Abstract
Heterocyclic compounds can specifically regulate bacterial development by targeting specific bacterial enzymes and metabolic pathways. The ESKAPE pathogens are multidrug-resistant and cause nosocomial infections, which is one of the greatest challenges in clinical practice. The search for novel agents to combat resistant bacteria has become one of the most important areas of antibacterial research today. Heterocyclic compounds offer a valuable strategy in the fight against resistance as they can be designed to interact with bacterial targets that are less prone to developing resistance mechanisms. Bacterial histidine kinases (HKs), which are a component of two-component bacterial systems, are a promising target for new antibacterial compounds. We have designed and synthesized novel indole derivatives as antibacterial agents. Among the series, indole-coumarin (4b) and bisindole (4e) have shown the best inhibitory activity against S. aureus. Further, in silico docking studies show that compounds 4b and 4e could target histidine kinases in bacteria.
1. Introduction
Heterocyclic compounds have broad-spectrum antibacterial activity and can target a variety of bacterial infections [1,2]. Heterocyclic compounds can specifically regulate bacterial development while causing little harm to human cells by targeting specific bacterial enzymes and metabolic pathways. Since penicillin’s discovery as an antibiotic by Alexander Fleming, the advantages of this “miracle drug” have been employed to treat infectious illnesses [3]. Later, a great number of small molecules were successfully synthesized for the treatment of bacterial infections [4]. Chemists have developed structure-based drug designs that concentrate on certain pathways [5]. Virtual docking, which permits in silico screening, can be utilized for the lead optimization of previously developed drug motifs [6,7]. Furthermore, de novo processes generate novel chemotypes for pharmaceutical compounds [8,9]. Several discoveries have been made based on natural products, such as teixobactin [10], acyldepsipeptides [11], and arylomycins [12], for antibiotics. Ciprofloxacin is a fluoroquinolone-based antibiotic used to treat a number of bacterial infections, and various reports describe bacterial resistance mechanisms to ciprofloxacin and novel techniques to improve its efficacy [13,14]. Almost all the marketed antibiotics target cell wall biosynthesis, DNA synthesis or protein biosynthesis and membrane integrity, all of which have already been counteracted via numerous resistance mechanisms [15,16].
Among many other bacteria, the ESKAPE pathogens (Enterococcus faecalis, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter cloacae) are multidrug-resistant and cause nosocomial infections, which is one of the greatest challenges in clinical practice. The effects include high mortality and morbidity rates, placing a significant burden on healthcare systems [17,18]. Understanding the resistance mechanisms of these bacteria is essential for the development of novel antibacterial agents to combat infections. There is a range of antimicrobial resistance mechanisms used by ESKAPE pathogens, including enzymatic inactivation, modification of the drug target site, changing cell permeability through porin loss, an increase in the expression of efflux pumps, and mechanical protection provided by biofilm formation [19]. Thus, the search for novel agents for resistant bacteria has become one of the most important areas of antibacterial research today.
Bacterial histidine kinases (HKs), which are a component of two-component bacterial systems, are a promising target for new antibacterial compounds [20]. The histidine kinase protein is made up of two primary domains, a catalytic domain and a sensory domain, which are each responsible for detecting different signals or stimuli in the cell’s environment, such as changes in temperature, osmolarity and pH or the presence of chemicals. On the other hand, the catalytic domain has kinase activity [21,22].
When histidine kinase is activated, it goes through a process called autophosphorylation, in which a phosphate group from ATP is transferred to a particular histidine residue inside the protein. The kinase domain is activated by this phosphorylation process, enabling it to work as a kinase enzyme [23,24]. The phosphorylated histidine residue is then transferred by the activated histidine kinase to a response regulator protein, which is normally found in the cytoplasm. This phosphorylation of the response regulator stimulates a series of subsequent signaling events that cause a cellular response, modulate gene expression and have an impact on cellular functions like adaptation to changing environmental conditions, metabolism regulation and cell division coordination [25,26].
Targeting histidine kinases is a promising approach to interfering with bacterial signaling pathways and disrupting critical processes required for bacterial survival and pathogenicity [27,28]. The evidence currently available suggests that various two-component systems and their related histidine kinases play distinct roles in the regulation of the critical bacterial life processes [29,30].
Heterocyclic compounds offer a valuable strategy in the fight against resistance as they can be designed to interact with bacterial targets that are less prone to developing resistance mechanisms. Isothiazolone and imidazolium salt are well-known inhibitors of histidine kinases [31]. Since the late 1940s, bacitracin, a polypeptide antibiotic made by the bacteria Bacillus licheniformis and Bacillus subtilis, has been used in clinical settings to treat staphylococcal infections, including those caused by S. aureus. Bacitracin inhibits cell wall production by binding to undecaprenyl pyrophosphate, and it has been effective when combined with other antibiotics to be used as a topical medication [32,33]. Daptomycin (DAP) is a lipopeptide antibiotic that has excellent antibacterial activity against most of the clinically significant Gram-positive bacteria [34]. Vancomycin is a branched, tricyclic, glycosylated peptide that was first isolated from the soil bacterium Streptomyces orientalis in 1956, and it is effective against Gram-positive microorganisms [35]. Luteolin and tetracycline are flavonoid compounds that are used as antibiotics [36,37].
Coumarin derivatives such as G4 poloxamer nanoparticles loaded with bioactive coumarin were evaluated for the inhibition of S. aureus [38], and coumarins extracted from the roots of Ferulago campestris showed potent antibacterial activity [39]. Synthetic coumarin derivatives like coumarin aminophosphonates [40] and various coumarin derivatives [41] have been reported for the inhibition of S. aureus.
Novobiocin and armillarisin A are well-known coumarin-containing antibiotics [42,43,44,45]. Many of the small molecules containing coumarin analogs, such as coumarin-triazoles (1) [46], coumarin-pyrazole (2) [47], pyranocoumarin and coumarin−sulphonamide hybrids (3) [48] and 3-amidocoumarins (4) [49], are found to inhibit S. aureus (Figure 1). Indoles play a vital role in antibacterial drug discovery [50], and verruculogen and tryprostatins A are commonly used antibiotics having indole motifs [51]. Indole diketopiperazine alkaloids (5) [52], benzothiophene–indole hybrids (6) [53] and deoxyribofuranosyl indoles (7) [54] are some of the reported indole derivatives that serve as inhibitors of S. aureus.
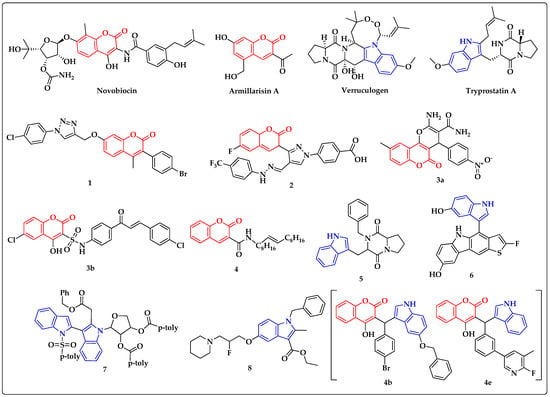
Figure 1.
Reported antibacterial and histidine kinase inhibitors containing coumarin (red) and indole (blue) motifs.
Given the major task of developing small molecules containing coumarin and indoles, we have developed novel bisindoles and heterodimers of indole-clubbed coumarin analogs that are active against S. aureus. Further, in silico docking analysis showed that compounds 4b and 4e can target histidine kinases in bacteria.
2. Materials and Methods
2.1. Chemistry
The headway of the response was distinguished utilizing thin layer chromatography (TLC). Analytical TLC was performed on precoated Merck silica gel 60 F254 plates involving ethyl acetate and hexane as eluents, and spots were recognized under UV light. 1H NMR and 13C NMR spectra were recorded on an Agilent NMR instrument in DMSO solvent. Chemical shifts were expressed in ppm comparative with TMS. Mass spectra were recorded on an Agilent LC-MS. All solvents and reagents were monetarily accessible and of reagent grade.
2.1.1. General Procedure for Synthesis of Indole Derivatives 4(a–m)
A mixture of aryl aldehyde (1) (1 mmol), substituted indoles (2) (1 mmol), substituted coumarin (3) (1 mmol) and glycine was stirred in water:chloroform (1:1) at 70 °C for about 12 h (Table 1). Further, the reaction mass was extracted to an ethyl acetate (25 mL × 3) layer, and the solvent was removed under reduced pressure. The crude products were purified through the column chromatography technique to obtain the desired compounds 4(a–m). The novel compounds were confirmed by 1H, 13C NMR and mass spectroscopy (Supplementary file contains spectral data of newly synthesized compounds).

Table 1.
Conditions of reaction.
2.1.2. 3-((4-bromophenyl)(1-methyl-1H-indol-3-yl)methyl)-4-hydroxy-2H-chromen-2-one (4a)
1H NMR (400 MHz, CDCl3) δ 7.65 (dd, J = 8.0, 1.0 Hz, 1H), 7.49 (dd, J = 8.6, 2.5 Hz, 4H), 7.34 (d, J = 1.9 Hz, 2H), 7.32 (s, 2H), 7.29 (s, 1H), 7.25–7.20 (m, 2H), 7.10 (dd, J = 10.9, 3.9 Hz, 1H), 6.53 (s, 1H), 5.95 (s, 1H), 3.75 (s, 3H); 13C NMR (100 MHz, CDCL3) δ 163.81, 162.70, 153.56, 140.21, 138.71, 132.78, 132.67, 130.74, 129.03, 127.30, 124.49, 124.08, 123.78, 121.73, 120.97, 120.06, 117.07, 116.32, 114.60, 110.39, 105.75, 39.61, 33.17; calculated mass for C25H18BrNO3 = 459.05; obtained mass = 460.79.
2.1.3. 3-((5-(benzyloxy)-1H-indol-3-yl)(4-bromophenyl)methyl)-4-hydroxy-2H-chromen-2-one (4b)
1H NMR (400 MHz, CDCl3) δ 8.25 (s, 1H), 7.56 (dd, J = 7.9, 1.4 Hz, 1H), 7.50–7.45 (m, 1H), 7.39 (d, J = 8.4 Hz, 2H), 7.26 (d, J = 8.4 Hz, 5H), 7.21 (d, J = 2.7 Hz, 2H), 7.17–7.12 (m, 4H), 7.03 (t, J = 7.4 Hz, 1H), 6.93 (dd, J = 8.9, 2.3 Hz, 1H), 6.84 (d, J = 2.1 Hz, 1H), 6.56 (s, 1H), 5.81 (s, 1H); 13C NMR (100 MHz, CDCl3) δ 163.86, 162.80, 154.49, 153.56, 140.08, 137.78, 132.92, 132.81, 132.67, 130.70, 129.04, 128.32, 127.94, 127.21, 125.05, 124.53, 123.87, 121.76, 117.11, 116.31, 116.07, 115.68, 113.15, 105.24, 102.85, 71.02, 39.62; calculated mass for C31H22BrNO4 = 552.42; observed mass = 552.14.
2.1.4. 3,3′-((4-bromophenyl) methylene) bis(2-methyl-1H-indole) (4c)
1H NMR (400 MHz, CDCl3) δ 5.89 (s, 1H), 6.64–6.66 (m, 2H), 7.03–7.07 (m, 2H), 7.19–7.23 (m, 2H), 7.26–7.31 (m, 4H), 7.36–7.40 (m, 4H), 7.92 (s, 2H), 2.35 (s, 6H); 13C NMR (100 MHz, CDCl3) δ 39.64, 111.04, 111.13, 119.21, 119.25, 119.38, 119.74, 119.83, 119.95, 121.94, 122.09, 123.61, 126.91, 128.23, 128.38, 128.74, 130.09, 131.81, 136.72, 142.59, 12.47.
2.1.5. 3-((3-(6-fluoro-5-methylpyridin-3-yl)phenyl)(1H-indol-3-yl)methyl)-4-hydroxy-7-meth-oxy-2H-chromen-2-one (4d)
1H NMR (400 MHz, DMSO) δ 10.95 (s, 1H), 8.21 (s, 1H), 8.03 (dd, J = 9.6, 1.7 Hz, 1H), 7.98 (d, J = 9.6 Hz, 1H), 7.65 (s, 1H), 7.42–7.40 (m, 4H), 7.37 (d, J = 8.1 Hz, 2H), 7.16 (d, J = 1.9 Hz, 1H), 7.09 (t, J = 7.1 Hz, 1H), 6.99 (d, J = 2.3 Hz, 1H), 6.97 (d, J = 4.0 Hz, 1H), 6.94 (d, J = 7.0 Hz, 1H), 6.18 (s, 1H), 3.87 (s, 3H), 2.30 (s, 3H); 13C NMR (100 MHz, DMSO) δ 162.75, 161.50, 154.53, 144.41, 142.73, 142.58, 140.94, 140.88, 136.53, 135.92, 128.97, 128.66, 127.65, 127.30, 125.12, 124.91, 124.76, 121.28, 119.86, 119.53, 119.00, 118.76, 114.77, 112.19, 111.96, 109.92, 106.08, 100.83, 56.35, 37.60, 14.44; calculated mass for C31H23FN2O4 = 506.16; obtained mass = 507.23.
2.1.6. 3-((3-(6-fluoro-5-methylpyridin-3-yl)phenyl)(1H-indol-3-yl)methyl)-4-hydroxy-2H-chromen-2-one (4e)
1H NMR (400 MHz, DMSO) δ 10.96 (s, 1H), 8.20 (s, 1H), 8.06 (d, J = 8.1 Hz, 1H), 8.02 (dd, J = 9.6, 1.9 Hz, 1H), 7.62 (dd, J = 11.2, 3.9 Hz, 3H), 7.39 (d, J = 7.8 Hz, 7H), 7.18 (d, J = 2.0 Hz, 1H), 7.08 (t, J = 7.4 Hz, 1H), 6.95 (t, J = 7.3 Hz, 1H), 6.21 (s, 1H), 2.29 (s, 3H); 13C NMR (100 MHz, DMSO) δ 162.72, 162.30, 160.93, 152.69, 144.12, 140.96, 140.90, 136.53, 135.97, 132.37, 129.00, 128.65, 127.64, 127.29, 124.99, 124.85, 124.25, 123.95, 121.33, 119.86, 119.53, 118.99, 118.81, 116.78, 116.69, 114.43, 111.99, 108.79, 37.75, 14.46.
2.1.7. 3-((3-(6-chloro-5-methylpyridin-3-yl)phenyl)(1H-indol-3-yl)methyl)-4-hydroxy-2H-chromen-2-one (4f)
1H NMR (400 MHz, DMSO) δ 10.83 (s, 1H), 8.36 (s, 1H), 8.09 (d, J = 67.2 Hz, 1H), 7.74 (dd, J = 27.9, 10.1 Hz, 3H), 7.53 (s, 2H), 7.36 (d, J = 17.2 Hz, 5H), 7.03 (s, 1H), 6.89 (s, 3H), 5.94 (s, 1H), 2.50 (s, 3H); 13C NMR (100 MHz, DMSO) δ 166.35, 146.28, 140.84, 139.96, 137.06, 135.98, 129.96, 129.61, 129.25, 129.19, 128.51, 128.18, 127.31, 127.07, 126.78, 125.96, 125.02, 124.08, 122.48, 121.38, 119.57, 118.67, 118.39, 111.95, 51.43, 32.59, 24.14.
2.1.8. 3-((3-(6-chloro-5-methylpyridin-3-yl)phenyl)(1H-indol-3-yl)methyl)-4-hydroxy-7-methoxy-2H-chromen-2-one (4g)
1H NMR (400 MHz, DMSO) δ 10.85 (s, 1H), 8.38 (d, J = 6.9 Hz, 1H), 8.02 (s, 1H), 7.79 (d, J = 7.4 Hz, 1H), 7.74 (s, 1H), 7.69 (d, J = 7.6 Hz, 1H), 7.37–7.33 (m, 4H), 7.04 (t, J = 7.4 Hz, 2H), 6.89 (d, J = 10.2 Hz, 3H), 5.96 (s, 1H), 3.40 (s, 3H), 1.89 (s, 3H); 13C NMR (100 MHz, DMSO) δ 166.38, 157.66, 146.28, 140.85, 139.97, 137.07, 135.99, 129.62, 129.25, 129.19, 128.20, 127.33, 127.08, 126.79, 125.98, 125.03, 124.09, 121.39, 119.59, 118.68, 118.41, 111.96, 51.45, 32.60, 24.15.
2.1.9. 3-((3-bromophenyl)(1-methyl-1H-indol-3-yl)methyl)-4-hydroxy-7-methoxy-2H-chromen-2-one (4h)
1H NMR (400 MHz, DMSO) δ 11.81 (s, 1H), 8.04 (s, 1H), 7.63 (s, 1H), 7.39 (d, J = 8.4 Hz, 5H), 7.31 (d, J = 0.5 Hz, 2H), 7.19 (d, J = 29.4 Hz, 3H), 6.97 (s, 1H), 6.10 (s, 1H), 3.83 (s, 3H), 3.76 (s, 3H); 13C NMR (100 MHz, DMSO) δ 162.24, 161.15, 152.70, 146.14, 136.89, 132.53, 131.20, 130.41, 129.17, 129.11, 127.84, 127.81, 124.33, 124.03, 121.64, 121.55, 119.06, 116.74, 116.71, 113.04, 110.21, 108.34, 55.82, 37.22, 32.84.
2.1.10. 3-((3-bromophenyl)(1-methyl-1H-indol-3-yl)methyl)-4-hydroxy-2H-chromen-2-one (4i)
1H NMR (400 MHz, DMSO) δ 11.81 (s, 1H), 8.04 (s, 1H), 7.63 (s, 1H), 7.39 (d, J = 8.4 Hz, 5H), 7.31 (d, J = 0.5 Hz, 2H), 7.19 (d, J = 29.4 Hz, 3H), 6.97 (s, 1H), 6.10 (s, 1H), 3.76 (s, 3H); 13C NMR (100 MHz, DMSO) δ 162.24, 161.15, 152.70, 146.14, 136.89, 132.53, 131.20, 130.41, 129.17, 129.11, 127.84, 127.81, 124.33, 124.03, 121.64, 121.55, 119.06, 116.74, 116.71, 113.04, 110.21, 108.34, 37.22, 32.84; calculated mass for C25H18BrNO3 = 459.05; obtained mass = 460.15.
2.1.11. 3-((3-(6-chloro-5-methylpyridin-3-yl)phenyl)(1-methyl-1H-indol-3-yl)methyl)-4-hydroxy-2H-chromen-2-one (4j)
1H NMR (400 MHz, DMSO) δ 8.41 (d, J = 2.2 Hz, 1H), 8.05 (d, J = 8.1 Hz, 1H), 7.99 (d, J = 2.0 Hz, 1H), 7.67 (s, 1H), 7.61 (t, J = 7.2 Hz, 1H), 7.40 (dd, J = 7.9, 4.9 Hz, 5H), 7.37–7.33 (m, 3H), 7.16–7.11 (m, 2H), 6.96 (t, J = 7.4 Hz, 1H), 6.19 (s, 1H), 3.76 (s, 3H), 2.36 (s, 3H); 13C NMR (100 MHz, DMSO) δ 162.29, 160.99, 152.70, 149.81, 145.34, 144.06, 138.50, 136.97, 136.08, 135.75, 132.57, 132.41, 129.17, 129.08, 127.93, 127.35, 125.12, 124.26, 123.97, 121.45, 119.20, 118.93, 116.77, 116.70, 113.74, 110.15, 108.63, 37.76, 32.81, 19.49.
2.1.12. 3-((4-(benzyloxy)-1H-indol-3-yl) (3-(6-chloro-5-methylpyridin-3-yl) phenyl) methyl)-4-hydroxy-2H-chromen-2-one (4k)
1H NMR (400 MHz, DMSO) δ 8.41 (d, J = 2.2 Hz, 1H), 8.05 (d, J = 8.1 Hz, 1H), 7.99 (d, J = 2.0 Hz, 1H), 7.67 (s, 1H), 7.61 (t, J = 7.2 Hz, 1H), 7.49 (dd, J = 8.6, 2.5 Hz, 3H, 7.40 (dd, J = 7.9, 4.9 Hz, 5H), 7.37–7.33 (m, 3H), 7.25–7.20 (m, 2H) 7.16–7.11 (m, 2H), 6.96 (t, J = 7.4 Hz, 1H), 6.19 (s, 1H), 4.9 (s, 2H) 3.76 (s, 3H), 2.36 (s, 3H); 13C NMR (100 MHz, DMSO) δ 162.29, 160.99, 152.70, 149.81, 145.34, 144.06, 138.50, 136.97, 136.08, 136.76, 135.75, 132.57, 132.41, 129.17, 129.08, 128.94, 127.93, 127.6, 127.35, 127.13, 125.12, 124.26, 123.97, 121.45, 119.20, 118.93, 116.77, 116.70, 113.74, 110.15, 108.63,70.65, 37.76, 32.81, 19.49; calculated mass for C38H28ClN2O4 = 598.17; obtained mass = 599.14.
2.1.13. 3-((4-(6-chloro-5-methylpyridin-3-yl)phenyl)(2-methyl-2,7a-dihydro-1H-indol-3-yl)-methyl)-4-hydroxy-2H-chromen-2-one (4l)
1H NMR (400 MHz, DMSO) δ 10.81 (s, 1H), 8.56 (d, J = 2.0 Hz, 1H), 8.12 (s, 1H), 8.05 (d, J = 7.8 Hz, 1H), 7.64 (d, J = 8.3 Hz, 2H), 7.60 (d, J = 7.4 Hz, 1H), 7.37 (d, J = 8.7 Hz, 2H), 7.32 (d, J = 8.2 Hz, 2H), 7.22 (d, J = 8.2 Hz, 2H), 6.92 (t, J = 7.6 Hz, 1H), 6.78 (t, J = 7.5 Hz, 1H), 6.13 (s, 1H), 2.40 (s, 3H), 2.19 (s, 3H); 13C NMR (100 MHz, DMSO) δ 162.52, 161.08, 152.60, 149.63, 145.18, 143.07, 138.22, 135.54, 135.43, 134.12, 133.33, 132.56, 132.38, 129.22, 128.80, 127.25, 126.72, 124.26, 123.81, 120.03, 119.55, 118.45, 116.73, 110.72, 110.37, 107.63, 37.76, 26.81, 19.52, 13.02.
2.1.14. 3,3′-((4-(pyrimidin-5-yl) phenyl) methylene)bis(2-methyl-1H-indole (4m)
1H NMR (400 MHz, cdcl3) δ 9.14 (s, 1H), 8.85 (s, 2H), 7.57 (s, 1H), 7.43–7.41 (m, 3H), 7.40–7.35 (m, 4H), 7.26 (s, 2H), 7.00 (dd, J = 16.2, 8.3 Hz, 4H), 5.98 (s, 1H), 2.61 (s, 6H); 13C NMR (100 MHz, CDCl3) δ 156.88, 154.83, 135.03, 134.60, 133.72, 132.70, 131.93, 129.88, 129.24, 128.65, 128.42, 124.63, 120.76, 119.18, 119.10, 112.70, 110.13, 39.14, 12.46.
2.2. Antibacterial Activity of Compounds 4(a–m)
The newly synthesized compounds were evaluated for their antibacterial activity against Gram-positive (Staphylococcus aureus MW2 and Enterococcus faecalis MTCC 439) and Gram-negative (Klebsiella pneumonia MTCC 661, Acinetobacter baumannii MTCC 1425, Pseudomonas aeruginosa MICC 2453 and Enterobacter cloacae MTCC 509) bacterial strains by the agar well diffusion method as mentioned above [19]. Briefly, trypticase soy agar (TSA), brain heart infusion agar (BHI agar) and nutrient agar (NA) (pH = 7.2–7.4) plates were used. The agar plates were then inoculated with 100 µL of the test bacteria, which were grown to 1.5 × 108 cfu/mL (overnight culture). The test compounds, which were dissolved at a concentration of 1 mg/mL of (1:1) ethanol:distilled water, were added to each well and the plates were incubated for 24 h at 37 °C. Ethanol:distilled water (1:1) was used as a negative control. All experiments were conducted in triplicate. Compounds with the largest zone of inhibition were selected for minimum inhibitory concentration (MIC) tests. The activity of each compound was compared with that of tetracycline as a standard.
2.3. Minimum Inhibitory Concentration (MIC) of the Active Compounds
In vitro, antibacterial activity was determined by their MIC values [55,56]. A stock solution of test compounds (1 mg/mL) was dissolved in (1:1) ethanol:distilled water as a stock solution. Further, serial broth dilution was carried out to achieve compounds ranging from 120 to 500 µg/mL in TSB broth; different concentrations of test compounds (12, 15, 18…50 µL) were added to a standardized suspension (~106 cfu/mL) of the test bacterium in a tube containing 1 mL of TSB broth and incubated for 24 h at 37 °C. The minimum inhibitory concentration (MIC) was noted by observing the growth of bacteria. The lowest concentration of the drug at which there was no visible growth was considered as the MIC. Compounds 4b and 4e were further investigated for their ZOI against S. aureus, which was expressed as the diameter of the inhibition zone according to the agar well diffusion method. All experiments were conducted in triplicate and the values are represented as mean ± SD.
2.4. Molecular Docking Studies
The three-dimensional structure of histidine kinase was obtained from a database of protein structures (RCBS) with PDB ID: 5IS1. The protein preparation was performed using the Discovery Studio software (version 21.1.0.20298), in which initially water and heteroatoms were removed and further hydrogens were added to the protein structure and later saved in a PDB format file. The structure was then prepared for docking simulations using AutoDock4 (v4.2.6) (accessed on 23 May 2023). Later, ligand preparation was performed for compound 4b and luteolin and used for docking simulation. The binding site of histidine kinase was identified by generating a grid in AutoDock4 having grid dimensions of 40 × 60 × 50 Å, with spacing of 1 Å for both 4b and luteolin. The ligands were then docked into the binding site using the Lamarckian Genetic Algorithm (LGA) as the search algorithm. The docking parameters were set to a population size of 150, a maximum number of 2,500,000 energy evaluations and a mutation rate of 0.02 and crossover the rate of 0.80, and 10 docking runs were performed for compound 4b and luteolin, respectively.
The output of the docking simulations was analyzed using the AutoDock4 Tools (ADT) [57] software. The docked complexes were also visualized using PyMOL [58]; the Discovery Studio [59] and UCSF Chimera 1.16 [60] software were used to analyze the interactions between the ligand and the protein.
3. Results
3.1. Synthesis of Bisindoles and Heterodimers of Indole-Clubbed Coumarin Derivatives 4a–m
Using MCR, the synthesis of the condensation products of the indole, aldehyde and coumarin was carried out in the presence of different catalysts (Scheme 1). The four-component reaction was primarily optimized between 4-bromobenzaldehyde (1), 4-hydroxy coumarin (2), 5-benzyloxyindole (3) and iron oxide (10 mol%) in ethanol at 70 °C for 12 h and afforded 4b with a 50% yield (Table 1, entry 1). Further, the reaction was carried out using different solvents, like DMSO and a mixture of water:chloroform (1:1); no product was formed after 12 h of reaction time (Table 1, entry 4), whereas, in the water:chloroform mixture, the product formed with a better yield (73%) (Table 1, entry 2). The same reaction was carried out by adding a glycine catalyst, resulting in the formation of the desired product in a significant yield (92%) (Table 1, entry 3). Moreover, increasing the amount of catalyst had no effect on the percentage yield. From the above observations we have synthesized derivatives 4(a–m) (Table 2) employing glycine as catalyst (Scheme 2) using various substituted indole, coumarin and aldehydes (Figure 2).
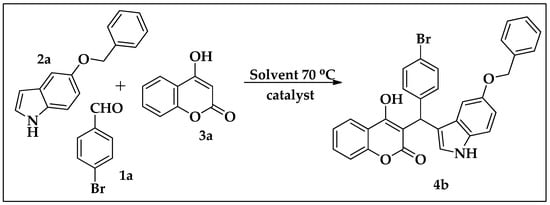
Scheme 1.
Optimized reaction scheme.

Table 2.
Bisindole and indole-clubbed coumarin derivatives.
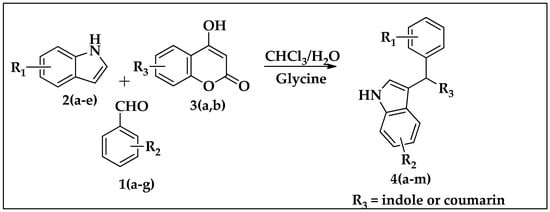
Scheme 2.
Synthesis of bisindoles and heterodimers of indole-clubbed coumarin derivatives 4(a–m).
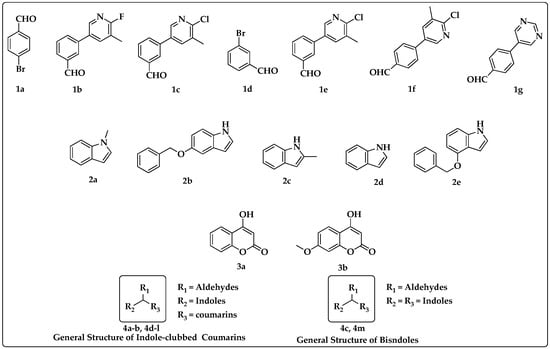
Figure 2.
Substituted functional groups and general structures of bisindoles and indole-clubbed coumarin derivatives.
A mixture of aryl aldehyde, substituted indoles, substituted coumarin, and glycine was stirred in water:chloroform (1:1) at 70 °C for about 12 h. After the completion of the reaction, the crude mass was extracted with ethyl acetate (25 mL × 3), distilled under high pressure, and purified through column chromatography to obtain the desired compounds 4(a–m). The novel compounds were confirmed by 1H, 13C NMR, and mass spectroscopy.
3.2. Antibacterial Activity of Newly Synthesized Heterodimers of Indole-Clubbed Coumarins
All synthesized compounds were screened for antibacterial activity. As 4b and 4e showed activity against S. aureus, these compounds were checked for their minimum inhibitory concentration (MIC) and zone of inhibition (ZOI).
MIC values were evaluated at a concentration range of 120–500 µg/mL, and the results are summarized in Table 3. The MIC value of compound 4b was 350 µg/mL and that of compound 4e was 160 µg/mL.

Table 3.
Minimum inhibitory concentration (MIC) of compounds 4b and 4e against S. aureus.
With this MIC value for 4b and 4e, the ZOI was calculated by the agar well diffusion method against S. aureus. The ZOI of 4b was 8 mm and that of compound 4e was 7.2 mm, as compared with tetracycline (30 µg/disc) as a positive control, showing 16.25 mm, as tabulated in Table 4.

Table 4.
Zone of inhibition (ZOI) of the synthesized compounds 4b and 4e against S. aureus in mm.
3.3. In Silico Molecular Interaction Studies of Novel Compound 4b in Inhibiting Histidine Kinase of S. aureus
To identify the potential targets for compound 4b, we herein used the Prediction of Activity Spectra for Substances (PASS) web tool [61], which predicts the likelihood of a compound to exhibit a particular type of biological activity based on its chemical structure. Therefore, compound 4b in smiles format was added to the PASS web tool and we obtained the results. The PASS web tool generated a list of predicted biological activity that the compound may exhibit. From the listed targets, histidine kinase was among the top six, with Pa and Pi values of 0.421 and 0.046, and was considered the target for the bacterium Staphylococcus aureus.
Therefore, we retrieved the histidine kinase protein (PDB ID: 5IS1) from RCBS and later bioinformatic studies were performed using AutoDock4 tools (ADT). In this study, we used AutoDock4 to simulate the binding of a novel compound to histidine kinase, a protein involved in the regulation of bacterial virulence and antibiotic resistance. Molecular docking studies revealed that our compound 4b had better binding affinity towards the active site of histidine kinase, showing −5.08 kcal/mol when compared to the compound luteolin, having binding affinity of −4.03 kcal/mol. The molecular interactions between compound 4b and its target site in histidine kinase involved hydrogen bonds, with the residue GLN-73 having a bond distance of 2.35 Å. π–cation and π–anion bonds were formed with the ASN-71, ASP-96 and ASP-98 amino acid residues. Hydrophobic interactions were observed in residues ALA-72 and ILE-100. These interactions make the protein and ligand complex stable. Meanwhile, the compound luteolin exhibited the formation of hydrogen bonds with SER-127, LYS-158 and VAL-160 with bond distances of 2.26 Å, 2.24 Å, 2.93 Å and 2.45 Å, respectively. Hydrophobic interactions were formed with the residues LYS-97, LEU-126 and LYS-159 (Figure 3A,B). Luteolin and tetracycline are both members of the flavonoid class of compounds. Tetracycline, a well-known antibiotic, is structurally related to luteolin, and luteolin is studied for its potential antibacterial properties [35]. They share similar core chemical structures but differ in functional groups. This structural similarity prompted us to explore their potential similarities in inhibitory activity against histidine kinase [30]. Thus, the docking simulations predicted a better interaction between the ligand (4b) and the histidine kinase protein, suggesting that the compound could be an effective inhibitor of histidine kinase, and compound 4b lies in close proximity to luteolin.
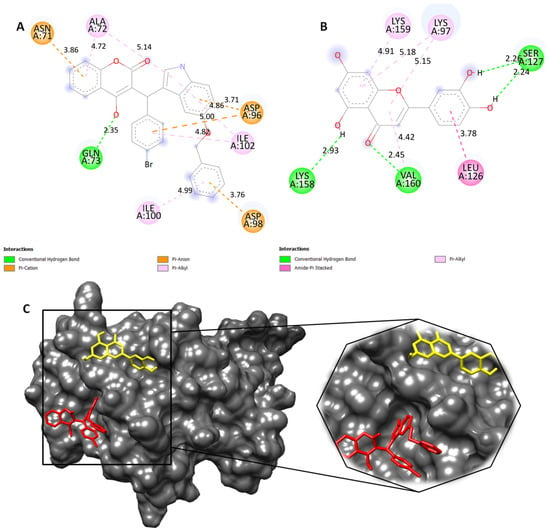
Figure 3.
(A) Two-dimensional structure of compound 4b (red) and (B) luteolin (yellow) showing molecular interactions with active site of histidine kinase. (C) Representation of 3D surface view of docked compounds in the groove of histidine kinase and its enlarged view for better visualization.
4. Discussion
Previously, it have been observed that the solvent and the presence of glycine played a significant part in the accomplishment of the reaction [62,63]. The outcomes propose that solvents likewise influenced the yield of compound 4b (Table 1). After the improvement of the reaction conditions, the extent of the technique was explored with a series of substituted aromatic aldehydes, coumarins and indoles. The outcomes are collated in Table 2. Several aldehydes were synthesized by the Suzuki coupling reaction. The coupling reaction was completed by utilizing 4-bromobenzaldehyde, with various boronic acids and 1,1′-bis(diphenylphosphino)ferrocenedichloropalladium (II) as the catalyst. As observed from Table 2, the indoles having both electron-withdrawing and electron-releasing functional groups underwent effective condensation with coumarin and aldehyde in the presence of a catalytic amount of glycine in water:chloroform (1:1) at 70 °C to afford the respected products 4(a–m) in good yields. It appears that the electronic effects and the nature of the substituents on the indole have a slight effect on both the reaction and reaction yield. The electron-donating group at the second position of the indole increased the formation of bisindoles, rather than the heteromeric product. It was found that the reaction pathway was directed towards the formation of bisindoles, as in 4c and 4m. In a few of the reactions, both homomeric and heteromeric products were observed, as in 4l.
The newly synthesized chemical compounds 4b and 4e exhibited promising antibacterial activity. These new data on the compounds might be helpful in their future development as novel antibacterial agents. From the present study, it may concluded that 4b and 4e have structural novelty and marked biological activity. This work explored on the structure and functions of biomolecules of drug development, as we previosuly reported [64,65,66,67,68,69,70,71,72,73]
5. Conclusions
Bacterial histidine kinases are components of two-component systems that serve as a promising target for new antibacterial compounds. In search of new chemical entities, we identified a new structure bearing a natural coumarin and indole rings that are attached to trigonal carbon derived from the benzene motif. The antibacterial study of the tested compounds revealed that two compounds were found to be active against pathogenic S. aureus, and our in silico docking study showed that the lead compound could target the histidine kinases of bacteria. In conclusion, we have identified selective antibacterial agents that could inhibit the histidine kinase of S. aureus.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/applmicrobiol3040084/s1, Figures in S2–S16 contain spectral data of newly synthesized compounds 4(a–m).
Author Contributions
L.K.P., R.R., A.R., A.M., P.M.U. and P.K.M., conceptualization, methodology, formal analysis and writing; S.L.G., formal analysis; S.G., P.B.S., N.S.S., A.Y.S., V.P. and B.B., conceptualization, methodology, software, data curation, original draft, validation, writing and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Vision Group on Science and Technology (CESEM) and the Government of Karnataka. This research was further supported by the National Natural Science Foundation of China (82172618); the Shenzhen Key Laboratory of Innovative Oncotherapeutics (ZDSYS20200820165400003) (Shenzhen Science and Technology Innovation Commission), China; Universities Stable Funding Key Projects (WDZC20200821150704001), China; the Shenzhen Bay Laboratory, Oncotherapeutics (21310031), China; and the Overseas Research Cooperation Project (HW2020008) (Tsinghua Shenzhen International Graduate School), China. L.K.P. thanks OBC Cell, University of Mysore, Mysuru, and A.R. thanks KSTEPS, DST, Govt. of Karnataka, India, for providing the fellowship.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are freely available within this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Campbell, I.B.; Macdonald, S.J.F.; Procopiou, P.A. Medicinal Chemistry in Drug Discovery in Big Pharma: Past, Present and Future. Drug Discov. Today 2018, 23, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Aatif, M.; Raza, M.A.; Javed, K.; Nashre-ul-Islam, S.M.; Farhan, M.; Alam, M.W. Potential Nitrogen-Based Heterocyclic Compounds for Treating Infectious Diseases: A Literature Review. Antibiotics 2022, 11, 1750. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, J.C.; Logan, K.R.H. A General Synthesis of the Penicillins. J. Am. Chem. Soc. 1959, 81, 5838–5839. [Google Scholar] [CrossRef]
- Chellat, M.F.; Raguž, L.; Riedl, R. Targeting Antibiotic Resistance. Angew. Chem. Int. Ed. 2016, 55, 6600–6626. [Google Scholar] [CrossRef] [PubMed]
- Grillone, K.; Riillo, C.; Rocca, R.; Ascrizzi, S.; Spanò, V.; Scionti, F.; Polerà, N.; Maruca, A.; Barreca, M.; Juli, G.; et al. The New Microtubule-Targeting Agent SIX2G Induces Immunogenic Cell Death in Multiple Myeloma. Int. J. Mol. Sci. 2022, 23, 10222. [Google Scholar] [CrossRef] [PubMed]
- Barreca, M.; Spanò, V.; Rocca, R.; Bivacqua, R.; Gualtieri, G.; Raimondi, M.V.; Gaudio, E.; Bortolozzi, R.; Manfreda, L.; Bai, R.; et al. Identification of Pyrrolo[3′,4′:3,4]Cyclohepta[1,2-d][1,2]Oxazoles as Promising New Candidates for the Treatment of Lymphomas. Eur. J. Med. Chem. 2023, 254, 115372. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Kim, D.G.; Lee, A.; Kim, Y.M.; Cui, L.; Kim, S.; Choi, I. Synthesis and Discovery of the First Potent Proteolysis Targeting Chimaera (PROTAC) Degrader of AIMP2-DX2 as a Lung Cancer Drug. J. Enzym. Inhib. Med. Chem. 2022, 38, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Hajduk, P.J.; Greer, J. A Decade of Fragment-Based Drug Design: Strategic Advances and Lessons Learned. Nat. Rev. Drug Discov. 2007, 6, 211–219. [Google Scholar] [CrossRef]
- Bivacqua, R.; Barreca, M.; Spanò, V.; Raimondi, M.V.; Romeo, I.; Alcaro, S.; Andrei, G.; Barraja, P.; Montalbano, A. Insight into Non-Nucleoside Triazole-Based Systems as Viral Polymerases Inhibitors. Eur. J. Med. Chem. 2023, 249, 115136. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A New Antibiotic Kills Pathogens without Detectable Resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]
- Sass, P.; Josten, M.; Famulla, K.; Schiffer, G.; Sahl, H.-G.; Hamoen, L.; Brötz-Oesterhelt, H. Antibiotic Acyldepsipeptides Activate ClpP Peptidase to Degrade the Cell Division Protein FtsZ. Proc. Natl. Acad. Sci. USA 2011, 108, 17474–17479. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.A.; Koehler, M.F.T.; Girgis, H.S.; Yan, D.; Chen, Y.; Chen, Y.; Crawford, J.J.; Durk, M.R.; Higuchi, R.I.; Kang, J.; et al. Optimized Arylomycins Are a New Class of Gram-Negative Antibiotics. Nature 2018, 561, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Shariati, A.; Arshadi, M.; Khosrojerdi, M.A.; Abedinzadeh, M.; Ganjalishahi, M.; Maleki, A.; Heidary, M.; Khoshnood, S. The Resistance Mechanisms of Bacteria against Ciprofloxacin and New Approaches for Enhancing the Efficacy of This Antibiotic. Front. Public Health 2022, 10, 1025633. [Google Scholar] [CrossRef] [PubMed]
- Shariati, A.; Noei, M.; Chegini, Z. Bacteriophages: The Promising Therapeutic Approach for Enhancing Ciprofloxacin Efficacy against Bacterial Infection. J. Clin. Lab. Anal. 2023, 37, e24932. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Staphylococcus aureus Infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, S.; Klinger-Strobel, M.; Bohnert, J.A.; Wendler, S.; Rödel, J.; Pletz, M.W.; Löffler, B.; Tuchscherr, L. Clinically Approved Drugs Inhibit the Staphylococcus aureus Multidrug NorA Efflux Pump and Reduce Biofilm Formation. Front. Microbiol. 2019, 10, 2762. [Google Scholar] [CrossRef] [PubMed]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [PubMed]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Bem, A.E.; Velikova, N.; Pellicer, M.T.; van Baarlen, P.; Marina, A.; Wells, J.M. Bacterial Histidine Kinases as Novel Antibacterial Drug Targets. ACS Chem. Biol. 2014, 10, 213–224. [Google Scholar] [CrossRef]
- Casino, P.; Rubio, V.; Marina, A. The Mechanism of Signal Transduction by Two-Component Systems. Curr. Opin. Struct. Biol. 2010, 20, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Kenney, L.J. How Important Is the Phosphatase Activity of Sensor Kinases? Curr. Opin. Microbiol. 2010, 13, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.C. Protein Histidine Kinases: Assembly of Active Sites and Their Regulation in Signaling Pathways. Curr. Opin. Microbiol. 2010, 13, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Wolanin, P.M.; Thomason, P.A.; Stock, J.B. Histidine protein kinases: Key signal transducers outside the animal kingdom. Genome Biol. 2002, 3, reviews3013.1. [Google Scholar] [CrossRef] [PubMed]
- Dikiy, I.; Edupuganti, U.R.; Abzalimov, R.R.; Borbat, P.P.; Srivastava, M.; Freed, J.H.; Gardner, K.H. Insights into Histidine Kinase Activation Mechanisms from the Monomeric Blue Light Sensor EL346. Proc. Natl. Acad. Sci. USA 2019, 116, 4963–4972. [Google Scholar] [CrossRef] [PubMed]
- Kenney, L.J. How Can a Histidine Kinase Respond to Mechanical Stress? Front. Microbiol. 2021, 12, 655942. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yu, C.; Wu, H.; Li, G.; Li, C.; Hong, W.; Yang, X.; Wang, H.; You, X. Recent Advances in Histidine Kinase-Targeted Antimicrobial Agents. Front. Chem. 2022, 10, 866392. [Google Scholar] [CrossRef] [PubMed]
- Vo, C.D.; Shebert, H.L.; Zikovich, S.; Dryer, R.A.; Huang, T.P.; Moran, L.J.; Cho, J.; Wassarman, D.R.; Falahee, B.E.; Young, P.D.; et al. Repurposing Hsp90 Inhibitors as Antibiotics Targeting Histidine Kinases. Bioorganic Med. Chem. Lett. 2017, 27, 5235–5244. [Google Scholar] [CrossRef]
- Mascher, T.; Helmann, J.D.; Unden, G. Stimulus Perception in Bacterial Signal-Transducing Histidine Kinases. Microbiol. Mol. Biol. Rev. 2006, 70, 910–938. [Google Scholar] [CrossRef]
- Singh, V.; Dhankhar, P.; Kumar, P. Bacterial Histidine Kinases as Potential Antibacterial Drug Targets. Protein Kinase Inhib. 2022, 26, 711–734. [Google Scholar] [CrossRef]
- Roychoudhury, S.; Zielinski, N.A.; Ninfa, A.J.; Allen, N.E.; Jungheim, L.N.; Nicas, T.I.; Chakrabarty, A.M. Inhibitors of Two-Component Signal Transduction Systems: Inhibition of Alginate Gene Activation in Pseudomonas Aeruginosa. Proc. Natl. Acad. Sci. USA 1993, 90, 965–969. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Anker, H.; Meleney, F.L. Bacitracin: A New Antibiotic Produced by a Member of the B. subtilis Group. Science 1945, 102, 376–377. [Google Scholar] [CrossRef] [PubMed]
- Stone, K.J.; Strominger, J.L. Mechanism of Action of Bacitracin: Complexation with Metal Ion and C55-Isoprenyl Pyrophosphate. Proc. Natl. Acad. Sci. USA 1971, 68, 3223–3227. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhang, D.; Li, G.; Liu, J.; He, G.; Zhang, P.; Yang, L.; Zhu, H.; Xu, N.; Liang, S. Antibacterial Mechanism of Daptomycin Antibiotic against Staphylococcus aureus Based on a Quantitative Bacterial Proteome Analysis. J. Proteom. 2017, 150, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Dinu, V.; Lu, Y.; Weston, N.; Lithgo, R.; Coupe, H.; Channell, G.; Adams, G.G.; Torcello Gómez, A.; Sabater, C.; Mackie, A.; et al. The Antibiotic Vancomycin Induces Complexation and Aggregation of Gastrointestinal and Submaxillary Mucins. Sci. Rep. 2020, 10, 960. [Google Scholar] [CrossRef] [PubMed]
- Shamsudin, N.F.; Ahmed, Q.U.; Mahmood, S.; Ali Shah, S.A.; Khatib, A.; Mukhtar, S.; Alsharif, M.A.; Parveen, H.; Zakaria, Z.A. Antibacterial Effects of Flavonoids and Their Structure-Activity Relationship Study: A Comparative Interpretation. Molecules 2022, 27, 1149. [Google Scholar] [CrossRef] [PubMed]
- Advances in Clinical Chemistry|Book Series|ScienceDirect.com by Elsevier. 1 January 2023. Available online: https://www.sciencedirect.com/bookseries/advances-in-clinical-chemistry (accessed on 20 July 2023).
- Foudah, A.I.; Alqarni, M.H.; Ross, S.A.; Alam, A.; Salkini, M.A.; Kumar, P. Site-Specific Evaluation of Bioactive Coumarin-Loaded Dendrimer G4 Nanoparticles against Methicillin Resistant Staphylococcus aureus. ACS Omega 2022, 7, 34990–34996. [Google Scholar] [CrossRef]
- Basile, A.; Sorbo, S.; Spadaro, V.; Bruno, M.; Maggio, A.; Faraone, N.; Rosselli, S. Antimicrobial and Antioxidant Activities of Coumarins from the Roots of Ferulago campestris (Apiaceae). Molecules 2009, 14, 939–952. [Google Scholar] [CrossRef]
- Yang, X.-C.; Zeng, C.-M.; Avula, S.R.; Peng, X.-M.; Geng, R.-X.; Zhou, C.-H. Novel Coumarin Aminophosphonates as Potential Multitargeting Antibacterial Agents against Staphylococcus aureus. Eur. J. Med. Chem. 2023, 245, 114891. [Google Scholar] [CrossRef]
- Ranjan Sahoo, C.; Sahoo, J.; Mahapatra, M.; Lenka, D.; Kumar Sahu, P.; Dehury, B.; Nath Padhy, R.; Kumar Paidesetty, S. Coumarin Derivatives as Promising Antibacterial Agent(s). Arab. J. Chem. 2021, 14, 102922. [Google Scholar] [CrossRef]
- Vickers, A.A.; Chopra, I.; O’Neill, A.J. Intrinsic Novobiocin Resistance in Staphylococcus Saprophyticus. Antimicrob. Agents Chemother. 2007, 51, 4484–4485. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.J.; Hansen, S.L.; Tatem, B.A.; Auger, F.; Standiford, H.C. Activity of Novobiocin against Methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 1985, 15, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Wishnow, R.M.; Strominger, J.L.; Birge, C.H.; Threnn, R.H. Biochemical Effects of Novobiocin on Staphylococcus aureus. J. Bacteriol. 1965, 89, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Kashman, Y.; Gustafson, K.R.; Fuller, R.W.; Cardellina, J.H.; McMahon, J.B.; Currens, M.J.; Buckheit, R.W.; Hughes, S.H.; Cragg, G.M.; Boyd, M.R. HIV Inhibitory Natural Products. Part 7. The Calanolides, a Novel HIV-Inhibitory Class of Coumarin Derivatives from the Tropical Rainforest Tree, Calophyllum lanigerum. J. Med. Chem. 1992, 35, 2735–2743. [Google Scholar] [CrossRef]
- Dharavath, R.; Nagaraju, N.; Reddy, M.R.; Ashok, D.; Sarasija, M.; Vijjulatha, M.; Vani, T.; Jyothi, K.; Prashanthi, G. Microwave-Assisted Synthesis, Biological Evaluation and Molecular Docking Studies of New Coumarin-Based 1,2,3-Triazoles. RSC Adv. 2020, 10, 11615–11623. [Google Scholar] [CrossRef] [PubMed]
- Alnufaie, R.; Raj KC, H.; Alsup, N.; Whitt, J.; Andrew Chambers, S.; Gilmore, D.; Alam, M.A. Synthesis and Antimicrobial Studies of Coumarin-Substituted Pyrazole Derivatives as Potent Anti Staphylococcus aureus Agents. Molecules 2020, 25, 2758. [Google Scholar] [CrossRef] [PubMed]
- Alshibl, H.M.; Al-Abdullah, E.S.; Haiba, M.E.; Alkahtani, H.M.; Awad, G.E.A.; Mahmoud, A.H.; Ibrahim, B.M.M.; Bari, A.; Villinger, A. Synthesis and Evaluation of New Coumarin Derivatives as Antioxidant, Antimicrobial, and AntiInflammatory Agents. Molecules 2020, 25, 3251. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.K.; Singh, V.; Tiwari, N.; Butcher, R.J.; Katiyar, D. Synthesis, Antimicrobial and Chitinase Inhibitory Activities of 3-Amidocoumarins. Bioorganic Chem. 2020, 98, 103700. [Google Scholar] [CrossRef]
- Lee, J.-H.; Cho, H.S.; Kim, Y.; Kim, J.-A.; Banskota, S.; Cho, M.H.; Lee, J. Indole and 7-Benzyloxyindole Attenuate the Virulence of Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2013, 97, 4543–4552. [Google Scholar] [CrossRef]
- Qin, H.-L.; Liu, J.; Fang, W.-Y.; Ravindar, L.; Rakesh, K.P. Indole-Based Derivatives as Potential Antibacterial Activity against Methicillin-Resistance Staphylococcus aureus (MRSA). Eur. J. Med. Chem. 2020, 194, 112245. [Google Scholar] [CrossRef]
- Jia, B.; Ma, Y.; Liu, B.; Chen, P.; Hu, Y.; Zhang, R. Synthesis, Antimicrobial Activity, Structure-Activity Relationship, and Molecular Docking Studies of Indole Diketopiperazine Alkaloids. Front. Chem. 2019, 7, 837. [Google Scholar] [CrossRef] [PubMed]
- Seethaler, M.; Hertlein, T.; Hopke, E.; Köhling, P.; Ohlsen, K.; Lalk, M.; Hilgeroth, A. Novel Effective Fluorinated Benzothiophene-Indole Hybrid Antibacterials against S. aureus and MRSA Strains. Pharmaceuticals 2022, 15, 1138. [Google Scholar] [CrossRef] [PubMed]
- Paudel, A.; Hamamoto, H.; Kobayashi, Y.; Yokoshima, S.; Fukuyama, T.; Sekimizu, K. Identification of Novel Deoxyribofuranosyl Indole Antimicrobial Agents. J. Antibiot. 2011, 65, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.M. Determination of Minimum Inhibitory Concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- Huey, R.; Morris, G.M.; Olson, A.J.; Goodsell, D.S. A Semiempirical Free Energy Force Field with Charge-Based Desolvation. J. Comput. Chem. 2007, 28, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L.L.C.; DeLano, W. PyMOL. 2020. Available online: http://www.pymol.org/pymol (accessed on 23 May 2023).
- BIOVIA Dassault Systèmes. Discovery Studio Visualizer, 21.1.0.20298; Dassault Systèmes: San Diego, CA, USA, 2020. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Lagunin, A.; Stepanchikova, A.; Filimonov, D.; Poroikov, V. PASS: Prediction of Activity Spectra for Biologically Active Substances. Bioinformatics 2000, 16, 747–748. [Google Scholar] [CrossRef]
- Datta, B.; Pasha, M.A. Glycine Catalyzed Convenient Synthesis of 2-Amino-4H-Chromenes in Aqueous Medium under Sonic Condition. Ultrason. Sonochem. 2012, 19, 725–728. [Google Scholar] [CrossRef]
- Valot, L.; Maumus, M.; Montheil, T.; Martinez, J.; Noël, D.; Mehdi, A.; Subra, G. Biocompatible Glycine-Assisted Catalysis of the Sol-Gel Process: Development of Cell-Embedded Hydrogels. ChemPlusChem 2019, 84, 1720–1729. [Google Scholar] [CrossRef]
- Sulaiman, N.B.; Mohan, C.D.; Basappa, S.; Pandey, V.; Rangappa, S.; Bharathkumar, H.; Kumar, A.P.; Lobie, P.E.; Rangappa, K.S. An azaspirane derivative suppresses growth and induces apoptosis of ER-positive and ER-negative breast cancer cells through the modulation of JAK2/STAT3 signaling pathway. Int J Oncol. 2016, 49, 1221–1229. [Google Scholar] [CrossRef]
- Bharathkumar, H.; Mohan, C.D.; Ananda, H.; Fuchs, J.E.; Li, F.; Rangappa, S.; Surender, M.; Bulusu, K.C.; Girish, K.S.; Sethi, G.; et al. Microwave-assisted synthesis, characterization and cytotoxic studies of novel estrogen receptor α ligands towards human breast cancer cells. Bioorg. Med. Chem. Lett. 2015, 25, 1804–1807. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, V.; Chevalier, F.; Imberty, A.; Leeflang, B.R.; Basappa; Sugahara, K.; Kamerling, J.P. Conformational studies on five octasaccharides isolated from chondroitin sulfate using NMR spectroscopy and molecular modeling. Biochemistry 2007, 46, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Rangappa, K.S.; Basappa. New cholinesterase inhibitors: Synthesis and structure–activity relationship studies of 1,2-benzisoxazole series and novel imidazolyl-d2-isoxazolines. J. Phys. Org. Chem. 2005, 18, 773–778. [Google Scholar] [CrossRef]
- Sebastian, A.; Pandey, V.; Mohan, C.D.; Chia, Y.T.; Rangappa, S.; Mathai, J.; Baburajeev, C.P.; Paricharak, S.; Mervin, L.H.; Bulusu, K.C.; et al. Novel Adamantanyl-Based Thiadiazolyl Pyrazoles Targeting EGFR in Triple-Negative Breast Cancer. ACS Omega 2016, 1, 1412–1424. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, V.; Mohan, C.D.; Baburajeev, C.P.; Rangappa, S.; Jagadish, S.; Fuchs, J.E.; Sukhorukov, A.Y.; Chandra; Mason, D.J.; Sharath; et al. Synthesis and characterization of novel oxazines and demonstration that they specifically target cyclooxygenase 2. Bioorg. Med. Chem. Lett. 2015, 25, 2931–2936. [Google Scholar] [CrossRef] [PubMed]
- Basappa; Sugahara, K.; Thimmaiah, K.N.; Bid, H.K.; Houghton, P.J.; Rangappa, K.S. Anti-tumor activity of a novel HS-mimetic-vascular endothelial growth factor binding small molecule. PLoS ONE 2012, 7, e39444. [Google Scholar] [CrossRef]
- Priya, B.S.; Swamy, S.N.; Tejesvi, M.V.; Basappa; Sarala, G.; Gaonkar, S.L.; Naveen, S.; Prasad, J.S.; Rangappa, K.S. Synthesis, characterization, antimicrobial and single crystal X-ray crystallographic studies of some new sulfonyl, 4-chloro phenoxy benzene and dibenzoazepine substituted benzamides. Eur. J. Med. Chem. 2006, 41, 1262–1270. [Google Scholar] [CrossRef]
- Rakesh, K.S.; Jagadish, S.; Vinayaka, A.C.; Hemshekhar, M.; Paul, M.; Thushara, R.M.; Sundaram, M.S.; Swaroop, T.R.; Mohan, C.D.; Basappa; et al. A new ibuprofen derivative inhibits platelet aggregation and ROS mediated platelet apoptosis. PLoS ONE 2014, 9, e107182, Erratum in PLoS ONE 2014, 9, e114675. [Google Scholar] [CrossRef]
- Basappa; Rangappa, K.S.; Sugahara, K. Roles of glycosaminoglycans and glycanmimetics in tumor progression and metastasis. Glycoconj. J. 2014, 31, 461–467. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).