Genome-Wide Association Mapping of Seedling Vigor and Regrowth Vigor in Winter Wheat
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotyping Seedling Vigor and Regrowth Vigor
2.3. Phenotypic Data Analysis
2.4. Genotyping and Population Structure
2.5. Genome-Wide Association Mapping Analysis
3. Results
3.1. Phenotypic Data Analysis
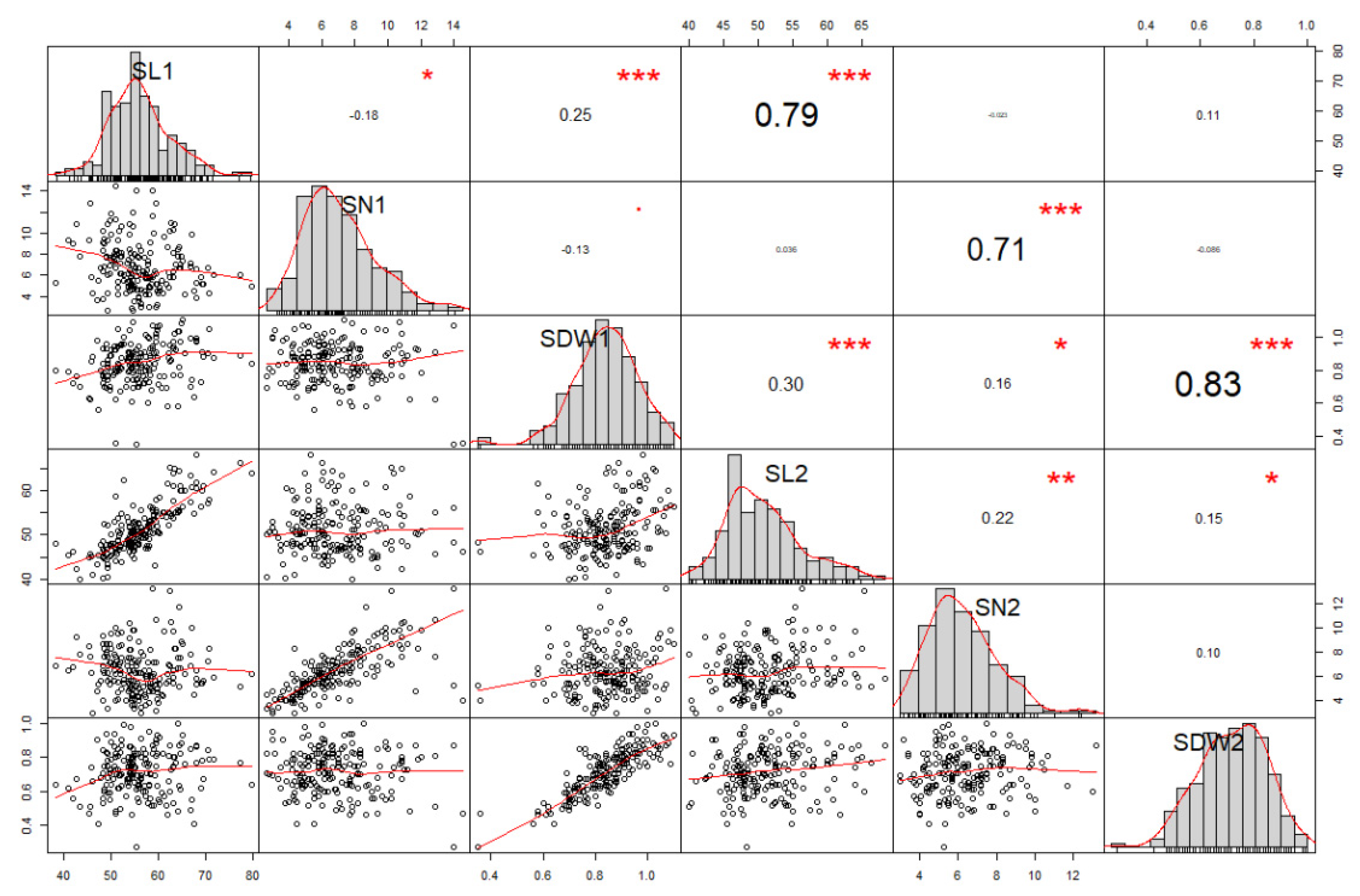
3.2. Correlations between Seedling Vigor and Regrowth Vigor Traits
3.3. Genome-Wide Association Analysis
3.3.1. QTL for Seedling Vigor
Shoot Length
Number of Shoots
Shoot Dry Weight
3.3.2. QTL for Regrowth Vigor
Shoot Length
Number of Shoots
Shoot Dry Weight
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Li, X.-M.; Chen, X.-M.; Xiao, Y.-G.; Xia, X.-C.; Wang, D.-S.; He, Z.-H.; Wang, H.-J. Identification of QTLs for seedling vigor in winter wheat. Euphytica 2014, 198, 199–209. [Google Scholar] [CrossRef]
- Botwright, T.; Condon, A.; Rebetzke, G.; Richards, R. Field evaluation of early vigour for genetic improvement of grain yield in wheat. Aust. J. Agric. Res. 2002, 53, 1137–1145. [Google Scholar] [CrossRef]
- ter Steege, M.W.; den Ouden, F.M.; Lambers, H.; Stam, P.; Peeters, A.J. Genetic and physiological architecture of early vigor in Aegilops tauschii, the D-genome donor of hexaploid wheat. A quantitative trait loci analysis. Plant Physiol. 2005, 139, 1078–1094. [Google Scholar] [CrossRef]
- Rebetzke, G.; Bruce, S.; Kirkegaard, J. Longer coleoptiles improve emergence through crop residues to increase seedling number and biomass in wheat (Triticum aestivum L.). Plant Soil 2005, 272, 87–100. [Google Scholar] [CrossRef]
- Maulana, F.; Anderson, J.D.; Butler, T.J.; Ma, X.-F. Improving dual-purpose winter wheat in the southern Great Plains of the United States. In Recent Advances in Grain Crops Research; Shah, F., Khan, Z., Iqbal, A., Turan, M., Olgun, M., Eds.; IntechOpen: London, UK, 2019; pp. 1–16. [Google Scholar]
- Kumssa, T.T.; Anderson, J.D.; Butler, T.J.; Ma, X.-F. Small grains as winter pasture in the Southern Great Plains of the United States. In Grasses and Grassland Aspects; Kindomihou, V.M., Ed.; IntechOpen: London, UK, 2019; pp. 1–13. [Google Scholar]
- Collard, B.C.; Jahufer, M.; Brouwer, J.; Pang, E. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Spielmeyer, W.; Hyles, J.; Joaquim, P.; Azanza, F.; Bonnett, D.; Ellis, M.; Moore, C.; Richards, R. A QTL on chromosome 6A in bread wheat (Triticum aestivum, L.) is associated with longer coleoptiles, greater seedling vigour and final plant height. Theor. Appl. Genet. 2007, 115, 59–66. [Google Scholar] [CrossRef]
- Landjeva, S.; Lohwasser, U.; Börner, A. Genetic mapping within the wheat D genome reveals QTL for germination, seed vigour and longevity, and early seedling growth. Euphytica 2010, 171, 129–143. [Google Scholar] [CrossRef]
- Yang, Y.; Wan, H.; Yang, F.; Xiao, C.; Li, J.; Ye, M.; Chen, C.; Deng, G.; Wang, Q.; Li, A. Mapping QTLs for enhancing early biomass derived from Aegilops tauschii in synthetic hexaploid wheat. PLoS ONE 2020, 15, 1–17. [Google Scholar] [CrossRef]
- Maydup, M.L.; Graciano, C.; Guiamet, J.J.; Tambussi, E.A. Analysis of early vigour in twenty modern cultivars of bread wheat (Triticum aestivum L.). Crop Pasture Sci. 2013, 63, 987–996. [Google Scholar] [CrossRef]
- Moore, C.; Rebetzke, G. Genomic regions for embryo size and early vigour in multiple wheat (Triticum aestivum L.) populations. Agronomy 2015, 5, 152–179. [Google Scholar] [CrossRef]
- Bai, C.; Liang, Y.; Hawkesford, M.J. Identification of QTLs associated with seedling root traits and their correlation with plant height in wheat. J. Exp. Bot. 2013, 64, 1745–1753. [Google Scholar] [CrossRef] [PubMed]
- Upadhyaya, H.D.; Wang, Y.-H.; Sastry, D.V.; Dwivedi, S.L.; Prasad, P.V.; Burrell, A.M.; Klein, R.R.; Morris, G.P.; Klein, P.E. Association mapping of germinability and seedling vigor in sorghum under controlled low-temperature conditions. Genome 2016, 59, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.M.; Yadav, S.; Dixit, S.; Ramayya, P.J.; Devi, M.N.; Raman, K.A.; Kumar, A. QTL hotspots for early vigor and related traits under dry direct-seeded system in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 286–300. [Google Scholar] [CrossRef]
- Landjeva, S.; Neumann, K.; Lohwasser, U.; Börner, A. Molecular mapping of genomic regions associated with wheat seedling growth under osmotic stress. Biol. Plant. 2008, 52, 259–266. [Google Scholar] [CrossRef]
- Iannucci, A.; Marone, D.; Russo, M.A.; De Vita, P.; Miullo, V.; Ferragonio, P.; Blanco, A.; Gadaleta, A.; Mastrangelo, A.M. Mapping QTL for root and shoot morphological traits in a Durum Wheat× T. dicoccum segregating population at seedling stage. Int. J. Genom. 2017, 2017, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-J.; Guo, Y.; Zhang, G.-Z.; Gao, M.-G.; Zhang, G.-H.; Kong, F.-M.; Zhao, Y.; Li, S.-S. QTL mapping for seedling traits under different nitrogen forms in wheat. Euphytica 2013, 191, 317–331. [Google Scholar] [CrossRef]
- Huang, X.; Han, B. Natural variations and genome-wide association studies in crop plants. Annu. Rev. Plant Biol. 2014, 65, 531–551. [Google Scholar] [CrossRef]
- Maulana, F.; Ayalew, H.; Anderson, J.D.; Kumssa, T.T.; Huang, W.; Ma, X.-F. Genome-wide association mapping of seedling heat tolerance in winter wheat. Front. Plant Sci. 2018, 9, 1272–1287. [Google Scholar] [CrossRef] [PubMed]
- Ayalew, H.; Liu, H.; Börner, A.; Kobiljski, B.; Liu, C.; Yan, G. Genome-wide association mapping of major root length QTLs under PEG induced water stress in wheat. Front. Plant Sci. 2018, 9, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Gahlaut, V.; Jaiswal, V.; Singh, S.; Balyan, H.; Gupta, P. Multi-locus genome wide association mapping for yield and its contributing traits in hexaploid wheat under different water regimes. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Maulana, F.; Huang, W.; Anderson, J.D.; Ma, X.-F. Genome-wide association mapping of seedling drought tolerance in winter wheat. Front. Plant Sci. 2020, 11, 573786. [Google Scholar] [CrossRef] [PubMed]
- Chopra, R.; Burow, G.; Burke, J.J.; Gladman, N.; Xin, Z. Genome-wide association analysis of seedling traits in diverse Sorghum germplasm under thermal stress. BMC Plant Biol. 2017, 17, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Marla, S.R.; Burow, G.; Chopra, R.; Hayes, C.; Olatoye, M.O.; Felderhoff, T.; Hu, Z.; Raymundo, R.; Perumal, R.; Morris, G.P. Genetic architecture of chilling tolerance in sorghum dissected with a nested association mapping population. G3 Genes Genomes Genet. 2019, 9, 4045–4057. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, Q.; Wang, C.-C.; Liu, Z.-X.; Jiang, Y.-J.; Zhai, L.-Y.; Zheng, T.-Q.; Xu, J.-L.; Li, Z.-K. Genetic dissection of seedling vigour in a diverse panel from the 3,000 Rice (Oryza sativa L.) Genome Project. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Cordero-Lara, K.I.; Kim, H.; Tai, T.H. Identification of seedling vigor-associated quantitative trait loci in temperate japonica rice. Plant Breed. Biotechnol. 2016, 4, 426–440. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, W.; Tong, W.; He, Q.; Yoon, M.-Y.; Li, F.-P.; Choi, B.; Heo, E.-B.; Kim, K.-W.; Park, Y.-J. A Genome-wide association study reveals candidate genes related to salt tolerance in rice (Oryza sativa) at the germination stage. Int. J. Mol. Sci. 2018, 19, 3145. [Google Scholar] [CrossRef]
- Abdel-Ghani, A.H.; Sharma, R.; Wabila, C.; Dhanagond, S.; Owais, S.J.; Duwayri, M.A.; Al-Dalain, S.A.; Klukas, C.; Chen, D.; Lübberstedt, T. Genome-wide association mapping in a diverse spring barley collection reveals the presence of QTL hotspots and candidate genes for root and shoot architecture traits at seedling stage. BMC Plant Biol. 2019, 19, 216–235. [Google Scholar] [CrossRef]
- Sharma, R.; Draicchio, F.; Bull, H.; Herzig, P.; Maurer, A.; Pillen, K.; Thomas, W.T.; Flavell, A.J. Genome-wide association of yield traits in a nested association mapping population of barley reveals new gene diversity for future breeding. J. Exp. Bot. 2018, 69, 3811–3822. [Google Scholar] [CrossRef]
- Wang, Q.J.; Yuan, Y.; Liao, Z.; Jiang, Y.; Wang, Q.; Zhang, L.; Gao, S.; Wu, F.; Li, M.; Xie, W. Genome-wide association study of 13 traits in Maize seedlings under low phosphorus stress. Plant Genome 2019, 12, 1–13. [Google Scholar] [CrossRef]
- Hu, G.; Li, Z.; Lu, Y.; Li, C.; Gong, S.; Yan, S.; Li, G.; Wang, M.; Ren, H.; Guan, H. Genome-wide association study identified multiple genetic loci on chilling resistance during germination in maize. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Zhang, Z.; Ersoz, E.; Lai, C.-Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Fu, Y.; Sun, R.; Wang, Y.; Wang, Q. Single-locus and multi-locus genome-wide association studies in the genetic dissection of fiber quality traits in upland cotton (Gossypium hirsutum L.). Front. Plant Sci. 2018, 9, 1083–1099. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-B.; Feng, J.-Y.; Ren, W.-L.; Huang, B.; Zhou, L.; Wen, Y.-J.; Zhang, J.; Dunwell, J.M.; Xu, S.; Zhang, Y.-M. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 2016, 6, 19444–19454. [Google Scholar] [CrossRef]
- Segura, V.; Vilhjálmsson, B.J.; Platt, A.; Korte, A.; Seren, Ü.; Long, Q.; Nordborg, M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 2012, 44, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Guttieri, M.J.; Baenziger, P.S.; Frels, K.; Carver, B.; Arnall, B.; Waters, B.M. Variation for grain mineral concentration in a diversity panel of current and historical Great Plains hard winter wheat germplasm. Crop Sci. 2015, 55, 1035–1052. [Google Scholar] [CrossRef]
- SAS Institute. The SAS System for Windows. 9.3. SAS Institute, Cary, NC, USA. Available online: http://support.sas.com/software/93/ (accessed on 16 May 2011).
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 20 August 2016).
- Peterson, B.G.; Carl, P.; Boudt, K.; Bennett, R.; Ulrich, J.; Zivot, E.; Cornilly, D.; Hung, E.; Lestel, M.; Balkissoon, K. Package ‘PerformanceAnalytics’. Available online: https://cran.r-project.org/web/packages/PerformanceAnalytics/ (accessed on 15 April 2018).
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotech. J. 2014, 12, 787–796. [Google Scholar] [CrossRef]
- IWGSC. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Sukumaran, S.; Reynolds, M.P.; Sansaloni, C. Genome-wide association analyses identify QTL hotspots for yield and component traits in durum wheat grown under yield potential, drought, and heat stress environments. Front. Plant Sci. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and Manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Alaux, M.; Rogers, J.; Letellier, T.; Flores, R.; Alfama, F.; Pommier, C.; Mohellibi, N.; Durand, S.; Kimmel, E.; Michotey, C. Linking the International Wheat Genome Sequencing Consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 2018, 19, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Lin, Y.; Tang, S.; Duan, S.; Wang, Q.; Wu, F.; Li, C.; Jiang, X.; Zhou, K.; Liu, Y. A genome-wide association study of coleoptile length in different Chinese wheat landraces. Front. Plant Sci. 2020, 11, 677. [Google Scholar] [CrossRef]
- Luo, H.; Hill, C.B.; Zhou, G.; Zhang, X.-Q.; Li, C. Genome-wide association mapping reveals novel genes associated with coleoptile length in a worldwide collection of barley. BMC Plant Biol. 2020, 20, 346–359. [Google Scholar] [CrossRef]
- Sidhu, J.S.; Singh, D.; Gill, H.S.; Brar, N.K.; Qiu, Y.; Halder, J.; Al Tameemi, R.; Turnipseed, B.; Sehgal, S.K. Genome-wide association study uncovers novel genomic regions associated with coleoptile length in hard winter wheat. Front. Genet. 2019, 10, 1345–1358. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Yang, W. E3 ubiquitin ligases: Ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef]
- Hosein, F.N.; Bandyopadhyay, A.; Peer, W.A.; Murphy, A.S. The catalytic and protein-protein interaction domains are required for APM1 function. Plant Physiol. 2010, 152, 2158–2172. [Google Scholar] [CrossRef][Green Version]
- Liu, X.; Cao, P.; Huang, Q.; Yang, Y.; Tao, D. Disruption of a rice chloroplast-targeted gene OsHMBPP causes a seedling-lethal albino phenotype. Rice 2020, 13, 1–11. [Google Scholar] [CrossRef]
- Fukuda, A.; Terao, T. QTLs for shoot length and chlorophyll content of rice seedlings grown under low-temperature conditions, using a cross between indica and japonica cultivars. Plant Prod. Sci. 2015, 18, 128–136. [Google Scholar] [CrossRef][Green Version]



| Trait | Seedling Vigor | Regrowth Vigor | ||||
|---|---|---|---|---|---|---|
| Mean | Range | SD | Mean | Range | SD | |
| Shoot length (cm) | 56.1 | 38.3–79.8 | 6.6 | 51.3 | 40.0–68.3 | 5.6 |
| Number of shoots per plant | 7 | 3–15 | 2.4 | 6 | 3–13 | 1.9 |
| Shoot dry weight per plant (g) | 0.84 | 0.35–1.10 | 0.12 | 0.71 | 0.27–1.00 | 0.13 |
| Trait | QTL Name | Peak SNP | Chr | Pos (cM) | MAF | MLM | MLMM | ||
|---|---|---|---|---|---|---|---|---|---|
| p-Value | R2 (%) | p-Value | |||||||
| Seedling vigor | Shoot length (cm) | QSLeV.nri-1A | IWB43660 | 1A | 29 | 0.36 | 6.25 × 10−4 | ||
| QSLeV.nri-2B | IWB36210 | 2B | 163 | 0.49 | 9.15 × 10−6 | 9.3 | 2.82 × 10−4 | ||
| QSLeV.nri-4B.1 | IWB31302 | 4B | 37 | 0.42 | 3.85 × 10−4 | ||||
| QSLeV.nri-4B.2 | IWB7508 | 4B | 55 | 0.07 | 2.43 × 10−5 | 8.4 | 2.47 × 10−4 | ||
| QSLeV.nri-5A | IWB43526 | 5A | 86 | 0.25 | 6.81 × 10−4 | ||||
| QSLeV.nri-7A | IWB9062 | 7A | 136 | 0.17 | 7.57 × 10−4 | 5.2 | 4.45 × 10−4 | ||
| Number of shoots | QSNeV.nri-2A | IWB28627 | 2A | 143 | 0.13 | 6.44 × 10−4 | 6.2 | 4.39 × 10−4 | |
| QSNeV.nri-5A | IWA2445 | 5A | 53 | 0.43 | 8.99 × 10−4 | ||||
| QSNeV.nri-7A | IWB8896 | 7A | 44 | 0.32 | 2.03 × 10−4 | 7.3 | 1.17 × 10−4 | ||
| Shoot dry weight (g) | QSDWeV.nri-6A | IWB50 | 6A | 141 | 0.21 | 9.59 × 10−4 | |||
| QSDWeV.nri-7A | IWB42636 | 7A | 228 | 0.11 | 6.72 × 10−5 | 8.4 | 3.21 × 10−4 | ||
| QSDWeV.nri-7D | IWB21364 | 7D | 138 | 0.13 | 6.86 × 10−4 | 6.0 | 4.71 × 10−4 | ||
| Regrowth vigor | Shoot length (cm) | QSLrV.nri-2A | IWB70425 | 2A | 126 | 0.12 | 7.67 × 10−4 | ||
| QSLrV.nri-2B | IWB36210 | 2B | 163 | 0.49 | 1.46 × 10−4 | 5.8 | 2.33 × 10−4 | ||
| QSLrV.nri-4B | IWB35611 | 4B | 57 | 0.42 | 2.72 × 10−6 | 9.1 | 4.77 × 10−4 | ||
| QSLrV.nri-5A | IWB38719 | 5A | 89 | 0.17 | 1.30 × 10−4 | ||||
| QSLrV.nri-6B.1 | IWB2837 | 6B | 67 | 0.07 | 2.95 × 10−4 | 5.3 | 6.76 × 10−4 | ||
| QSLrV.nri-6B.2 | IWB59306 | 6B | 114 | 0.33 | 3.64 × 10−4 | ||||
| QSLrV.nri-7B | IWB8019 | 7B | 54 | 0.20 | 3.21 × 10−4 | 5.2 | 8.44 × 10−4 | ||
| Number of shoots | QSNrV.nri-4B | IWB27735 | 4B | 105 | 0.15 | 7.29 × 10−4 | 6.0 | 5.04 × 10−4 | |
| QSNrV.nri-6A | IWB71326 | 6A | 141 | 0.13 | 6.88 × 10−4 | 6.0 | 4.72 × 10−4 | ||
| QSNrV.nri-7B | IWA4802 | 7B | 134 | 0.45 | 4.80 × 10−4 | 6.4 | 3.15 × 10−4 | ||
| Shoot dry weight (g) | QSDWrV.nri-1B | IWB66973 | 1B | 118 | 0.15 | 7.79 × 10−4 | 6.1 | 5.42 × 10−4 | |
| QSDWrV.nri-2B | IWB4865 | 2B | 134 | 0.06 | 1.86 × 10−4 | 7.6 | 1.06 × 10−4 | ||
| QSDWrV.nri-4A | IWA5968 | 4A | 109 | 0.15 | 8.64 × 10−4 | 6.0 | 6.10 × 10−4 | ||
| QSDWrV.nri-4B | IWB24289 | 4B | 78 | 0.39 | 9.35 × 10−4 | ||||
| QSDWrV.nri-6A | IWB12052 | 6A | 26 | 0.22 | 9.85 × 10−4 | ||||
| QSDWrV.nri-7A | IWB42636 | 7A | 228 | 0.11 | 7.76 × 10−4 | 6.1 | 5.40 × 10−4 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maulana, F.; Huang, W.; Anderson, J.D.; Kumssa, T.T.; Ma, X.-F. Genome-Wide Association Mapping of Seedling Vigor and Regrowth Vigor in Winter Wheat. Crops 2021, 1, 153-165. https://doi.org/10.3390/crops1030015
Maulana F, Huang W, Anderson JD, Kumssa TT, Ma X-F. Genome-Wide Association Mapping of Seedling Vigor and Regrowth Vigor in Winter Wheat. Crops. 2021; 1(3):153-165. https://doi.org/10.3390/crops1030015
Chicago/Turabian StyleMaulana, Frank, Wangqi Huang, Joshua D. Anderson, Tadele T. Kumssa, and Xue-Feng Ma. 2021. "Genome-Wide Association Mapping of Seedling Vigor and Regrowth Vigor in Winter Wheat" Crops 1, no. 3: 153-165. https://doi.org/10.3390/crops1030015
APA StyleMaulana, F., Huang, W., Anderson, J. D., Kumssa, T. T., & Ma, X.-F. (2021). Genome-Wide Association Mapping of Seedling Vigor and Regrowth Vigor in Winter Wheat. Crops, 1(3), 153-165. https://doi.org/10.3390/crops1030015






