Phenotypic and Draft Genome Sequence Analyses of a Paenibacillus sp. Isolated from the Gastrointestinal Tract of a North American Gray Wolf (Canis lupus)
Abstract
:1. Introduction
2. Materials and Methods
3. Results
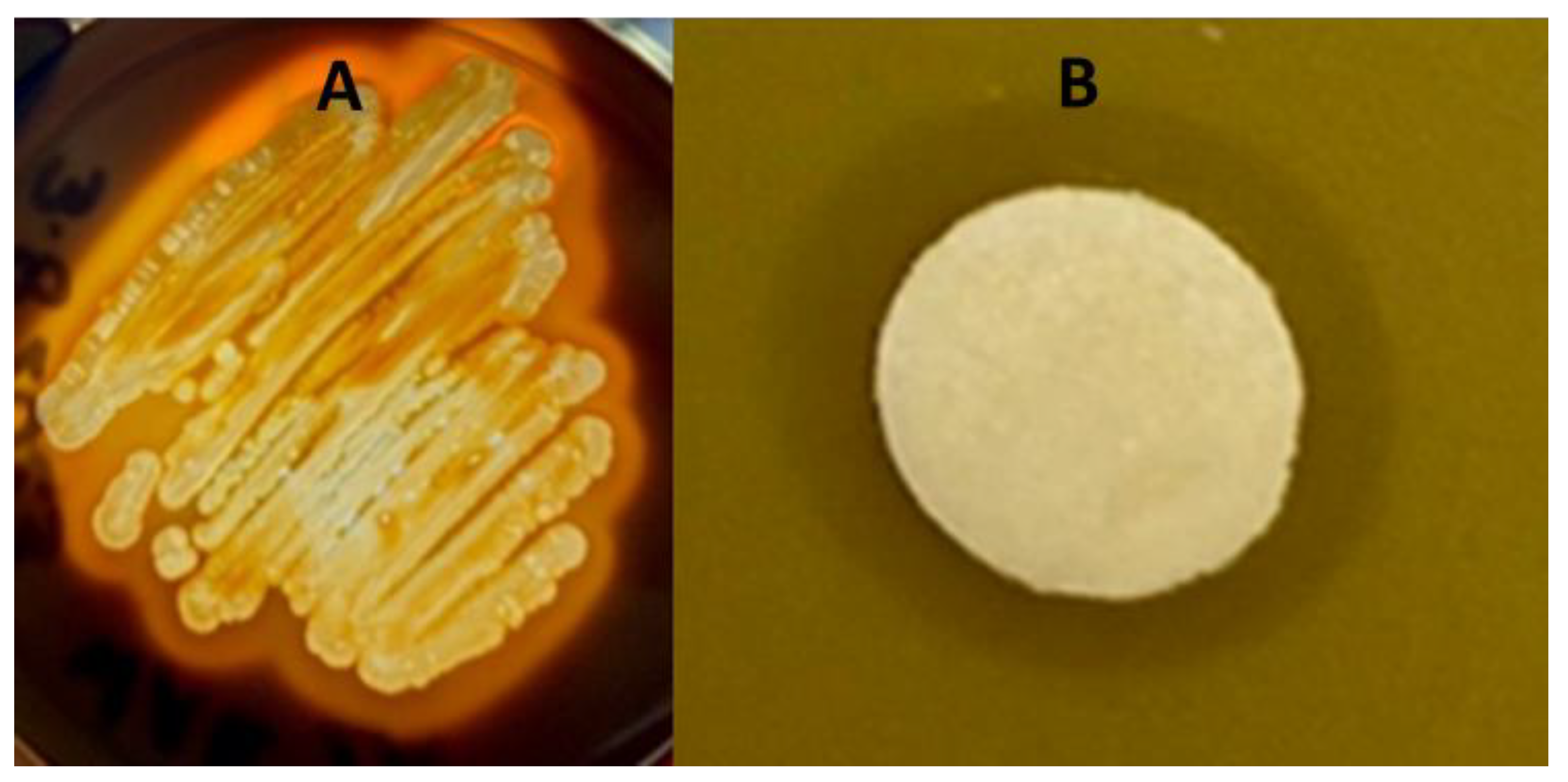
3.1. The Isolation of Bacteria from a Gray Wolf’s Gastointestinal Tract and Its Phenotypic Characterisitcs
3.2. Growth Inhibition of the Target Bacteria
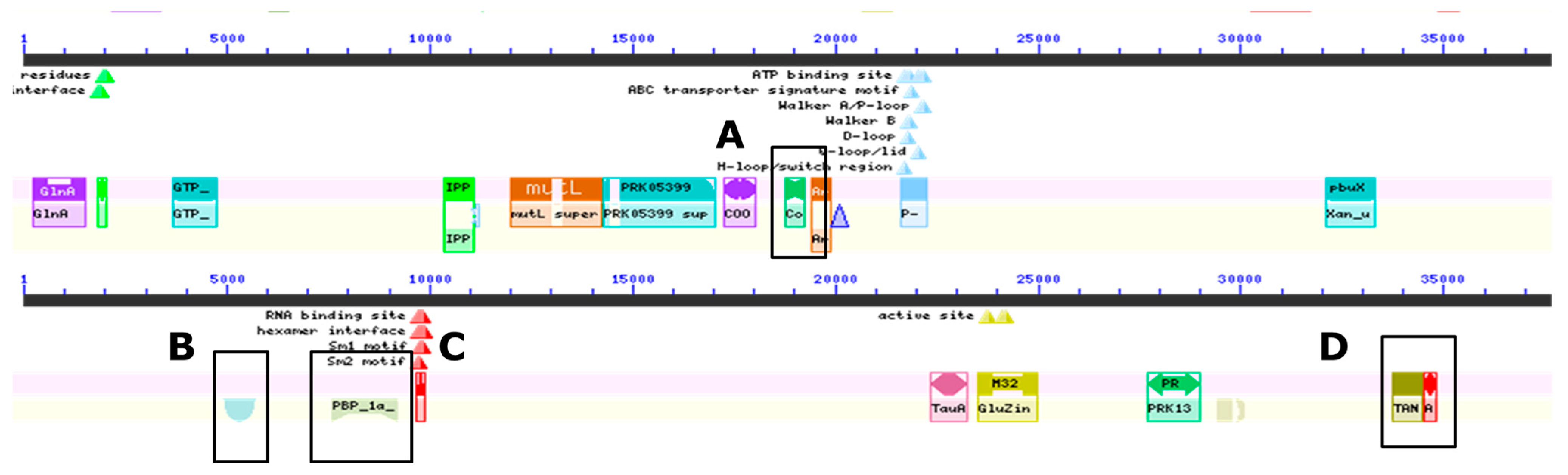
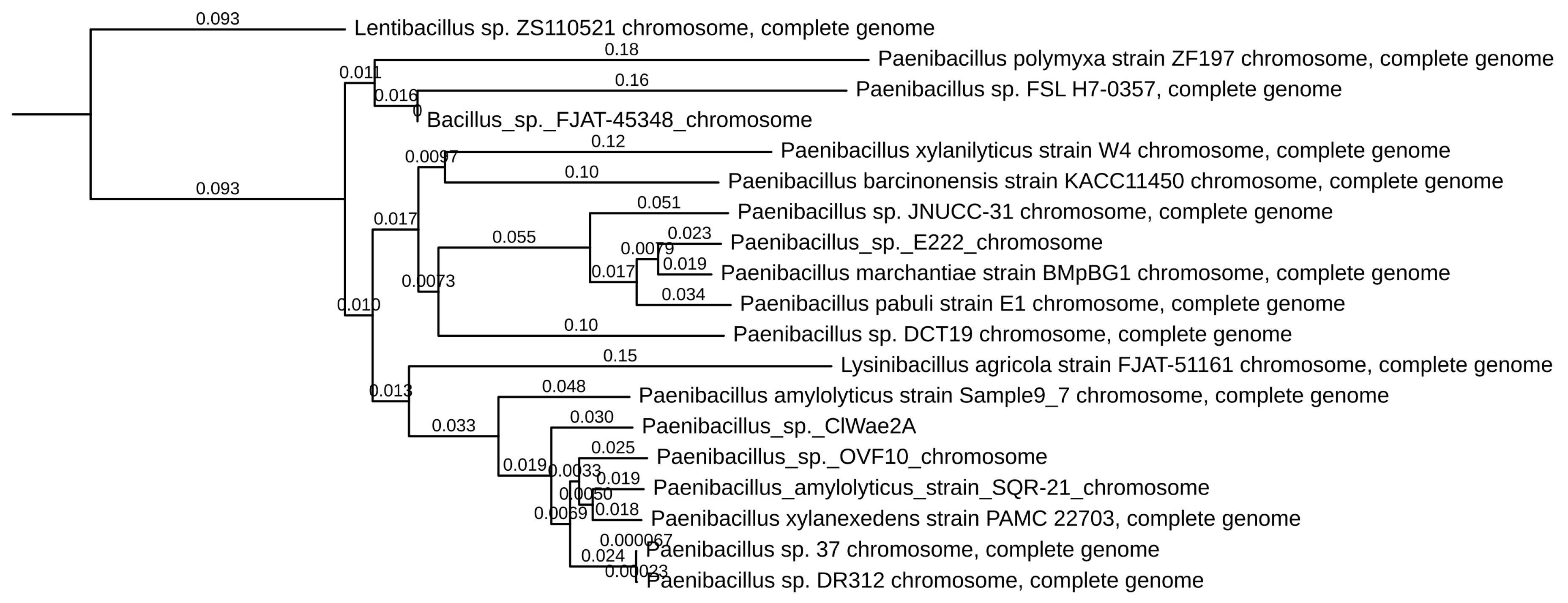
3.3. Genomic Analyses of the Gray Wolf Bacterium
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vázquez-Baeza, Y.; Hyde, E.R.; Suchodolski, J.S.; Knight, R. Dog and human inflammatory bowel disease rely on overlapping yet distinct dysbiosis networks. Nat. Microbiol. 2016, 1, 16177. [Google Scholar] [CrossRef] [PubMed]
- Kopper, J.J.; Iennarella-Servantez, C.; Jergens, A.E.; Sahoo, D.K.; Guillot, E.; Bourgois-Mochel, A.; Martinez, M.N.; Allenspach, K.; Mochel, J.P. Harnessing the biology of canine intestinal organoids to heighten understanding of inflammatory bowel disease pathogenesis and accelerate drug discovery: A one health approach. Front. Toxicol. 2021, 3, 773953. [Google Scholar] [CrossRef] [PubMed]
- Rhimi, S.; Kriaa, A.; Mariaule, V.; Saidi, A.; Drut, A.; Jablaoui, A.; Akermi, N.; Maguin, E.; Hernandez, J.; Rhimi, M. The Nexus of Diet, Gut Microbiota, and Inflammatory Bowel Diseases in Dogs. Metabolites 2022, 12, 1176. [Google Scholar] [CrossRef]
- Doulidis, P.G.; Galler, A.I.; Hausmann, B.; Berry, D.; Rodríguez-Rojas, A.; Burgener, I.A. Gut microbiome signatures of Yorkshire Terrier enteropathy during disease and remission. Sci. Rep. 2023, 13, 4337. [Google Scholar] [CrossRef]
- Minamoto, Y.; Minamoto, T.; Isaiah, A.; Sattasathuchana, P.; Buono, A.; Rangachari, V.R.; McNeely, I.H.; Lidbury, J.; Steiner, J.M.; Suchodolski, J.S. Fecal short-chain fatty acid concentrations and dysbiosis in dogs with chronic enteropathy. J. Vet. Intern. Med. 2019, 33, 1608–1618. [Google Scholar] [CrossRef]
- Bansal, T.; Alaniz, R.C.; Wood, T.K.; Jayaraman, A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. USA 2010, 107, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Hand, D.; Wallis, C.; Colyer, A.; Penn, C.W. Pyrosequencing the canine faecal microbiota: Breadth and depth of biodiversity. PLoS ONE 2013, 8, e53115. [Google Scholar] [CrossRef] [PubMed]
- Pilla, R.; Suchodolski, J.S. The gut microbiome of dogs and cats, and the influence of diet. Vet. Clin. N. Am. Small Anim. Pract. 2021, 51, 605–621. [Google Scholar] [CrossRef]
- Garcia-Mazcorro, J.F.; Lanerie, D.J.; Dowd, S.E.; Paddock, C.G.; Grützner, N.; Steiner, J.M.; Ivanek, R.; Suchodolski, J.S. Effect of a multi-species synbiotic formulation on fecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing. FEMS Microbiol. Ecol. 2011, 78, 542–554. [Google Scholar] [CrossRef]
- Bermingham, E.N.; Maclean, P.; Thomas, D.G.; Cave, N.J.; Young, W. Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ 2017, 2, e3019. [Google Scholar] [CrossRef]
- Halfvarson, J.; Brislawn, C.J.; Lamendella, R.; Vázquez-Baeza, Y.; Walters, W.A.; Bramer, L.M.; D’Amato, M.; Bonfiglio, F.; McDonald, D.; Gonzalez, A.; et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2017, 2, 17004. [Google Scholar] [CrossRef]
- Moeller, A.H.; Sanders, J.G. Roles of the gut microbiota in the adaptive evolution of mammalian species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190597. [Google Scholar] [CrossRef]
- Bergström, A.; Frantz, L.; Schmidt, R.; Ersmark, E.; Lebrasseur, O.; Girdland-Flink, L.; Lin, A.T.; Storå, J.; Sjögren, K.G.; Anthony, D.; et al. Origins and genetic legacy of prehistoric dogs. Science 2020, 370, 557–564. [Google Scholar] [CrossRef]
- Bergström, A.; Stanton, D.W.G.; Taron, U.H.; Frantz, L.; Sinding, M.S.; Ersmark, E.; Pfrengle, S.; Cassatt-Johnstone, M.; Lebrasseur, O.; Girdland-Flink, L.; et al. Grey wolf genomic history reveals a dual ancestry of dogs. Nature 2022, 607, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Perri, A.R.; Feuerborn, T.R.; Frantz, L.A.F.; Larson, G.; Malhi, R.S.; Meltzer, D.J.; Witt, K.E. Dog domestication and the dual dispersal of people and dogs into the Americas. Proc. Natl. Acad. Sci. USA 2021, 118, e2010083118. [Google Scholar] [CrossRef] [PubMed]
- Arendt, M.; Cairns, K.M.; Ballard, J.W.; Savolainen, P.; Axelsson, E. Diet adaptation in dog reflects spread of prehistoric agriculture. Heredity 2016, 117, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Corsato Alvarenga, I.; Aldrich, C.G. Starch characterization of commercial extruded dry pet foods. Transl. Anim. Sci. 2020, 4, txaa018. [Google Scholar] [CrossRef] [PubMed]
- Richards, T.L.; Rankovic, A.; Cant, J.P.; Shoveller, A.K.; Adolphe, J.L.; Ramdath, D.; Verbrugghe, A. Effect of total starch and resistant starch in commercial extruded dog foods on gastric emptying in Siberian huskies. Animals 2021, 11, 2928. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 8, 506–514. [Google Scholar] [CrossRef]
- Schmitz, S.S. Value of probiotics in canine and feline gastroenterology. Vet. Clin. N. Am. Small. Anim. Pract. 2021, 1, 171–217. [Google Scholar] [CrossRef]
- Lee, D.; Goh, T.W.; Kang, M.G.; Choi, H.J.; Yeo, S.Y.; Yang, J.; Huh, C.S.; Kim, Y.Y.; Kim, Y. Perspectives and advances in probiotics and the gut microbiome in companion animals. J. Anim. Sci. Technol. 2022, 64, 197–217. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Chapin, K.; Murray, P. Manual of Clinical Microbiology, 7th ed.; Murray, P.R., Baron, E.J., Pfaller, M.A., Tenover, F.C., Yolken, R.H., Eds.; American Society for Microbiology: Washington, DC, USA, 1999. [Google Scholar]
- Mahon, C.R.; Lehman, D.C.; Manuselis, G. Textbook of Diagnostic Microbiology, 5th ed.; W. B Saunders Co.: Philadelphia, PA, USA, 2014. [Google Scholar]
- Hardy, B.L.; Bansal, G.; Hewlett, K.H.; Arora, A.; Schaffer, S.D.; Kamau, E.; Bennett, J.W.; Merrell, D.S. Antimicrobial Activity of Clinically Isolated Bacterial Species Against Staphylococcus aureus. Front. Microbiol. 2020, 10, 2977. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdom. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Frederiksen, W.; Garrity, G.M.; Grimont, P.A.D.; Kämpfer, P.; Maiden, M.C.J.; Nesme, X.; Rosselló-Mora, R.; Swings, J.; Trüper, H.G.; et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 2002, 52, 1043–1047. [Google Scholar] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Chen, I.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.J.; Webb, C.; Wu, D.; et al. The IMG/M data management and analysis system v.7: Content updates and new features. Nucleic Acids Res. 2023, 51, D723–D732. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Bianchini, G.; Sánchez-Baracaldo, P. TreeViewer—Cross-Platform Software to Draw Phylogenetic Trees (Version 2.1.0); Zenodo: Geneva, Switzerland, 2023. [Google Scholar] [CrossRef]
- Angiuoli, S.V.; Salzberg, S.L. Mugsy: Fast multiple alignment of closely related whole genomes. Bioinformatics 2011, 27, 334–342. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Rambaut, A. Figtree; v1.3.1; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2010; Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 19 September 2023).
- Reese, A.T.; Chadaideh, K.S.; Diggins, C.E.; Schell, L.D.; Beckel, M.; Callahan, P.; Ryan, R.; Emery Thompson, M.; Carmody, R.N. Effects of domestication on the gut microbiota parallel those of human industrialization. eLife 2021, 10, e60197. [Google Scholar] [CrossRef]
- Lyu, T.; Liu, G.; Zhang, H.; Wang, L.; Zhou, S.; Dou, H.; Pang, B.; Sha, W.; Zhang, H. Changes in feeding habits promoted the differentiation of the composition and function of gut microbiotas between domestic dogs (Canis lupus familiaris) and gray wolves (Canis lupus). AMB Express 2018, 8, 123. [Google Scholar] [CrossRef]
- Xu, J.; Becker, A.A.M.J.; Luo, Y.; Zhang, W.; Ge, B.; Leng, C.; Leng, C.; Wang, G.; Ding, L.; Wang, J.; et al. The Fecal Microbiota of Dogs Switching to a Raw Diet Only Partially Converges to That of Wolves. Front. Microbiol. 2021, 12, 701439. [Google Scholar] [CrossRef]
- Chen, L.; Sun, M.; Xu, D.; Gao, Z.; Shi, Y.; Wang, S.; Zhou, Y. Gut microbiome of captive wolves is more similar to domestic dogs than wild wolves indicated by metagenomics study. Front. Microbiol. 2022, 13, 1027188. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 2016, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.K.; Barbetti, M.J.; Lamichhane, J.R. Paenibacillus polymyxa. Trends Microbiol. 2023, 31, 657–659. [Google Scholar] [CrossRef] [PubMed]
- Ekim, B.; Calik, A.; Ceylan, A.; Saçakli, P. Effects of Paenibacillus xylanexedens on growth performance, intestinal histomorphology, intestinal microflora, and immune response in broiler chickens challenged with Escherichia coli K88. Poult. Sci. 2020, 99, 214–223. [Google Scholar] [CrossRef]
- Jung, H.I.; Park, S.; Niu, K.M.; Lee, S.W.; Kothari, D.; Yi, K.J.; Kim, S.K. Complete genome sequence of Paenibacillus konkukensis sp. nov. SK3146 as a potential probiotic strain. J. Anim. Sci. Technol. 2021, 63, 666–670. [Google Scholar] [CrossRef]
- Huang, E.; Guo, Y.; Yousef, A.E. Draft genome sequence of Paenibacillus sp. strain OSY-SE, a bacterium producing the novel broad-spectrum lipopeptide antibiotic paenibacterin. J. Bacteriol. 2012, 78, 3156–3165. [Google Scholar] [CrossRef]
- Pajor, M.; Sogin, J.; Worobo, R.W.; Szweda, P. Draft genome sequence of antimicrobial producing Paenibacillus alvei strain MP1 reveals putative novel antimicrobials. BMC Res. Notes 2020, 13, 280. [Google Scholar] [CrossRef]
- Eltokhy, M.A.; Saad, B.T.; Eltayeb, W.N.; Yahia, I.S.; Aboshanab, K.M.; Ashour, M.S.E. Exploring the nature of the antimicrobial metabolites produced by Paenibacillus ehimensis soil isolate MZ921932 using a metagenomic nanopore sequencing coupled with LC-mass analysis. Antibiotics 2021, 1, 12. [Google Scholar] [CrossRef] [PubMed]
- Keggi, C.; Doran-Peterson, J. Paenibacillus amylolyticus 27C64 has a diverse set of carbohydrate-active enzymes and complete pectin deconstruction system. J. Ind. Microbiol. Biotechnol. 2019, 46, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Liu, P.; Liao, W.; Miao, L. Complete Genome of the Chitin-Degrading Bacterium, Paenibacillus xylanilyticus W4. Genome Biol. Evol. 2019, 10, 2044. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.-J.; Son, S.; Kim, J.-A.; Jung, M.Y.; Choi, Y.-j.; Kim, D.-H.; Lee, H.K.; Shin, D.; Kim, Y. Characterization and functional test of canine probiotics. Front. Microbiol. 2021, 12, 625562. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, B.; Liu, C.; Hu, Y.; Liu, C.; Li, X.; Li, X.; Zhang, X.; Irwin, D.M.; Wu, Z.; et al. Differences in the gut microbiomes of dogs and wolves: Roles of antibiotics and starch. BMC Vet. Res. 2021, 17, 112. [Google Scholar] [CrossRef]
- Veiga, P.; Suez, J.; Derrien, M.; Elinav, E. Moving from probiotics to precision probiotics. Nat. Microbiol. 2020, 5, 878–880. [Google Scholar] [CrossRef]
- Collado, M.C.; Morelli, L.; Montoya, G.A.; Szajewska, H.; Tancredi, D.J.; Sanders, M.E. Emerging issues in probiotic safety: 2023 perspectives. Gut Microbes 2023, 15, 2185034. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
| Gene Category | Search Terms | Hits | Total Genes | Percent |
|---|---|---|---|---|
| Antimicrobials | “bacteriocin” | 5 | 6543 | 0.08% |
| Antimicrobials | “lantibiotic” | 6 | 6543 | 0.09% |
| Antimicrobials | “chitinase” | 2 | 6543 | 0.03% |
| Antimicrobials | “lysin” | 22 | 6543 | 0.34% |
| Antimicrobials | “amidase” | 42 | 6543 | 0.64% |
| Antimicrobials | Total | 77 | 6543 | 1.18% |
| Phage-derived | “phage” | 40 | 6543 | 0.61% |
| Phage-derived | “terminase” | 4 | 6543 | 0.06% |
| Phage-derived | “tail” | 4 | 6543 | 0.06% |
| Phage-derived | Total | 48 | 6543 | 0.73% |
| Spore-formation genes | “spore” | 14 | 6543 | 0.21% |
| Spore-formation genes | “sporulation” | 85 | 6543 | 1.30% |
| Spore-formation genes | Total | 99 | 6543 | 1.51% |
| Metabolic | “lactate” | 13 | 6543 | 0.20% |
| Metabolic | “lactic” | 0 | 6543 | 0.00% |
| Metabolic | “Cytochrome c” | 69 | 6543 | 1.05% |
| Metabolic | “catalase” | 6 | 6543 | 0.09% |
| Metabolic | “oxidase” | 74 | 6543 | 1.13% |
| Metabolic | Total | 162 | 6543 | 2.48% |
| Germination | Total | 34 | 6543 | 0.52% |
| Antitoxin | Total | 12 | 6543 | 0.18% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McCabe, J.; Bryant, J.L.; Klews, C.C.; Johnson, M.; Atchley, A.N.; Cousins, T.W.; Dominguez, A.; Gabriel, M.; Middleton, K.; Bowles, N.A.; et al. Phenotypic and Draft Genome Sequence Analyses of a Paenibacillus sp. Isolated from the Gastrointestinal Tract of a North American Gray Wolf (Canis lupus). Appl. Microbiol. 2023, 3, 1120-1129. https://doi.org/10.3390/applmicrobiol3040077
McCabe J, Bryant JL, Klews CC, Johnson M, Atchley AN, Cousins TW, Dominguez A, Gabriel M, Middleton K, Bowles NA, et al. Phenotypic and Draft Genome Sequence Analyses of a Paenibacillus sp. Isolated from the Gastrointestinal Tract of a North American Gray Wolf (Canis lupus). Applied Microbiology. 2023; 3(4):1120-1129. https://doi.org/10.3390/applmicrobiol3040077
Chicago/Turabian StyleMcCabe, Jennifer, Jessika L. Bryant, C. Cristoph Klews, MiCayla Johnson, Ariel N. Atchley, Thomas W. Cousins, Analiska Dominguez, Marie Gabriel, Katie Middleton, Natasha A. Bowles, and et al. 2023. "Phenotypic and Draft Genome Sequence Analyses of a Paenibacillus sp. Isolated from the Gastrointestinal Tract of a North American Gray Wolf (Canis lupus)" Applied Microbiology 3, no. 4: 1120-1129. https://doi.org/10.3390/applmicrobiol3040077
APA StyleMcCabe, J., Bryant, J. L., Klews, C. C., Johnson, M., Atchley, A. N., Cousins, T. W., Dominguez, A., Gabriel, M., Middleton, K., Bowles, N. A., Broughton, H. M., Smith, K. M., Ackermann, M. R., Bildfell, R., Ball, P. N., Forsythe, E. S., & Seal, B. S. (2023). Phenotypic and Draft Genome Sequence Analyses of a Paenibacillus sp. Isolated from the Gastrointestinal Tract of a North American Gray Wolf (Canis lupus). Applied Microbiology, 3(4), 1120-1129. https://doi.org/10.3390/applmicrobiol3040077







