plant-parasitic nematodes, however, will require an improved understanding of symbiont occurrence and their functional effects, as well as their genomic diversity. Accordingly, this study focused on uncovering the range of plant-parasitic nematode species infected by Wolbachia and Cardinium endosymbionts, followed by a comparative analysis of the symbiont genomes.
Historically, the main endosymbiont discovery methods involved transmission electron microscopy (TEM), Fluorescent In Situ Hybridization (FISH), and Polymerase Chain Reaction (PCR) [16,17,21,22,27]. These methods are often effective, but can be time and labor-intensive (particularly with TEM and FISH methods) and can also be biased and potentially lead to false negatives (especially for PCR-based approaches). To determine the presence of bacteria in eukaryotic tissues via TEM requires fixed and stained preparations of host cells [28,29]. FISH requires surface sterilization and the fixing of the samples, followed by probe hybridization and confocal microscopy; all utilize sophisticated techniques [16,17]. Both TEM and FISH approaches require significant effort when scaling to many samples. One constraint associated with PCR-based approaches is the trade-off between primer sensitivity and specificity; more sensitive primers lack specificity [30]. The PCR bias could be due to primer sensitivity, where some primers occasionally fail to amplify endosymbiont DNA by a standard PCR, leading to false negatives. Thus, an improved understanding of endosymbiont occurrence and distribution across many nematode species requires alternative strategies that are easier to scale and less prone to bias.
Genome skimming has been demonstrated as an effective and affordable approach to endosymbiont discovery that relies on high-throughput DNA sequencing technologies (next-generation sequencing) [1,2]. In this method, all samples undergo low-coverage whole genome sequencing (~10× coverage of the host genome), followed by an assembly of DNA sequence reads into contigs (contiguous sequences). The resulting contigs are then analyzed using tools such as blob-plots which display key DNA sequence parameters (e.g., GC content and n-fold coverage) and BLAST-based similarity metrics as tools for characterizing different DNA sequences present in the original sample. Thus, blob plots offer effective bioinformatics tools for a range of applications including the “cleanup” of contaminated DNA samples, DNA sources in difficult-to-disentangle host–parasite systems, and the isolation of intracellular bacterial symbiont genomes from within a whole-organism dataset [2]. In fact, Wolbachia and Cardinium endosymbionts in P. penetrans were discovered utilizing the genome skimming approach with blob tools [1]. However, the extent and diversity of Wolbachia and Cardinium endosymbionts in plant-parasitic nematodes remains unclear. Knowledge on the range of plant-parasitic nematode species infected by bacterial endosymbionts and their genomic diversity will advance our understanding of plant-parasitic nematode–bacterial endo-symbioses and its potential for application in biocontrol strategies.
In this study, a genome skimming strategy was applied to 52 samples representing 12 different plant-parasitic nematode species, including samples from North American, South American, and African continents. The objectives of this study were: (1) to provide insights into the range of plant-parasitic nematode species infected by Wolbachia and Cardinium endosymbionts and (2) better understand the genomic diversity of bacterial endosymbionts in plant-parasitic nematodes.
2. Materials and Methods
2.1. Nematode Populations and Sampling
A total of 52 plant-parasitic nematode populations that consisted of 12 species in eight genera were collected (Figure 1; Table 1). To obtain nematodes, multiple root and soil samples were collected from different areas in a field randomly and then combined into a single composite sample. Some soil and root samples were collected from greenhouse pot cultures. Pratylenchus species were extracted from roots by intermittent mist [31]; root samples were washed free of soil, cut into small pieces (<5 cm), placed on screens over funnels draining into test tubes, and misted for 15 s at 2 min intervals for 5 days. To extract nematodes from soil, Baermann funnels were used; 50 g of soil was placed on a Baermann funnel and kept for 5 days. To extract eggs of some nematode species from roots, roots were shaken in 0.05% NaOCl for 3 min. The solution was then poured over nested 170- and 500-mesh sieves, with eggs being retained on the latter. For Globodera pallida and Heterodera glycines, eggs were liberated from cysts by cutting the cysts open under a microscope. For all extractions, nematodes were collected in water and stored at 4 °C until DNA extraction. The sampling locations include USA, Uganda, Nigeria, Costa Rica, Colombia, and Chile. Samples from locations outside the USA were sent in DESS solution (dimethyl sulfoxide, disodium EDTA, and saturated NaCl) and washed three times with deionized water before extracting DNA [32,33].
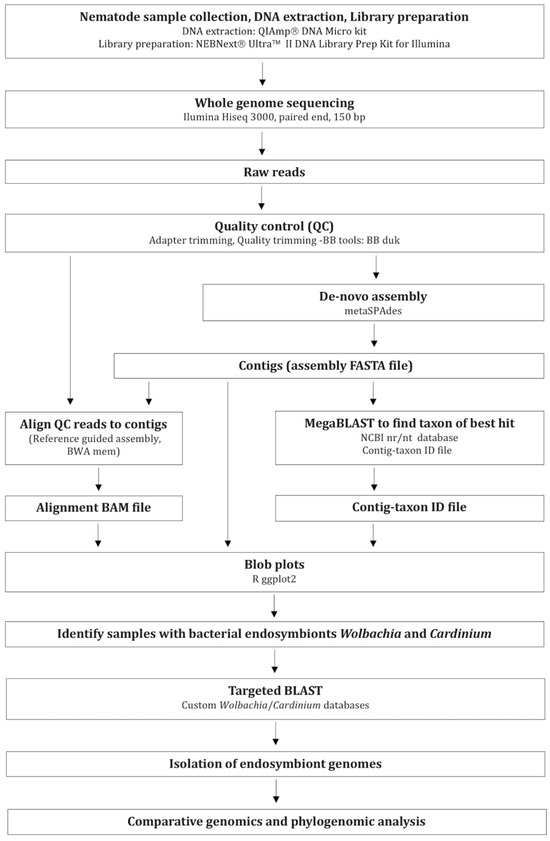
Figure 1.
Workflow of data collection, bioinformatics, and analysis methods.

Table 1.
Plant-parasitic nematode samples collected for DNA extraction. Each sample consisted of 100–250 adults and/or juveniles or 1000 eggs, all obtained from different sites.
2.2. DNA Extraction, Library Preparation, and Genome Sequencing
Samples consisting of >100 nematodes of the same species were used for DNA extraction. DNA isolation was performed using QIAampDNA Micro kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. The total genomic DNA was sheared for 50 s using a Diagenode Bioruptor Pico (Diagenode, Inc., Denville, NJ, USA) to obtain peak library fragment sizes of ~500 bp, and genomic libraries were prepared using the NEBNext_Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s instructions. Whole genome sequencing was performed using Illumina HiSeq 3000 for 2 × 150 bp reads (paired-end) at the Center for Quantitative Life Sciences at Oregon State University. Raw reads are available from NCBI’s Sequence Read Archive (SRA) under BioProject numbers PRJNA1026095, PRJNA679610, and PRJNA541590.
2.3. Metagenomic Assembly and Blob Plot Generation
Following the genome sequencing, raw reads were trimmed to remove adaptors and filtered for quality using bbduk—BBtools, (base calls with Phred quality score < 20 were excluded from read ends) (http://jgi.doe.gov/data-and-tools/bbtools, accessed on 29 January 2022). Quality-filtered reads were assembled de novo using metaSPAdes version 3.12.0 [34] to obtain an assembly FASTA file. Contigs in the assembly that were less than 300 bp were removed. Putative taxonomic assignments of the resulting filtered contigs were investigated by comparing to NCBI non-redundant nucleotide database (nt) using a mega-blast [2]. Here, individual contigs were assigned a phylum/genus based on BLAST similarity (E-value < 1 × 10−25) to obtain a Contig-taxon ID file. Next, quality-controlled reads were mapped back to the preliminary de novo assembly using BWA mem (Burrows-Wheeler Aligner, version 0.7.12-r1039, Reference guided assembly) [35] to produce an alignment BAM file. These three types of input files (assembly FASTA files, alignment BAM files, and Contig-taxon ID files) were collated using a custom Perl script and gc_cov_annotate.pl, and generated the blob plots using a ggplot2 package in R-studio [2] (Figure 1). A total of 52 blob plots were analyzed for sub sets of DNA-sequenced data that corresponded to single species’ genomes based on BLAST results.
2.4. Screening Metagenomic Assemblies for Bacterial Endosymbionts
Blob plots and blob plot tables were used to identify and visualize endosymbiont DNA in each de novo assembly resulted from each nematode sample. The de novo assemblies that had contigs identified as belonging to Wolbachia or Cardinium genera were sorted for further analysis. One nematode sample (greenhouse P. penetrans) that was previously demonstrated to carry Wolbachia and Cardinium was used as a positive control and a point of reference to validate the endosymbiont occurrence in other samples [20,21]. For the greenhouse P. penetrans sample, the organisms to be separated were the nematode, its bacterial endosymbionts Wolbachia/Cardinium, and any other extra-cellular microorganisms.
2.5. Targeted Blasting and Endosymbiont Genome Isolation
Once the bacterial endosymbionts were identified in a nematode sample, the corresponding de novo assemblies were subjected to BLAST search against custom BLAST databases. The custom BLAST databases were built utilizing the publicly available NCBI reference genomes of Wolbachia and Cardinium (one custom database per genus). Contigs in each assembly that matched the expected bacterial endosymbiont genera and had the expected GC% (~35%) were isolated. Next, QUAST was used to calculate genome assembly statistics [36]. The endosymbiont genome assembly statistics (size, GC content, N50, and number of contigs) were then compared with the publicly available reference genomes (NCBI accession numbers: ASM175266v1-Wolbachia, ASM317691v1-Cardinium) to assess the assembly quality and completeness. Further, to assess assembly completeness in terms of gene content, BUSCO (Benchmarking Universal Single Copy Orthologs, version 5.4.3) analyses were conducted per assembly using 124 universal single-copy bacterial orthologs (shared by 4085 bacterial species) [37]. This analysis was extended to the class level. In the case of Wolbachia, 432 single-copy orthologous genes highly conserved among alphaproteobacteria were employed. For Cardinium, 768 single-copy orthologous genes highly conserved among cytophagia were utilized. Finally, a conservative method was used to distinguish “strong evidence” from “weak evidence” for the existence of an endosymbiont in a given sample. This was conducted by setting threshold values based on the positive control and publicly available endosymbiont reference genomes as follows:
- -
- “No evidence” = no contigs in the assembly matched the target endosymbiont.
- -
- “Weak evidence” = some contigs in the assembly matched the target endosymbiont, the total assembly size was >10%, but <65% of expected genome size and the missing BUSCO percentage was >25%.
- -
- “Strong evidence” = contigs in the assembly matched the target endosymbiont, with a total assembly size > 65% of expected genome size; the missing BUSCO percentage was <25%.
Only the assemblies that met both thresholds (genome size and BUSCO score) were used for further analysis.
2.6. Genomic Analyses of Bacterial Endosymbionts
Once bacterial endosymbiont(s) were detected in a certain plant-parasitic nematode sample based on strong evidence, the assembled endosymbiont genome(s) were isolated for comparative analyses. To investigate the population level variability among the endosymbionts, multiple genome alignment and pairwise genome comparisons were performed using Mauve software version 2.4.0 [38]. For Wolbachia comparisons, genome assemblies of Wolbachia from different P. penetrans and R. similis populations were used. For Cardinium comparisons, genome assemblies of Cardinium from different H. glycines populations were used.
The contigs from the endosymbiont assemblies were first ordered against a reference genome obtained from NCBI. This included a reference Wolbachia genome from P. penetrans, hereafter known as Pp_Wol_Ref (NCBI accession number ASM175266v1), and a reference Cardinium genome from H. glycines, hereafter known as Hg_Car_Ref (NCBI accession number ASM317691v1). A second Cardinium reference genome, with one obtained from P. penetrans, hereafter known as Pp_Car_Ref (NCBI accession number ASM378869v1), was included to investigate the Cardinium diversity between different plant-parasitic nematode species.
Single-copy full length (complete) BUSCO genes were used as reliable markers for the phylogenomic inference of the bacterial endosymbionts identified in this study. First, the number of shared bacterial orthologous genes in endosymbiont genomes was computed. Then, each individual gene corresponding to each genome was aligned and trimmed using the ClustalW function of MEGA6 software; the IUB DNA weight matrix was used and the gap-opening and extension penalties were set to 20 and 6.66 (default settings) [39]. Next, a concatenated matrix of all single-copy full length BUSCO genes was created and aligned using the ClustalW function of MEGA6. To evaluate the diversity of endosymbiont lineages in different nematode populations, phylogenomic trees were reconstructed by the maximum likelihood method with 1000 bootstrap replicates in MEGA6.
3. Results
3.1. Metagenomic Assembly Statistics
First, we examined unrefined meta-genomic assemblies for 52 plant-parasitic nematode populations, representing 12 different species from USA, Uganda, Nigeria, Costa Rica, Colombia, and Chile (Table 1). Meta-assemblies yielded between ~5000 and ~320,000 contigs per sample, with N50 values ranging from ~1000 bp to 26,000 bp (Table S1). The GC content of the meta-assemblies ranged from ~30% to ~60% with an average GC content of ~45%. As bacterial DNA was a predominant component of the metagenome assemblies, the observed GC content was higher than that typically observed for nematodes (typical nematode genomes consist of ~35% of GC) [40]. The samples yielded genome assembly sizes between ~15 to 400 Mb (nematode and other DNA, together) with an average coarse genome size estimate of 154 Mb (Table S1). The overall plant-parasitic nematode genome assembly patterns were consistent with known nematode genome size ranges of 20 to 500 Mb [40].
3.2. Detection of Bacterial Endosymbionts
Once meta-assemblies were analyzed using blob plot approaches, nematode DNA was distinguished from bacterial DNA. For the greenhouse P. penetrans, evidence for its known bacterial endosymbionts Wolbachia and Cardinium [20,21,41] was observed as expected (Figure 2a). Based on blob plots from our initial nt BLAST, we found evidence for Wolbachia in 18/52, and Cardinium in 12/52 nematode populations obtained from USA, Costa Rica, Colombia, Uganda, and Nigeria (Table S2). Wolbachia was detected in 8/12 nematode species, while Cardinium was detected in 5/12 nematode species investigated (Figure 3).
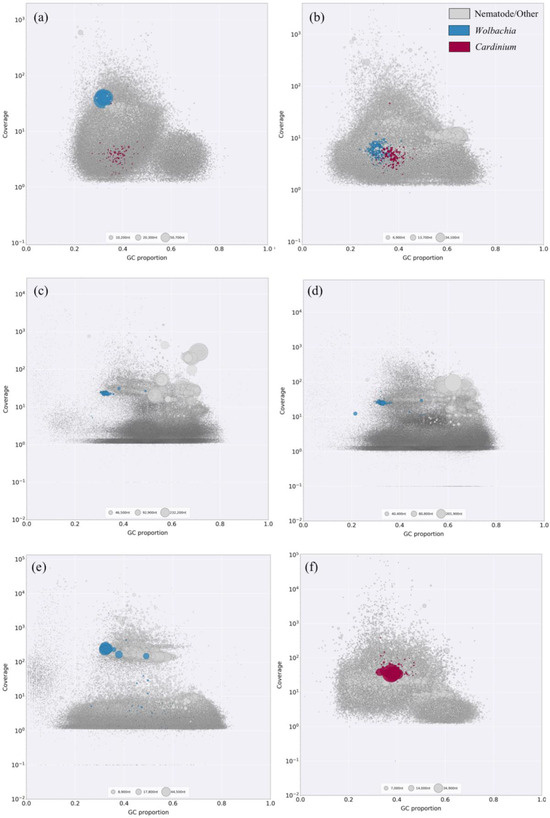
Figure 2.
Taxon-annotated blob plots for meta-genomic assemblies from six nematode samples: (a) Pratylenchus penetrans, Oregon; (b) Pratylenchus penetrans, Costa Rica; (c) Radopholus similis, Uganda; (d) R. similis, Colombia; (e) R. similis, Nigeria; (f) Heterodera glycines, Alabama. Sequences are represented by circles in the plot with diameters proportional to sequence length and colored by taxonomic affiliation. Gray: nematode and other; Blue: Wolbachia; Red: Cardinium. There is GC proportion on the x-axis with coverage on the y-axis.
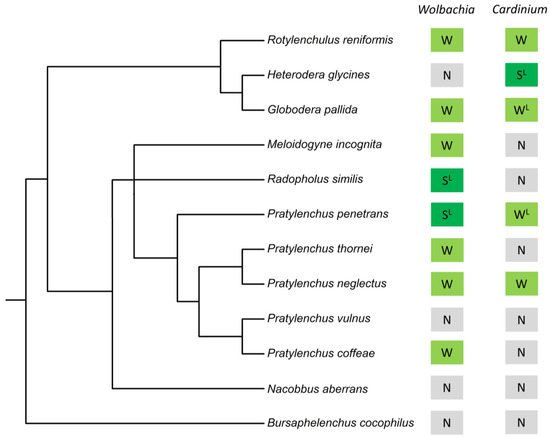
Figure 3.
Evidence for bacterial endosymbiont occurrence (Wolbachia and Cardinium) in 12 plant-parasitic nematode species. “S” indicates strong evidence for the endosymbiont, “W” indicates weak evidence for the endosymbiont, “N” indicates no evidence for the endosymbiont, and “L” indicates evidence for the endosymbiont from scientific literature. The phylogeny is based on 18S rRNA; Bayesian small subunit (SSU) rDNA tree of the Tylenchida [42].
3.3. Endosymbiont Assembly Isolation and Assessment—Wolbachia
Once the metagenomic assemblies underwent BLAST searches against custom Wolbachia BLAST database, 18 Wolbachia genome assemblies were isolated. Five of the Wolbachia assemblies had the expected genome size (~0.9 Mb) based on the publicly available Wolbachia reference genome, Pp_Wol_Ref (Table 2), and were found in P. penetrans and R. similis populations. These five Wolbachia assemblies yielded between 62 and 606 contigs per sample, with N50 values ranging from ~2500 bp to 147,000 bp, and a GC content of ~32% (Table 2). The remaining 13 isolated Wolbachia genome assemblies were between ~10% to 65% of the expected genome size.

Table 2.
Genomic assembly statistics of Wolbachia and Cardinium bacterial endosymbionts obtained from plant-parasitic nematode populations.
To assess the completeness of endosymbiont genome assemblies in terms of gene content, BUSCO analysis was performed. Based on 124 single-copy orthologous genes that are highly conserved among bacterial species, the BUSCO analysis resulted in fractions (%) of single-copy (complete) genes, duplicated (complete) genes, fragmented genes, and missing genes per endosymbiont genome (Table 3). For the five Wolbachia genome assemblies, the percentage of complete genes, fragmented genes and missing genes ranged from 50.8 to 83.1%, 4 to 29%, and 12 to 20%, respectively (Table 3).

Table 3.
BUSCO analysis results of Wolbachia and Cardinium genome assemblies based on 124 single-copy orthologous genes that are highly conserved among bacterial species.
The genome assemblies were further assessed utilizing the BUSCO reference sets curated to be specific to the classes. Based on 432 single-copy orthologous genes that are highly conserved among alphaproteobacteria, the percentage of complete genes, fragmented genes and missing genes ranged from 53.7 to 73.1%, 2.3 to 15.7%, and 24.5 to 30.6%, respectively (Table 4).

Table 4.
BUSCO analysis results of Wolbachia and Cardinium genome assemblies based on 432 and 768 single-copy orthologous genes that are highly conserved among alphaproteobacteria and Cytophagia, respectively.
Based on the assembly quality, we categorized endosymbiont detection into “strong evidence” and “weak evidence” (see Section 2). Of the 18 nematode populations in which Wolbachia DNA was detected, five had strong evidence while thirteen had weak evidence (Table S2). Excluding the positive control, strong evidence for the Wolbachia occurrence was observed in one newly analyzed P. penetrans population from Costa Rica and three newly analyzed R. similis populations from Uganda, Colombia, and Nigeria (Figure 2b–e). Four other P. penetrans populations and one other R.similis population showed weak evidence for carrying Wolbachia (Table S2). Although P. penetrans and R. similis were expected to carry Wolbachia [21,22,41], one P. penetrans population and three R. similis populations included in this study did not show any evidence of the endosymbiont DNA in the meta-assemblies (Table S2). Wolbachia was also detected in G. pallida, M. incognita, R. reniformis, P. coffeae, P. neglectus, and P. thornei; however, these populations had weak evidence for the endosymbiont presence (Figure 3, Table S2).
3.4. Endosymbiont Assembly Isolation and Assessment—Cardinium
Once the metagenomic assemblies underwent BLAST searches against custom Cardinium BLAST databases, 12 Cardinium genome assemblies were isolated. Two of the Cardinium assemblies obtained from H. glycines had the expected genome size (~1.2 Mb) based on the publicly available Cardinium reference genome, Hg_Car_Ref (Table 2). Another two Cardinium assemblies obtained from P. penetrans had a genome size between 0.78–1.2 Mb which accounted for more than 65% of the expected genome size. The four Cardinium assemblies mentioned above yielded between 56 and 739 contigs per sample, with N50 values ranging from ~1200 bp to 1,200,000 bp, and a GC content of ~35% (Table 2). The remaining eight isolated Cardinium assemblies had genome sizes that were between ~10% to 65% of the expected genome size.
According to the BUSCO analysis which was performed to assess the completeness of Cardinium genome assemblies in terms of gene content, the following results were obtained. Based on 124 single-copy orthologous genes that are highly conserved among bacterial species, the percentage of complete genes, fragmented genes, and missing genes in the Cardinium assemblies ranged from 34.7% to 71.8%, 4 to 25%, and 24.2 to 42.7%, respectively (Table 3). The genome assemblies were further assessed utilizing the BUSCO reference sets curated to be specific to the classes. Based on 768 single-copy orthologous genes that are highly conserved among cytophagia, the percentage of complete genes, fragmented genes, and missing genes ranged from 7.3 to 38.8%, 1 to 6.4%, and 60.2 to 87.9%, respectively (Table 4).
According to our “strong evidence” and “weak evidence” criteria to assess overall assembly quality, the majority of the Cardinium assemblies were categorized as weak evidence. From the 12 populations in which Cardinium DNA was detected, only one had strong evidence, while 11 had weak evidence (Table S2). Cardinium was detected with strong evidence in H. glycines population, from Alabama, (Figure 2f). Two other H. glycines sample showed weak evidence for carrying the Cardinium endosymbiont (Table S2). Although H. glycines, P. penetrans, and G. pallida were expected to carry Cardinium [18,19,20], one H. glycines population, five P. penetrans populations, and six G. pallida populations investigated in this study did not show any evidence of Cardinium DNA in the meta-assemblies (Table S2). Weak evidence for Cardinium was observed in P. penetrans, G. pallida, P. neglectus, and R. reniformis (Figure 3, Table S2). Neither endosymbiont was detected in B. cocophilus, N. aberrans, and P. vulnus populations investigated in this study (Figure 3).
3.5. Genomic Analyses of Bacterial Endosymbionts
Based on strong evidence, we further analyzed five Wolbachia and one Cardinium genome assemblies isolated from three plant-parasitic nematode species, i.e., two Wolbachia assemblies from P. penetrans, three Wolbachia assemblies from R. similis, and one Cardinium assembly from H. glycines. Compared to the publicly available genomes, these newly sequenced and assembled genomes had comparable degrees of completeness (reference data; genome size: Wolbachia = ~0.97 Mb and Cardinium = ~1.12 Mb; complete BUSCOs: Wolbachia = ~80% and Cardinium = ~70%) (Table 3).
Of the 124 universal single-copy bacterial orthologs used for BUSCO analysis, the Wolbachia reference genome, Pp_Wol_Ref, lacked 15 orthologs. The remaining 109 orthologs were used for a comparative analysis of Wolbachia. Accordingly, 95/109 housekeeping genes (complete and fragmented BUSCOs) were shared between the six Wolbachia genomes obtained from plant-parasitic nematodes (including the reference genome). Wolbachia obtained from three different R. similis populations shared 108/109 housekeeping genes, of which 100 were complete BUSCOs. Wolbachia obtained from three different P. penetrans populations (including the reference genome) shared 96/109 housekeeping genes, of which 60 were complete (Figure 4). These 60 complete genes were common to all six Wolbachia genomes analyzed. Maximum-likelihood phylogeny based on concatenation of the 60 complete genes placed Wolbachia obtained from plant-parasitic nematodes in a separate clade from Wolbachia obtained from filarial nematodes (Figure 5). Wolbachia obtained from P. penetrans formed a sister clade at the root of the tree, while Wolbachia obtained from R. similis formed a separate clade with high bootstrap support (Figure 5).
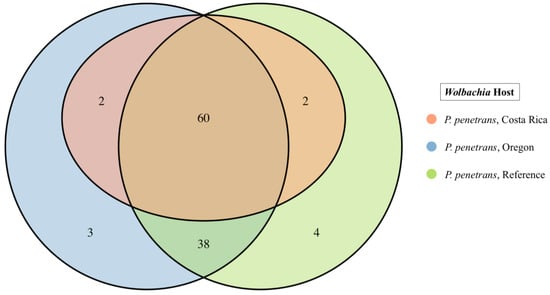
Figure 4.
Single copy orthologous genes (BUSCOs) present in Wolbachia and obtained from three different Pratylenchus penetrans populations (based on “Bacteria_odb10”).
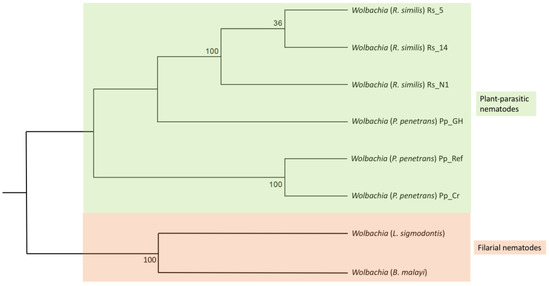
Figure 5.
Maximum likelihood phylogeny of Wolbachia in nematode hosts based on 60 concatenated single copy orthologous genes. Color indicates Wolbachia hosts: green—plant-parasitic nematodes; red—filarial nematodes. Corresponding host species are indicated within brackets.
To investigate the overall genome structure and content among the six Wolbachia genomes, a multiple genome alignment was performed. Wolbachia obtained from R. similis (Rs_Wol_N1, Rs_Wol_UG, Rs_Wol_CO) shared a high similarity in synteny profile (gene order) and gene content. In contrast, synteny was not conserved between Wolbachia genomes from R. similis and P. penetrans (Pp_Wol_Ref) (Figure 6). Based on genome structure, Wolbachia in the same host species seems to be more similar to each other compared to Wolbachia from different host species (Pratylenchus and Radopholus).
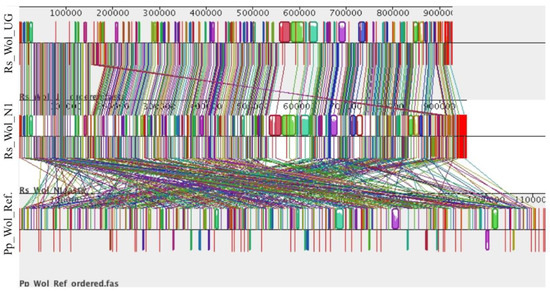
Figure 6.
Multiple genome comparisons of Wolbachia from Radopholus and Pratylenchus. Each horizontal panel represents a Wolbachia genome. Colored blocks represent homology among genomes. Vertical red lines indicate contig boundaries. Rs_Wol_UG: Wolbachia in R. similis from Uganda, Rs_Wol_N1: Wolbachia in R. similis from Nigeria, and Pp_Wol_Ref: Wolbachia reference from Pratylenchus (NCBI accession number ASM175266v1).
Of the 124 universal single-copy bacterial orthologs used for BUSCO analysis, the Cardinium reference genome (NCBI accession number ASM317691v1) lacked 31 orthologs. The remaining 93 orthologs were used for comparative analysis of Cardinium. When the newly sequenced and assembled Cardinium genome obtained from H. glycines was compared with the Cardinium reference genome, all the (93/93) housekeeping genes were shared between them and 88 were complete BUSCO genes. These two genomes (Hg_Car_Al and Hg_Car_Ref), shared high similarity in synteny profile (gene order) and gene content (Figure 7). In contrast, synteny was not conserved between Cardinium genomes from H. glycines (Hg_Car_Ref) and P. penetrans (Pp_Car_Ref) (Figure 7). Based on genome structure, Cardinium in the same host species (Heterodera) seems to be more similar to each other compared to Cardinium from different host species (Heterodera and Pratylenchus).
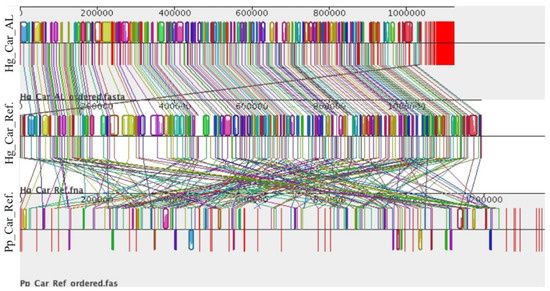
Figure 7.
Multiple genome comparisons of Cardinium from Heterodera and Pratylenchus. Each horizontal panel represents a Cardinium genome. Colored blocks represent homology among genomes. Vertical red lines indicate contig boundaries. Hg_Car_AL: Cardinium in H. glycines from Alabama; Hg_Car_Ref: Cardinium reference from Heterodera (NCBI accession number ASM317691v1); Pp_Car_Ref: Cardinium reference from Pratylenchus (NCBI accession number ASM378869v1).
4. Discussion
The intricate interplay between microorganisms and their hosts is of significant scientific interest, having profound implications in agriculture, human health, and the environment. Deepening our understanding of these symbiotic relationships reveals new avenues for addressing global challenges and fostering sustainable practices. This study was primarily centered on elucidating microbial symbiosis in plant-parasitic nematodes, with the methodology being universally applicable to a wide range of organisms. Knowledge of bacterial endosymbiont occurrence in plant-parasitic nematodes is essential for crafting effective microbe-based biocontrol strategies against them. Accordingly, we utilized a rapid and simple genomic screen for well-known bacterial endosymbionts, Wolbachia and Cardinium, in plant-parasitic nematodes. Our first goal was to uncover the range of plant-parasitic nematode species infected by Wolbachia and Cardinium endosymbionts. The approach for detecting endosymbionts categorized genomic verification into “strong”, “weak”, and “no” evidence. This study revealed a limited distribution of Wolbachia and Cardinium endosymbionts in plant-parasitic nematode species (n = 12); based on strong evidence, only 16% of the species investigated carried Wolbachia while 8% carried Cardinium. This value was observed to be low considering the number of nematode populations investigated (n = 52), and resulted in 10% and 2% of the populations being positive for Wolbachia and Cardinium, respectively (based on strong evidence).
Wolbachia has been reported in just two plant-parasitic nematode genera, Pratylenchus and Radopholus, encompassing three species: P. penetrans, R. similis, and R. arabocoffeae [15]. This study demonstrated a discontinuous distribution of Wolbachia across plant-parasitic populations belonging to Pratylenchus and Radopholus, indicating a non-essential function of the endosymbiont within these nematodes. So far, Wolbachia occurrence in plant-parasitic nematodes has been noted in Asia, Africa, and North America [21,22]. This study confirmed this observation and further demonstrated Wolbachia occurrence in South America as well.
Cardinium has been reported in three plant-parasitic nematode genera, Heterodera, Globodera, and Pratylenchus, encompassing five species: H. glycines, H. avenae, H. goettingiana, Globodera rostochiensis, and Pratylenchus penetrans [15]. Cardinium occurrence in plant-parasitic nematodes has been noted in Asia, Europe, North America, and South America, supporting our observation of Cardinium in North and South America. This study demonstrated a discontinuous distribution of Cardinium across plant-parasitic populations belonging to Heterodera, Globodera, and Pratylenchus, indicating a non-essential function of the endosymbiont within these nematodes. This observation is supported by previous studies, where both Wolbachia and Cardinium endosymbionts in plant-parasitic nematodes were reported to act as potential parasites rather than obligate mutualists [18,21]. The complete absence of these endosymbionts in certain populations of the same plant-parasitic nematode species and the potential for these endosymbionts to act as parasites may offer a novel avenue for plant-parasitic nematode biocontrol through the introduction of these endosymbionts into nematodes.
Wolbachia has been artificially transferred, both intraspecifically and interspecifically, in many arthropod species utilizing embryo or adult microinjection techniques [43]. Once transinfection is successful, the host was able to vertically transmit the endosymbionts to their progeny. Other techniques such as the co-rearing of the recipient and donor species have achieved the successful transfer of Wolbachia into new hosts, but these techniques are only suitable for a limited number of arthropods [43]. There is recent evidence that Wolbachia has the capacity to transmit horizontally through plants. In Bemisia whiteflies, for example, after infected individuals fed on leaves, Wolbachia was detected in the plant’s phloem. When Wolbachia-free whiteflies subsequently fed on the infected plant leaves, they became infected and were able to vertically transmit endosymbionts to their progeny [44].
Building upon our findings of the presence of Wolbachia and Cardinium endosymbionts within plant-parasitic nematodes, as well as their potential parasitic roles, our work paves the way for similar studies to explore targeted biocontrol strategies. Of particular interest is the introduction of Wolbachia into uninfected nematode populations by mixing a small number of infected nematodes with uninfected populations. This could be conducted through controlled greenhouse pot cultures, allowing for the monitoring of Wolbachia establishment and spread, as well as its potential impact on nematode fitness and its ability to reduce parasitic effects on host plants. The insights gained from these studies can inform the development of sustainable biocontrol measures, aligning with the broader goals of eco-friendly and responsible agriculture practices.
Based on sequence similarity search results, Wolbachia was reported in six plant-parasitic nematode species that were previously unknown to carry the endosymbiont with weak evidence. Similarly, Cardinium was reported in two plant-parasitic nematode species that were previously unknown to carry the endosymbiont with weak evidence. The observation of “weak” evidence could be due to many reasons: (1) the presence of endosymbionts in very low abundance, (2) environmental contamination, and (3) Palaeosymbiosis (presence of ancient horizontally transferred endosymbiont DNA fragments in the nematode genome) [45]. Given the assumption of low-titer infections in nematode populations, it is important not to completely dismiss the possibility that a plant-parasitic nematode species, previously not known to host these endosymbionts, might indeed carry them. However, in order to gain more clarity, additional experiments utilizing FISH and/or PCR techniques are required. Environmental contamination of the samples is less likely because Wolbachia and Cardinium are obligate intracellular bacteria and they cannot survive in the environment. The horizontal transfer of endosymbiont DNA to the host genome is common when the host is infected by the endosymbiont [45]. However, the presence of endosymbiont DNA in the host genome does not necessarily provide evidence that a host carries a live infection; instead, the endosymbiont could have been infected in the past and lost at some point of their evolution. The DNA fragments were unlikely derived from a live endosymbiont infection if their genes were disabled. Future studies involving experiments to test for the expression of endosymbiont-derived genes could determine the presence of live infections.
While we have rigorously categorized the strength of our endosymbiont evidence as weak or strong based on the data obtained, it is crucial to recognize that genome skimming, like any sequencing method, is subject to sequencing depth constraints. When sequencing depth is insufficient, it can lead to false negatives, implying that certain low-abundance endosymbionts may go undetected. Further, the variation in endosymbiont abundance is relative to the host, particularly across different developmental stages and sexes. For instance, in filarial nematodes and certain arthropod species, Wolbachia endosymbionts are typically the most abundant in adult females but are expected to vary depending on the life stages [46,47]. These dynamics highlight the need for caution when interpreting negative results obtained through genome skimming, as the absence of endosymbiont sequences in a given sample may reflect genuine absence or simply a limitation in sequencing depth at that particular life stage.
Our next goal was to better understand the genomic diversity of bacterial endosymbionts in plant-parasitic nematodes. Accordingly, we compared the assembled genomes of the endosymbionts obtained from different nematode populations/species. Our results indicated >87% gene conservation in Wolbachia genome assemblies. Wolbachia obtained from the three R. similis populations shared 99% of these homologous genes, while it was 88% among Wolbachia obtained from the three P. penetrans populations (including the reference). The homology values indicate overall genetic similarity among Wolbachia occurring in different plant-parasitic nematode species. However, according to the genome structure analysis, the synteny profile was not shared in Wolbachia occurring in different host species. Future studies involving high coverage (>200×) genome sequencing, leading to high quality endosymbiont genome assemblies, will provide more information on deeper chromosomal dynamics and synteny/rearrangement rates between Wolbachia strains in plant-parasitic nematodes.
The maximum-likelihood phylogeny based on concatenation of the 60 complete homologs placed Wolbachia in separate clades based on its host nematode species. Similar topology was observed in a previous analysis based on 16S ribosomal RNA, ftsZ, and groEL genes [41]. At a larger scale, Wolbachia in plant-parasitic nematodes forms a separate clade from Wolbachia in filarial nematodes. This observation was expected based on the genetic differences in Wolbachia strains in these two nematode groups. In fact, the two nematode groups carry Wolbachia strains with functional differences, i.e., Wolbachia in plant-parasitic nematodes seems to act as a parasite while Wolbachia in filarial nematodes acts as an obligate mutualist [21,26].
The newly sequenced and assembled Cardinium genome obtained from H. glycines shared 100% of homologous genes with the Cardinium reference genome, indicating a high degree of genetic similarity of Cardinium endosymbionts occurring in different H. glycines populations. We also identified Cardinium in P. penetrans, but did not have enough DNA sequence data to produce a better genome assembly. Therefore, we compared the publicly available Cardinium reference genome from P. penetrans with that of H. glycines and observed changes in the structure of the genomes which lacked conserved synteny (gene order). Some bacterial endosymbiont strains such as Pasteuria or Xiphinematobacter are species-specific, where different host species are infected by different endosymbiont strains [13,14]. Species-specific endosymbionts account for a high degree of genetic variability and do not elicit similar phenotypes [42]. For example, phenotypes desirable for parasite/pest control might not be shared by species-specific endosymbiont strains. This might be true for Cardinium as well. Future studies that focus on the phenotypic effects of Cardinium on different plant-parasitic nematode species will provide important insights into Cardinium-nematode co-evolution and their potential to be used as biocontrol agents.
5. Conclusions
This study explored the utility of genome skimming in applied microbiology, with a primary emphasis on revealing previously unknown microbial endosymbiosis and providing further insights into endosymbiont diversity in plant parasitic nematodes. Genome skimming offers a rapid and efficient avenue for discovering previously unknown microbial associations across a broad spectrum of organisms, species, and populations, generating data for prospective research studies. Our study presents evidence supporting the existence of a limited occurrence of Wolbachia and Cardinium bacterial endosymbionts among plant-parasitic nematode species. The study revealed that both “Wolbachia—P. penetrans” and “Wolbachia—R. similis” symbioses prevail in South America. The study also confirmed the “Wolbachia—P. penetrans” and “Wolbachia—R. similis” symbioses in North America, and Africa, respectively. Further, the presence of Cardinium in H. glycines was confirmed in North America. Based on the occurrence patterns, both bacterial endosymbionts appear to serve a non-obligatory function within plant-parasitic nematodes and display a potential to act in a species-specific manner based on the differences in genomic structure and content. These findings contribute to our understanding of the intricate microbial interactions in nematodes and shed light on potential avenues for further research and the development of targeted biocontrol strategies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/applmicrobiol3040085/s1, Table S1: Metagenomic genome assembly statistics for 52 nematode populations consist of twelve plant-parasitic nematode species. All statistics are based on contigs of size ≥500 bp. Table S2: Wolbachia and Cardinium bacterial endosymbiont occurrence in 52 nematode populations.
Author Contributions
Conceptualization, S.K.W., D.R.D. and I.A.Z.; methodology, S.K.W. and D.R.D.; software, S.K.W. and C.H.; validation, S.K.W., C.H. and C.L.W.; formal analysis, S.K.W. and C.H.; investigation, S.K.W., C.L.W. and D.K.H.; resources, I.A.Z.; data curation, S.K.W. and C.H.; writing—original draft preparation, S.K.W.; writing—review and editing, S.K.W., C.H., C.L.W., D.K.H., I.A.Z. and D.R.D.; visualization, S.K.W.; supervision, D.R.D.; project administration, D.R.D. and I.A.Z.; funding acquisition, D.R.D. and I.A.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by USDA-ARS CRIS project 2072-22000-046-00D.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data generated in this study are openly available in NCBI’s Sequence Read Archive (SRA) under the BioProject numbers PRJNA1026095, PRJNA679610, and PRJNA541590.
Acknowledgments
The authors would like to thank OSU CQLS for genome sequencing assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Denver, D.R.; Brown, A.M.V.; Howe, D.K.; Peetz, A.B.; Zasada, I.A. Genome skimming: A rapid approach to gaining diverse biological insights into multicellular pathogens. PLoS Pathog. 2016, 12, e1005713. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Jones, M.; Koutsovoulos, G.; Clarke, M.; Blaxter, M. Blobology: Exploring raw genome data for contaminants, symbionts, and parasites using taxon-annotated GC-coverage plots. Front. Genet. 2013, 4, 237. [Google Scholar] [CrossRef] [PubMed]
- Abad, P.; Gouzy, J.; Aury, J.M.; Castagnone-Sereno, P.; Danchin, E.G.J.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2008, 26, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Bird, D.M.; Kaloshian, I. Are roots special? Nematodes have their say. Physiol. Mol. Plant Pathol. 2003, 62, 115–123. [Google Scholar] [CrossRef]
- Decraemer, W.; Hunt, D.J. Structure and classification. In Plant Nematology; Perry, R.N., Moens, M., Eds.; CAB International: Wallingford, UK, 2006; pp. 3–32. [Google Scholar]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M.; et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef]
- Chitwood, D.J. Research on plant-parasitic nematode biology conducted by the United States Department of Agriculture—Agricultural Research Service. Pest Manag. Sci. 2003, 59, 748–753. [Google Scholar] [CrossRef]
- Zasada, I.A.; Halbrendt, J.M.; Kokalis-burelle, N.; LaMondia, J.; Mckenry, M.V.; Noling, J.W. Managing nematodes without methyl bromide. Annu. Rev. Phytopathol. 2010, 48, 311–328. [Google Scholar] [CrossRef]
- Atibalentja, N.; Noel, G.R. Bacterial endosymbionts of plant-parasitic nematodes. Symbiosis 2008, 46, 87–93. [Google Scholar]
- Bekal, S.; Borneman, J.; Springer, M.S.; Giblin-Davis, R.M.; Becker, J.O. Phenotypic and molecular analysis of a Pasteuria strain parasitic to the sting nematode 1. J. Nematol. 2001, 33, 110–115. [Google Scholar]
- Sayre, R.M.; Starr, M.P. Pasteuria penetrans (ex Thome, 1940) nom. rev., comb, n., sp. n., a mycelial and endospore-forming bacterium parasitic in plant-parasitic nematodes. Proc. Helm. Soc. Wash. 1985, 52, 149–165. [Google Scholar]
- Bird, D.M.C.K.; Opperman, C.H.; Davies, K.G. Interactions between bacteria and plant-parasitic nematodes: Now and then. Int. J. Parasitol. 2003, 33, 1269–1276. [Google Scholar] [CrossRef] [PubMed]
- Atibalentja, N.; Noel, G.R.; Domier, L.L. Phylogenetic position of the North American isolate of Pasteuria that parasitizes the soybean cyst nematode, Heterodera glycines, as inferred from 16S rDNA sequence analysis. J. Syst. Evol. Biol. 2000, 50, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Gives, P.M.; Davies, K.G.; Morgan, M.; Behnke, J.M. Attachment tests of Pasteuria panetrans to the cuticle of plant and animal parasitic nematodes, free living nematodes and srf mutants of Caenorhabditis elegans. J. Helminthol. 1999, 73, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.M.V. Endosymbionts of plant-parasitic nematodes. Annu. Rev. Phytopathol. 2018, 56, 225–242. [Google Scholar] [CrossRef]
- Vandekerckhove, T.T.M.; Coomans, A.; Cornelis, K.; Baert, P.; Gillis, M. Use of the Verrucomicrobia-specific probe EUB338-III and fluorescent in situ hybridization for detection of “Candidatus Xiphinematobacter” cells in nematode hosts. Appl. Environ. Microbiol. 2002, 68, 3121–3125. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.M.V.; Howe, D.K.; Wasala, S.K.; Peetz, A.B.; Zasada, I.A.; Denver, D.R. Comparative genomics of a plant-parasitic nematode endosymbiont suggest a role in nutritional symbiosis. Genome Biol. Evol. 2015, 7, 2727–2746. [Google Scholar] [CrossRef]
- Walsh, J.A.; Shepherd, A.M.; Lee, D.L. The distribution and effect of intracellular Rickettsia-like microorganisms infecting second-stage juveniles of the potato cyst-nematode Globodera rostochiensis. J. Zool. Lond. 1983, 199, 395–419. [Google Scholar] [CrossRef]
- Endo, B.Y. The ultrastructure and distribution of an intracellular bacterium-like microorganism in tissue of larvae of the soybean cyst nematode Heterodera glycines. J. Ultrastruct. Res. 1979, 67, 1–14. [Google Scholar] [CrossRef]
- Brown, A.M.V.; Wasala, S.K.; Howe, D.K.; Peetz, A.B.; Zasada, I.A.; Denver, D.R. Comparative genomics of Wolbachia–Cardinium dual endosymbiosis in a plant-parasitic nematode. Front. Microbiol. 2018, 9, 2482. [Google Scholar] [CrossRef]
- Wasala, S.K.; Brown, A.M.V.; Kang, J.; Howe, D.K.; Peetz, A.B.; Zasada, I.A.; Denver, D.R. Variable abundance and distribution of Wolbachia and Cardinium endosymbionts in plant-parasitic nematode field populations. Front. Microbiol. 2019, 10, 964. [Google Scholar] [CrossRef]
- Haegeman, A.; Vanholme, B.; Jacob, J.; Vandekerckhove, T.T.M.; Claeys, M.; Borgonie, G.; Gheysen, G. An endosymbiotic bacterium in a plant-parasitic nematode: Member of a new Wolbachia supergroup. Int. J. Parasitol. 2009, 39, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.; Mediannikov, O.; Raoult, D.; Greub, G. Endosymbiotic bacteria associated with nematodes, ticks and amoebae. Immunol. Med. Microbiol. 2012, 64, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Kajtoch, L.; Kolasa, M.; Kubisz, D.; Gutows, J.M.; Scibior, R.; Mazur, M.A.; Holecová, M. Using host species traits to understand the Wolbachia infection distribution across terrestrial beetles. Sci. Rep. 2019, 9, 847. [Google Scholar] [CrossRef] [PubMed]
- Flores, H.A.; O’Neill, S.L. Controlling vector-borne diseases by releasing modified mosquitoes. Nat. Rev. Microbiol. 2018, 16, 508–518. [Google Scholar] [CrossRef]
- Turner, J.D.; Sharma, R.; Al Jayoussi, G.; Tyrer, H.E.; Gamble, J.; Hayward, L.; Priestley, R.S.; Murphy, E.A.; Davies, J.; Waterhouse, D.; et al. Albendazole and antibiotics synergize to deliver short-course anti-Wolbachia curative treatments in preclinical models of filariasis. Proc. Natl. Acad. Sci. USA 2017, 114, E9712–E9721. [Google Scholar] [CrossRef] [PubMed]
- Bandi, C.; Anderson, T.J.C.; Genchi, C.; Blaxter, M.L.; Celoria, V. Phylogeny of Wolbachia in filarial nematodes. Proc. R. Soc. 1998, 265, 2407–2413. [Google Scholar] [CrossRef]
- Hise, A.G.; Gillette-Ferguson, I.; Pearlman, E. Microreview The role of endosymbiotic Wolbachia bacteria in filarial disease. Cell. Microbiol. 2004, 6, 97–104. [Google Scholar] [CrossRef][Green Version]
- Konecka, E.; Olszanowski, Z. A screen of maternally inherited microbial endosymbionts in oribatid mites (Acari: Oribatida). Microbiology 2015, 161, 1561–1571. [Google Scholar] [CrossRef]
- Simoes, P.M.; Mialdea, G.; Reiss, M.; Sagot, M.-F.; Charlat, S. Wolbachia detection: An assessment of standard PCR Protocols. Mol. Ecol. Resour. 2011, 11, 567–572. [Google Scholar] [CrossRef]
- Gigot, J.; Walters, T.W.; Zasada, I.A. Impact and occurrence of Phytophthora rubi and Pratylenchus penetrans in commercial red raspberry (Rubus ideaus) fields in Northwestern Washington. Int. J. Fruit Sci. 2013, 13, 357–372. [Google Scholar] [CrossRef]
- Beknazarova, M.; Millsteed, S.; Robertson, G.; Whiley, H.; Ross, K. Validation of DESS as a DNA preservation method for the detection of Strongyloides spp. in Canine Feces. Int. J. Environ. Res. Public Health 2017, 14, 624. [Google Scholar] [CrossRef] [PubMed]
- Yoder, M.; De Ley, I.T.A.; King, I.W.K.; Mundo-Ocampo, M.; Mann, J.; Blaxter, M.; Poiras, L.; De Ley, P. DESS: A versatile solution for preserving morphology and extractable DNA of nematodes. Nematology 2006, 8, 367–376. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. Progressive Mauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Kumar, S.; Koutsovoulos, G.; Kaur, G.; Blaxter, M. Toward 959 nematode genomes Landes Bioscience. Worm 2012, 1, 42–50. [Google Scholar] [CrossRef]
- Brown, A.M.V.; Wasala, S.K.; Howe, D.K.; Peetz, A.B.; Zasada, I.A.; Denver, D.R. Genomic evidence for plant-parasitic nematodes as the earliest Wolbachia hosts. Sci. Rep. 2016, 6, 34955. [Google Scholar] [CrossRef]
- Holterman, M.; Karssen, G.; Van Den Elsen, S.; Van Megen, H.; Bakker, J.; Helder, J. Small subunit rDNA-based phylogeny of the Tylenchida sheds light on relationships among some high-impact plant-parasitic nematodes and the evolution of plant feeding. Nematology 2009, 99, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Hughes, G.L.; Rasgon, J.L. Transinfection: A method to investigate Wolbachia-host interactions and control arthropod-borne disease. Insect Mol. Biol. 2014, 23, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Ahmed, M.; Lv, N.; Shi, P.Q.; Wang, X.M.; Huang, J.L.; Qiu, B.L. Plantmediated horizontal transmission of Wolbachia between whiteflies. ISME J. 2017, 11, 1019–1028. [Google Scholar] [CrossRef]
- Koutsovoulos, G.; Makepeace, B.; Tanya, V.N.; Blaxter, M. Palaeosymbiosis revealed by genomic fossils of Wolbachia in a Strongyloidean nematode. PLoS Genet. 2014, 10, e1004397. [Google Scholar] [CrossRef] [PubMed]
- Diouf, M.; Miambi, E.; Mora, P.; Frechault, S.; Robert, A.; Rouland-Lefèvre, C.; Hervé, V. Variations in the relative abundance of Wolbachia in the gut of Nasutitermes arborum across life stages and castes. FEMS Microbiol. Lett. 2018, 365, fny046. [Google Scholar] [CrossRef] [PubMed]
- Fenn, K.; Blaxter, M. Quantification of Wolbachia bacteria in Brugia malayi through the nematode lifecycle. Mol. Biochem. Parasitol. 2004, 137, 361–364. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).