Bacillus velezensis and Paenibacillus peoriae Strains Effective as Biocontrol Agents against Xanthomonas Bacterial Spot
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Environmental Samples for the Isolation of Microorganisms
2.3. Isolation of Microorganisms from Environmental Samples
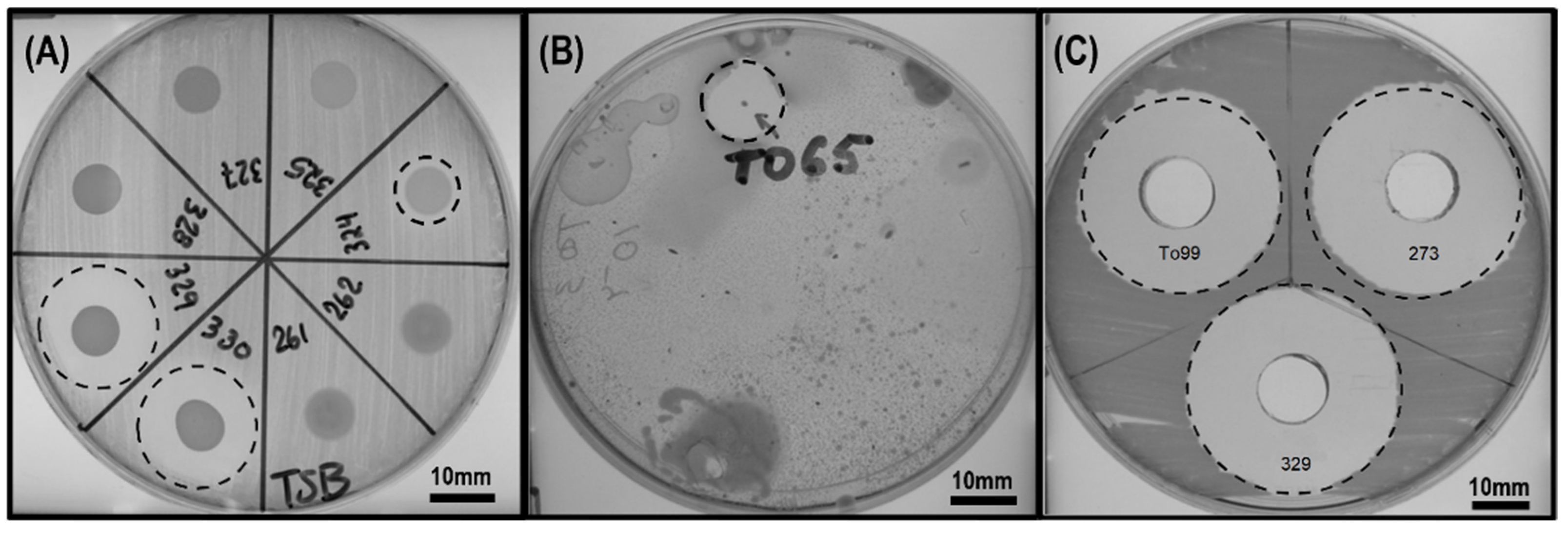
2.4. Screening the In Vitro Antagonism of Isolates against X. campestris and X. perforans T4
2.5. 16S rRNA Gene Sequence Analyses
2.6. Whole-Genome Sequencing and In Silico Analyses of Secondary Metabolite Biosynthetic Gene Clusters
2.7. Growth Chamber Trials
2.7.1. Plant Materials
2.7.2. Plant Pathogens
2.7.3. Preparation of Bacterial Cell-Free Supernatants and Cell Suspensions
2.7.4. Curative and Preventive Effects of Bacterial Cell-Free Supernatants and Cell Suspensions
2.7.5. Microbial Viability of Bacterial Isolates on Tomato Leaves
2.8. Greenhouse Trial Assay
2.9. Bioassay-Guided Fractionation and Isolation of Active Metabolites
2.10. LC-ESI-MS Analysis
3. Results
3.1. Screening of Isolated Microorganisms with Antimicrobial Activity against Xanthomonas Species
3.2. Identification of Isolates
3.3. Curative Effect of B. velezensis 71 and P. peoriae To99 Cell-Free Supernatants in Controlling Tomato Bacterial Spot Disease Caused by X. perforans T4
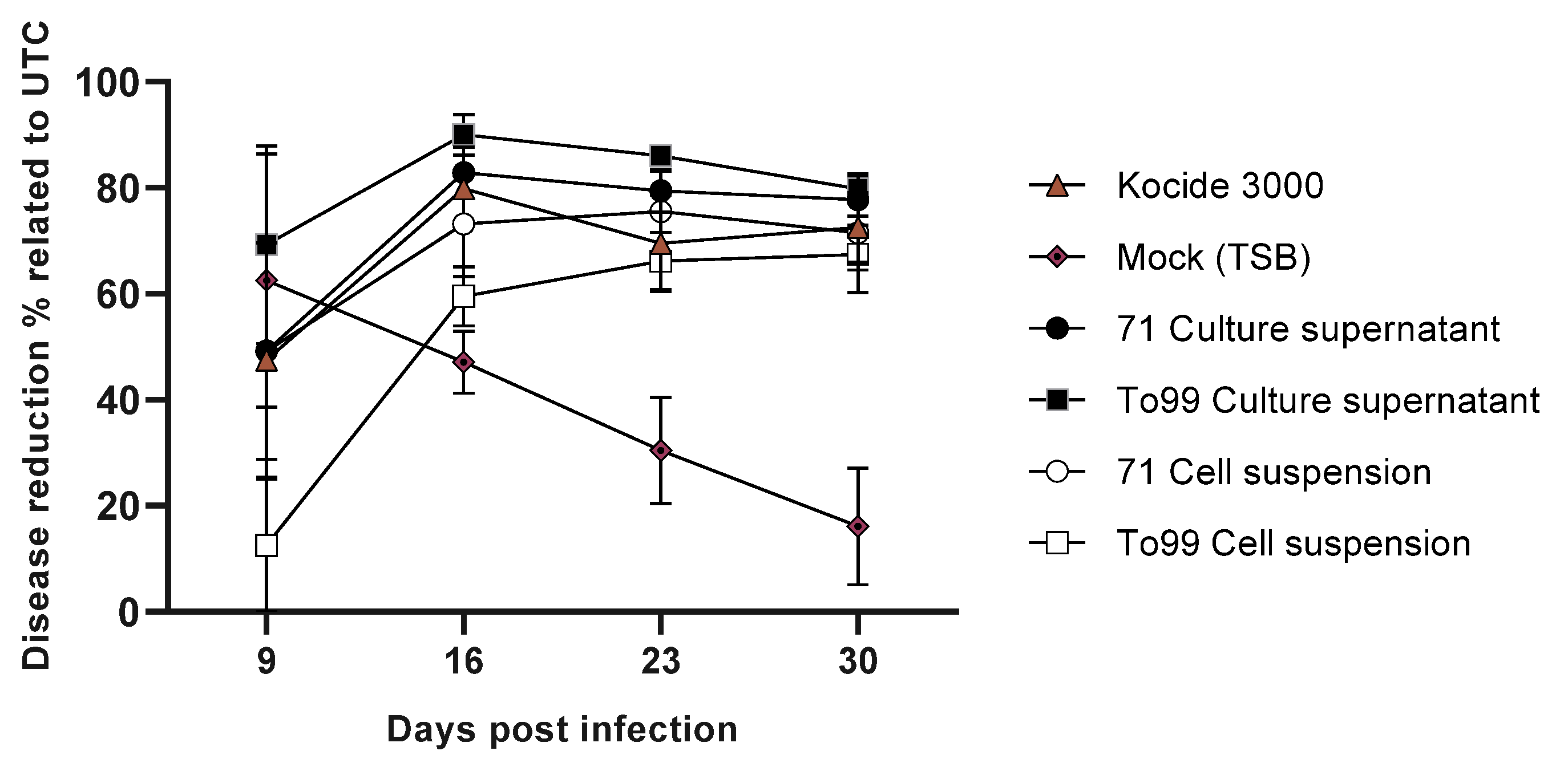
3.4. Preventive Effects of Bacillus and Paenibacillus Cells and Their Cell-Free Supernatants in Controlling Bacterial Spot Disease Caused by X. gardneri DC00T7A
3.5. Microbial Viability of Bacillus and Paenibacillus Isolates on Tomato Leaves
3.6. Greenhouse Trials
3.7. Gene Clusters for Secondary Metabolites
3.8. Bioassay-Guided Fractionation and Identification of Active Metabolites
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, J.B.; Lacy, G.H.; Bouzar, H.; Stall, R.E.; Schaad, N.W. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst. Appl. Microbiol. 2004, 27, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.B.; Lacy, G.H.; Bouzar, H.; Minsavage, G.V.; Stall, R.E.; Schaad, N.W. Bacterial Spot-Worldwide Distribution, Importance and Review. In I International Symposium on Tomato Diseases; ISHS: Leuven, Belgium, 2005; pp. 27–34. [Google Scholar]
- Potnis, N.; Krasileva, K.; Chow, V.; Almeida, N.F.; Patil, P.B.; Ryan, R.P.; Sharlach, M.; Behlau, F.; Dow, J.M.; Momol, M.; et al. Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genom. 2011, 12, 146. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. Xanthomonas spp. (Xanthomonas euvesicatoria, Xanthomonas gardneri, Xanthomonas perforans, Xanthomonas vesicatoria) causing bacterial spot of tomato and sweet pepper. EPPO Bull. 2013, 43, 7–20. [Google Scholar] [CrossRef]
- Toussaint, V. Bacterial leaf spot, a new disease of lettuce in Quebec caused by Xanthomonas campestris pv. vitians. Phytoprotection 1999, 80, 121–125. [Google Scholar] [CrossRef]
- Conover, R.A.; Gerhold, N.R. Mixtures of Copper and Maneb or Mancozeb for Control of Bacterial Spot of Tomato and Their Compatibility for Control of Fungus Diseases. Fla. State Hortic. Soc. Proc. 1981, 94, 154–156. [Google Scholar]
- Strayer-Scherer, A.; Liao, Y.Y.; Young, M.; Ritchie, L.; Vallad, G.E.; Santra, S.; Freeman, J.H.; Clark, D.; Jones, J.B.; Paret, M.L. Advanced Copper Composites Against Copper-Tolerant Xanthomonas perforans and Tomato Bacterial Spot. Phytopathology 2018, 108, 196–205. [Google Scholar] [CrossRef]
- Basim, H.; Minsavage, G.V.; Stall, R.E.; Wang, J.F.; Shanker, S.; Jones, J.B. Characterization of a unique chromosomal copper resistance gene cluster from Xanthomonas campestris pv. vesicatoria. Appl. Environ. Microbiol. 2005, 71, 8284–8291. [Google Scholar] [CrossRef]
- Lee, Y.A.; Hendson, M.; Panopoulos, N.J.; Schroth, M.N. Molecular cloning, chromosomal mapping, and sequence analysis of copper resistance genes from Xanthomonas campestris pv. juglandis: Homology with small blue copper proteins and multicopper oxidase. J. Bacteriol. 1994, 176, 173–188. [Google Scholar] [CrossRef]
- Voloudakis, A.E.; Reignier, T.M.; Cooksey, D.A. Regulation of resistance to copper in Xanthomonas axonopodis pv. vesicatoria. Appl. Environ. Microbiol. 2005, 71, 782–789. [Google Scholar] [CrossRef]
- Bull, C.T.; Koike, S.T. Evaluating the efficacy of commercial products for management of bacterial leaf spot on lettuce. Plant Health Prog. 2005, 6, 3. [Google Scholar] [CrossRef]
- Baldi, E.; Miotto, A.; Ceretta, C.A.; Quartieri, M.; Sorrenti, G.; Brunetto, G.; Toselli, M. Soil-applied phosphorous is an effective tool to mitigate the toxicity of copper excess on grapevine grown in rhizobox. Sci. Hortic. 2018, 227, 102–111. [Google Scholar] [CrossRef]
- Weller, D.M. Biological-Control of Soilborne Plant-Pathogens in the Rhizosphere with Bacteria. Annu. Rev. Phytopathol. 1988, 26, 379–407. [Google Scholar] [CrossRef]
- Compant, S.; Duffy, B.; Nowak, J.; Clement, C.; Barka, E.A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef]
- Eljounaidi, K.; Lee, S.K.; Bae, H. Bacterial endophytes as potential biocontrol agents of vascular wilt diseases—Review and future prospects. Biol. Control 2016, 103, 62–68. [Google Scholar] [CrossRef]
- Chavan, N.P.; Pandey, R.; Nawani, N.; Nanda, R.K.; Tandon, G.D.; Khetmalas, M.B. Biocontrol potential of actinomycetes against Xanthomonas axonopodis pv. punicae, a causative agent for oily spot disease of pomegranate. Biocontrol. Sci. Technol. 2016, 26, 351–372. [Google Scholar] [CrossRef]
- El-Hendawy, H.H.; Osman, M.E.; Sorour, N.M. Biological control of bacterial spot of tomato caused by Xanthomonas campestris pv. vesicatoria by Rahnella aquatilis. Microbiol. Res. 2005, 160, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Massomo, S.M.S.; Mortensen, C.N.; Mabagala, R.B.; Newman, M.A.; Hockenhull, J. Biological control of black rot (Xanthomonas campestris pv. campestris) of cabbage in Tanzania with Bacillus strains. J. Phytopathol. 2004, 152, 98–105. [Google Scholar] [CrossRef]
- Monteiro, L.; Mariano, R.D.R.; Souto-Maior, A.M. Antagonism of Bacillus spp. against Xanthomonas campestris pv. campestris. Braz. Arch. Biol. Technol. 2005, 48, 23–29. [Google Scholar] [CrossRef]
- Lee, G.; Lee, S.H.; Kim, K.M.; Ryu, C.M. Foliar application of the leaf-colonizing yeast Pseudozyma churashimaensis elicits systemic defense of pepper against bacterial and viral pathogens. Sci. Rep. 2017, 7, 39432. [Google Scholar] [CrossRef]
- Wu, L.; Wu, H.; Chen, L.; Yu, X.; Borriss, R.; Gao, X. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 2015, 5, 12975. [Google Scholar] [CrossRef]
- Marin, V.R.; Ferrarezi, J.H.; Vieira, G.; Sass, D.C. Recent advances in the biocontrol of Xanthomonas spp. World J. Microbiol. Biotechnol. 2019, 35, 72. [Google Scholar] [CrossRef] [PubMed]
- Besset-Manzoni, Y.; Joly, P.; Brutel, A.; Gerin, F.; Soudière, O.; Langin, T.; Prigent-Combaret, C. Does in vitro selection of biocontrol agents guarantee success in planta? A study case of wheat protection against Fusarium seedling blight by soil bacteria. PLoS ONE 2019, 14, e0225655. [Google Scholar] [CrossRef] [PubMed]
- Collinge, D.B.; Jensen, D.F.; Rabiey, M.; Sarrocco, S.; Shaw, M.W.; Shaw, R.H. Biological control of plant diseases—What has been achieved and what is the direction? Plant Pathol. 2022, 71, 1024–1047. [Google Scholar] [CrossRef]
- Clough, S.E.; Jousset, A.; Elphinstone, J.G.; Friman, V.-P. Combining in vitro and in vivo screening to identify efficient Pseudomonas biocontrol strains against the phytopathogenic bacterium Ralstonia solanacearum. Microbiologyopen 2022, 11, e1283. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.N.; Wilhelm, J.J.; Tresner, H.D. Method for the preferential isolation of Actinomycetes from soil. Appl. Microbiol. 1960, 8, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Henry, D.A.; Campbell, M.E.; LiPuma, J.J.; Speert, D.P. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J. Clin. Microbiol. 1997, 35, 614–619. [Google Scholar] [CrossRef]
- Fromin, N.; Achouak, W.; Thiéry, J.M.; Heulin, T. The genotypic diversity of Pseudomonas brassicacearum populations isolated from roots of Arabidopsis thaliana: Influence of plant genotype. FEMS Microbiol. Ecol. 2001, 37, 21–29. [Google Scholar] [CrossRef]
- Rashid, A.; Khan, M.A. Evaluation of antagonistic organisms against Xanthomonas campestris pv. malvacearum in vitro and on the inoculated cotton plant for the control of bacterial blight disease. Pak. J. Agric. Sci. 2000, 37, 135–139. [Google Scholar]
- Velmurugan, N.; Choi, M.S.; Han, S.S.; Lee, Y.S. Evaluation of antagonistic activities of Bacillus subtilis and Bacillus licheniformis against wood-staining fungi: In vitro and in vivo experiments. J. Microbiol. 2009, 47, 385–392. [Google Scholar] [CrossRef]
- Obradovic, A.; Jones, J.B.; Momol, M.T.; Balogh, B.; Olson, S.M. Management of Tomato Bacterial Spot in the Field by Foliar Applications of Bacteriophages and SAR Inducers. Plant Dis. 2004, 88, 736–740. [Google Scholar] [CrossRef]
- Lindow, S.E.; Brandl, M.T. Microbiology of the Phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef] [PubMed]
- Priest, F.G.; Goodfellow, M.; Shute, L.A.; Berkeley, R.C.W. Bacillus amyloliquefaciens sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 1987, 37, 69–71. [Google Scholar] [CrossRef]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Assefa, S.; Keane, T.M.; Otto, T.D.; Newbold, C.; Berriman, M. ABACAS: Algorithm-based automatic contiguation of assembled sequences. Bioinformatics 2009, 25, 1968–1969. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Chen, X.H.; Vater, J.; Piel, J.; Franke, P.; Scholz, R.; Schneider, K.; Koumoutsi, A.; Hitzeroth, G.; Grammel, N.; Strittmatter, A.W.; et al. Structural and Functional Characterization of Three Polyketide Synthase Gene Clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 2006, 188, 4024–4036. [Google Scholar] [CrossRef]
- Schneider, K.; Chen, X.H.; Vater, J.; Franke, P.; Nicholson, G.; Borriss, R.; Sussmuth, R.D. Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J. Nat. Prod. 2007, 70, 1417–1423. [Google Scholar] [CrossRef]
- Jangra, M.; Randhawa, H.K.; Kaur, M.; Srivastava, A.; Maurya, N.; Patil, P.P.; Jaswal, P.; Arora, A.; Patil, P.B.; Raje, M.; et al. Purification, Characterization and in vitro Evaluation of Polymyxin A From Paenibacillus dendritiformis: An Underexplored Member of the Polymyxin Family. Front. Microbiol. 2018, 9, 2864. [Google Scholar] [CrossRef] [PubMed]
- Olishevska, S.; Nickzad, A.; Déziel, E. Bacillus and Paenibacillus secreted polyketides and peptides involved in controlling human and plant pathogens. Appl. Microbiol. Biotechnol. 2019, 103, 1189–1215. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Fazle Rabbee, M.; Baek, K.-H. Antimicrobial Activities of Lipopeptides and Polyketides of Bacillus velezensis for Agricultural Applications. Molecules 2020, 25, 4973. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Choi, S.-K.; Ryu, C.-M.; Park, S.-H. Chronicle of a Soil Bacterium: Paenibacillus polymyxa E681 as a Tiny Guardian of Plant and Human Health. Front. Microbiol. 2019, 10, 467. [Google Scholar] [CrossRef]
- Fan, B.; Wang, C.; Song, X.; Ding, X.; Wu, L.; Wu, H.; Gao, X.; Borriss, R. Bacillus velezensis FZB42 in 2018: The Gram-Positive Model Strain for Plant Growth Promotion and Biocontrol. Front. Microbiol. 2018, 9, 2491. [Google Scholar] [CrossRef]
- Zeriouh, H.; Romero, D.; Garcia-Gutierrez, L.; Cazorla, F.M.; de Vicente, A.; Perez-Garcia, A. The iturin-like lipopeptides are essential components in the biological control arsenal of Bacillus subtilis against bacterial diseases of cucurbits. Mol. Plant-Microbe Interact. 2011, 24, 1540–1552. [Google Scholar] [CrossRef]
- Beric, T.; Kojic, M.; Stankovic, S.; Topisirovic, L.; Degrassi, G.; Myers, M.; Venturi, V.; Fira, D. Antimicrobial Activity of Bacillus sp Natural Isolates and Their Potential Use in the Biocontrol of Phytopathogenic Bacteria. Food Technol. Biotechnol. 2012, 50, 25–31. [Google Scholar]
- da Silva, R.S.; Moutinho, B.L.; dos Santos, D.R.; Vasconcelo-Rodrigues, I.S.; Talamini, V.; Fernandes, M.F.; Fernandes, R.P.M. Using antagonistic soil bacteria and their cell-free filtrates to control the black rot pathogen Xanthomonas campestris pv. campestris. J. Phytopathol. 2018, 166, 494–501. [Google Scholar] [CrossRef]
- Ghazalibiglar, H.; Hampton, J.G.; van ZijlldeJong, E.; Holyoake, A. Evaluation of Paenibacillus spp. isolates for the biological control of black rot in Brassica oleracea var. capitata (cabbage). Biocontrol. Sci. Techn. 2016, 26, 504–515. [Google Scholar] [CrossRef]
- Wilson, K.E.; Flor, J.E.; Schwartz, R.E.; Joshua, H.; Smith, J.L.; Pelak, B.A.; Liesch, J.M.; Hensens, O.D. Difficidin and oxydifficidin: Novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. II. Isolation and physico-chemical characterization. J. Antibiot. 1987, 40, 1682–1691. [Google Scholar] [CrossRef] [PubMed]
- Dagher, F.; Nickzad, A.; Zheng, J.; Hoffmann, M.; Déziel, E. Characterization of the biocontrol activity of three bacterial isolates against the phytopathogen Erwinia amylovora. Microbiologyopen 2021, 10, e1202. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.H.; Scholz, R.; Borriss, M.; Junge, H.; Mogel, G.; Kunz, S.; Borriss, R. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J. Biotechnol. 2009, 140, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Aleti, G.; Sessitsch, A.; Brader, G. Genome mining: Prediction of lipopeptides and polyketides from Bacillus and related Firmicutes. Comput. Struct. Biotechnol. J. 2015, 13, 192–203. [Google Scholar] [CrossRef]
- Chakraborty, K.; Kizhakkekalam, V.K.; Joy, M.; Dhara, S. Difficidin class of polyketide antibiotics from marine macroalga-associated Bacillus as promising antibacterial agents. Appl. Microbiol. Biotechnol. 2021, 105, 6395–6408. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Hwang, B.-S.; Baek, K.-H. Bacillus velezensis: A Beneficial Biocontrol Agent or Facultative Phytopathogen for Sustainable Agriculture. Agronomy 2023, 13, 840. [Google Scholar] [CrossRef]
- Brown, P.; Dawson, M.J. Development of new polymyxin derivatives for multi-drug resistant Gram-negative infections. J. Antibiot. 2017, 70, 386–394. [Google Scholar] [CrossRef]
- Katznelson, H.; Sutton, M.D. Inhibition of plant pathogenic bacteria in vitro by antibiotics and quaternary ammonium compounds. Can. J. Bot. 1951, 29, 270–278. [Google Scholar] [CrossRef]
- Niu, B.; Vater, J.; Rueckert, C.; Blom, J.; Lehmann, M.; Ru, J.-J.; Chen, X.-H.; Wang, Q.; Borriss, R. Polymyxin P is the active principle in suppressing phytopathogenic Erwinia spp. by the biocontrol rhizobacterium Paenibacillus polymyxa M-1. BMC Microbiol. 2013, 13, 137. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Kim, B.K.; Chung, J.-h.; Kim, S.-Y.; Jeong, H.; Kang, S.G.; Kwon, S.-K.; Lee, C.H.; Song, J.Y.; Yu, D.S.; Ryu, C.-M.; et al. Genome Sequence of the Leaf-Colonizing Bacterium Bacillus sp. Strain 5B6, Isolated from a Cherry Tree. J. Bacteriol. 2012, 194, 3758–3759. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ryu, C.-M.; Shin, J.-N.; Qi, W.; Ruhong, M.; Kim, E.-J.; Pan, J.-G. Potential for augmentation of fruit quality by foliar application of bacilli spores on apple tree. Plant Pathol. J. 2011, 27, 164–169. [Google Scholar] [CrossRef]


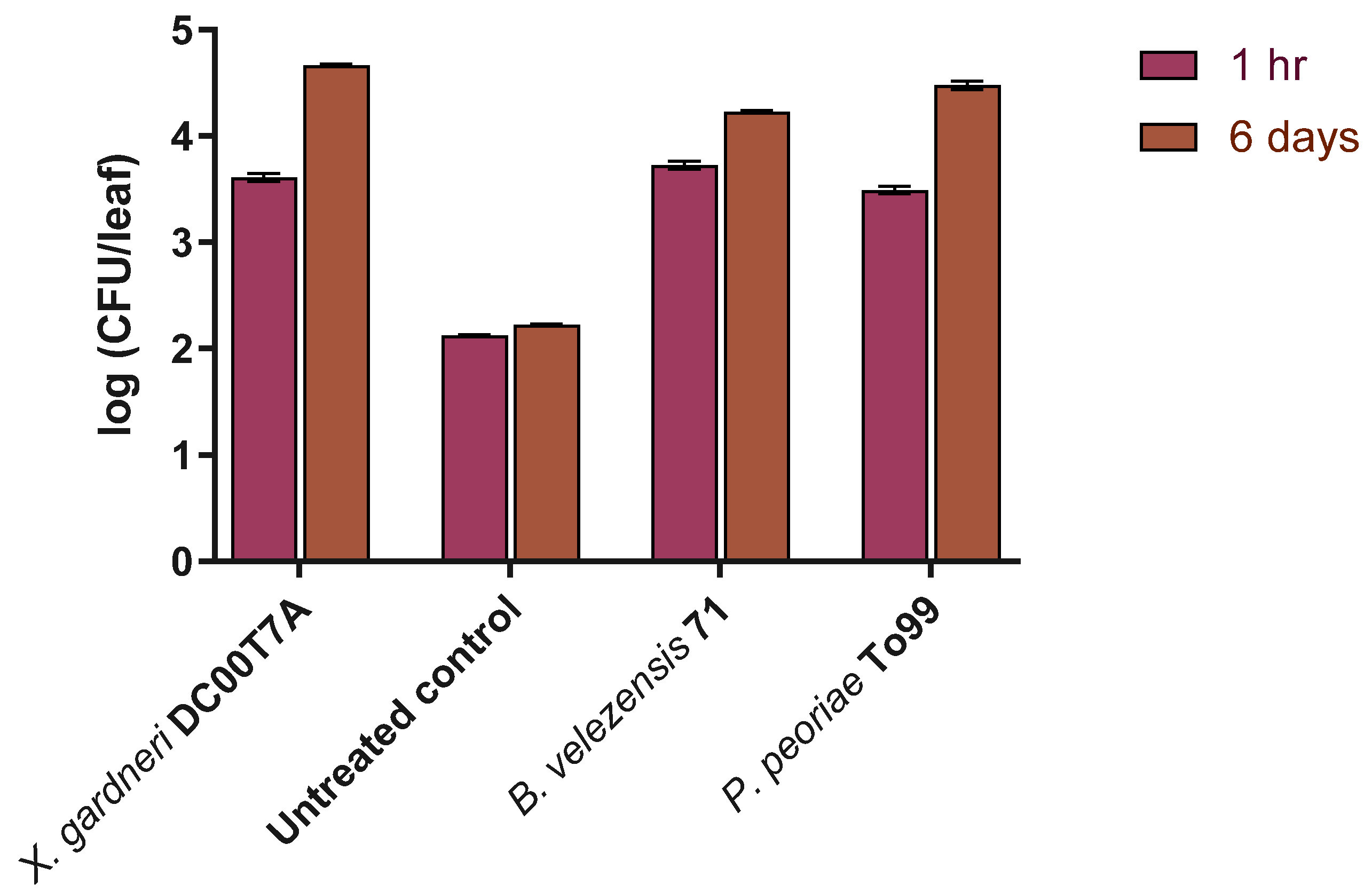


| Sample | Location and Date of Sampling | Quantity |
|---|---|---|
| Soil from agricultural fields | Laval (Québec, Canada), November 2012 | 60 |
| Soil from agricultural fields | Sherrington (Québec, Canada), November 2011 | 11 |
| Soil from vegetable garden of INRS | Laval (Québec, Canada), November 2012 | 2 |
| Soil from a strawberry field | Florida Area (USA), April 2013 | 2 |
| Dead plant leaves, stems, and roots from agricultural fields | Laval (Québec, Canada), November 2012 | 12 |
| Dead plant leaves, stems, and roots from agricultural fields | Sherrington (Québec, Canada), November 2011 | 4 |
| Fruits and leaves of fresh plants from vegetable garden of INRS | Laval (Québec, Canada), June 2012 | 6 |
| Fruits and leaves of fresh plants | Sherrington (Québec, Canada), November 2011 | 4 |
| Fruits and leaves of tomatoes | Florida Area (USA), June and July 2012 | 4 |
| Fruits and leaves of citrus | Florida Area (USA), June 2012 | 2 |
| Leaves of garden strawberries | Florida Area (USA), July 2012 | 2 |
| Vegetables seeds (tomatoes, peppers, onions) | Provided by farmers from Laval (Québec, Canada), November 2012 | 8 |
| Ditch water | Sherrington (Québec, Canada), November 2011 | 2 |
| Mud from a river | Rivière des Prairies, Laval (Québec, Canada), June 2012 | 2 |
| Soil from rhizosphere from vegetable garden of INRS | Laval (Québec, Canada), June 2012 | 2 |
| Total | 123 | |
| Isolates | 71 | To99 |
| Genome coverage (%) | 97 | 110 |
| N50 (bp) | 93,762 | 92,187 |
| Number of contigs | 105 | 203 |
| Unclassified reads (%) | 2.06 | 4.53 |
| Type Strains | Bacillus amyloliquefaciens subsp. amyloliquefaciens DSM7 | Bacillus velezensis FZB42 | Paenibacillus peoriae HS311 | Paenibacillus polymyxa SC2 | |
| Isolate | |||||
| 71 | 94.17 | 98.22 | - | - | |
| To99 | - | - | 99.03 | 89.24 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olishevska, S.; Nickzad, A.; Restieri, C.; Dagher, F.; Luo, Y.; Zheng, J.; Déziel, E. Bacillus velezensis and Paenibacillus peoriae Strains Effective as Biocontrol Agents against Xanthomonas Bacterial Spot. Appl. Microbiol. 2023, 3, 1101-1119. https://doi.org/10.3390/applmicrobiol3030076
Olishevska S, Nickzad A, Restieri C, Dagher F, Luo Y, Zheng J, Déziel E. Bacillus velezensis and Paenibacillus peoriae Strains Effective as Biocontrol Agents against Xanthomonas Bacterial Spot. Applied Microbiology. 2023; 3(3):1101-1119. https://doi.org/10.3390/applmicrobiol3030076
Chicago/Turabian StyleOlishevska, Snizhana, Arvin Nickzad, Concetta Restieri, Fadi Dagher, Yan Luo, Jie Zheng, and Eric Déziel. 2023. "Bacillus velezensis and Paenibacillus peoriae Strains Effective as Biocontrol Agents against Xanthomonas Bacterial Spot" Applied Microbiology 3, no. 3: 1101-1119. https://doi.org/10.3390/applmicrobiol3030076
APA StyleOlishevska, S., Nickzad, A., Restieri, C., Dagher, F., Luo, Y., Zheng, J., & Déziel, E. (2023). Bacillus velezensis and Paenibacillus peoriae Strains Effective as Biocontrol Agents against Xanthomonas Bacterial Spot. Applied Microbiology, 3(3), 1101-1119. https://doi.org/10.3390/applmicrobiol3030076







