Reliable Methodologies and Impactful Tools to Control Fruit Tree Viruses
Abstract
:1. Introduction
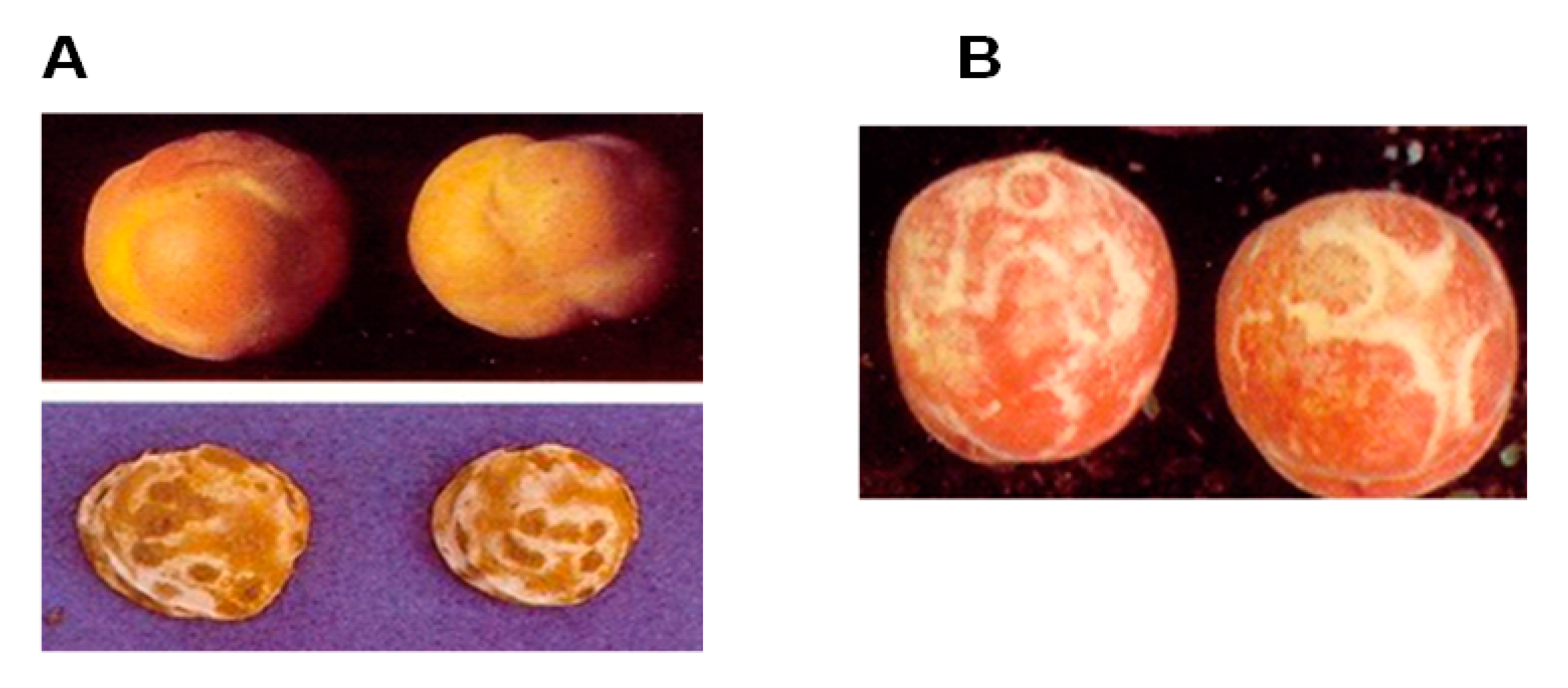
2. Fruit Tree Virus Disease Spread
3. Molecular Tools to Control Fruit Tree Viruses and Impactful High Throughput-Sequencing (HTS) Methodologies to Detect Hidden Fruit Tree Viruses
4. Maintenance of Virus Collection
5. Genetic Engineering of Viral RNAi in the Sustainability of Fruit Trees
6. Gene Editing: A New Technology for Challenging Fruit Tree Viruses
7. Conclusions
Funding
Conflicts of Interest
References
- Singh, R.; Srivastava, A. Prevention and control of viral diseases of crops. In Applied Plant Virology, Advances, Detection and Antivral Strategy; Academic Press: Cambridge, MA, USA, 2020; pp. 593–599. [Google Scholar] [CrossRef]
- EFSA REGULATION (EU) 2016/2031 OF THE EUROPEAN PARLIAMENT OF THE COUNCIL of 26 October 2016 on pro-tective measures against pests of plants, amending Regulations (EU) No 228/2013, (EU) No 652/2014 and (EU) No 1143/2014 of the European Parliament and of the Council and repealing Council Directives 69/464/EEC, 74/647/EEC, 93/85/EEC, 98/57/EC, 2000/29/EC, 2006/91/EC and 2007/33/EC. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:02016R2031-20191214 (accessed on 26 October 2016).
- Gottwald, T. Epidemiology of Sharka disease in North America. Eur. Mediter. Plant Prot. Org. Bull. 2006, 36, 269–286. [Google Scholar] [CrossRef]
- Rimbaud, L.; Dallot, S.; Gottwald, T.; Decroocq, V.; Jacquot, E.; Soubeyrand, S.; Thébaud, G. Sharka Epidemiology and Worldwide Management Strategies: Learning Lessons to Optimize Disease Control in Perennial Plants. Annu. Rev. Phytopathol. 2015, 53, 357–378. [Google Scholar] [CrossRef] [Green Version]
- Constable, F.; Rodoni, B. Review of the Post Entry Quarantine Conditions for Imports of Almond Germplasm; DEPI: East Melbourne, VIC, Australia, 2011. [Google Scholar] [CrossRef] [Green Version]
- Capote, N.; Bertolini, E.; Olmos, A.; Vidal, E.; Martinez, M.C.; Cambra, M. Direct sample preparation methods for the detection of Plum pox virus by real-time RT-PCR. Int. Microbiol. 2009, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Barba, M.; Hadidi, A.; Candresse, T.; Cambra, M. Plum pox virus. In Virus and Virus-Like Diseases of Pome and Stone Fruit; Hadidi, A., Barba, M., Candresse, T., Jelkmann, W., Eds.; American Phytopathological Society: St. Paul, MN, USA, 2011; pp. 185–197. [Google Scholar]
- Stobbs, L.; Van Driel, L.; Whybourne, K.; Carlsion, C. Distribution of Plum pox virus in residential sites, commercial nurseries and native plant species in Niagara Region, Ontario Canada. Plant Dis. 2005, 89, 822–827. [Google Scholar] [CrossRef] [Green Version]
- Atanasoff, D. Plum pox. A new virus disease. Ann. Univ. Sofía Fac. Agric. Silvic. 1932, 11, 49–69. [Google Scholar]
- Clark, M.F.; Adams, A.N.; Graham, F.L.; Smiley, J.; Russell, W.C.; Nairn, R. Characteristics of the Microplate Method of Enzyme-Linked Immunosorbent Assay for the Detection of Plant Viruses. J. Gen. Virol. 1977, 34, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Varga, A.; James, D. Use of reverse transcription loop-mediated isothermal amplification for the detection of Plum pox virus. J. Virol. Methods 2006, 138, 184–190. [Google Scholar] [CrossRef]
- Zhang, S.; Ravelonandro, M.; Russell, P.; McOwen, N.; Briard, P.; Bohannon, S.; Vrient, A. Rapid diagnostic detection of plum pox virus in Prunus plants by isothermal AmplifyRP® using reverse transcription-recombinase polymerase amplification. J. Virol. Methods 2014, 207, 114–120. [Google Scholar] [CrossRef]
- MacKenzie, D.J.; McLean, M.A.; Mukerji, S.; Green, M. Improved RNA Extraction from Woody Plants for the Detection of Viral Pathogens by Reverse Transcription-Polymerase Chain Reaction. Plant Dis. Rep. 1997, 81, 222–226. [Google Scholar] [CrossRef] [Green Version]
- Dallot, S.; Gottwald, T.; LaBonne, G.; Quiot, J.-B. Spatial Pattern Analysis of Sharka Disease (Plum pox virus Strain M) in Peach Orchards of Southern France. Phytopathology 2003, 93, 1543–1552. [Google Scholar] [CrossRef] [Green Version]
- Coetzee, B.; Freeborough, M.-J.; Maree, H.J.; Celton, J.-M.; Rees, J.; Burger, J.T. Deep sequencing analysis of viruses infecting grapevines: Virome of a vineyard. Virology 2010, 400, 157–163. [Google Scholar] [CrossRef] [Green Version]
- Giampetruzzi, A.; Roumi, V.; Roberto, R.; Malossini, U.; Yoshikawa, N.; La Notte, P.; Terlizzi, F.; Credi, R.; Saldarelli, P. A new grapevine virus discovered by deep sequencing of virus- and viroid-derived small RNAs in Cv ‘Pinot gris’. Virus Res. 2012, 163, 262–268. [Google Scholar] [CrossRef]
- Lu, Y.; Yao, B.; Wang, G.; Hong, N. The detection of ACLSV and ASPV in pear plants by RT-LAMP assays. J. Virol. Methods 2018, 252, 80–85. [Google Scholar] [CrossRef]
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Amari, K.; Sanchez-Pina, M.A.; Myrta, A.; Sanchez-Navarro, J.A. Ilarviruses of Prunus spp.: A Continued Concern for Fruit Trees. Phytopatholgy 2012, 102, 1108–1120. [Google Scholar] [CrossRef] [Green Version]
- Kominek, P.; Glasa, M.; Kominkova, M. Analysis of multiple virus-infected grapevine plant reveals persistence but uneven virus distribution. Acta Virol. 2009, 53, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Sela, N.; Luria, N.; Yaari, M.; Prusky, D.; Dombrovsky, A. Genome Sequence of a Potential New Benyvirus Isolated from Mango RNA-seq Data. Genome Announc. 2016, 4, e01250-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nassuth, A.; Pollari, E.; Helmeczy, K.; Stewart, S.; KoFalvi, S.A. Improved RNA extraction and one-tube RT-PCR assay for simultaneous detection of control plant RNA plus several viruses in plant extracts. J. Virol. Methods 2000, 90, 37–49. [Google Scholar] [CrossRef]
- Ravelonandro, M.; Scorza, R.; Bachelier, J.C.; LaBonne, G.; Levy, L.; Damsteegt, V.; Callahan, A.M.; Dunez, J. Resistance of Transgenic Prunus domestica to Plum Pox Virus Infection. Plant Dis. 1997, 81, 1231–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olmos, A.; Bertolini, E.; Gil, M.; Cambra, M. Real-time assay for quantitative detection of non-persistently transmitted Plum pox virus RNA targets in single aphids. J. Virol. Methods 2005, 128, 151–155. [Google Scholar] [CrossRef]
- Gottwald, T.R.; Wierenga, E.; Luo, W.; Parnell, S. Epidemiology of Plum pox ‘D’ strain in Canada and the USA. Can. J. Plant Pathol. 2013, 35, 442–457. [Google Scholar] [CrossRef]
- Gentit, P. Detection of Plum pox virus: Biological methods. EPPO Bull. 2006, 36, 251–253. [Google Scholar] [CrossRef]
- Damsteegt, V.D.; Stone, A.L.; Mink, G.I.; Howell, W.E.; Waterworth, H.E.; Levy, L. The versatility of Prunus tomentosa as a biondicator of viruses. Acta Hortic. 1998, 472, 143–146. [Google Scholar] [CrossRef]
- García, J.A.; Glasa, M.; Cambra, M.; Candresse, T. Plum pox virusand sharka: A model potyvirus and a major disease. Mol. Plant Pathol. 2014, 15, 226–241. [Google Scholar] [CrossRef] [PubMed]
- Kondratenjo, P.; Udovychenko, V. Plum pox virus in Ukraine. EPPO Bull. 2006, 36. [Google Scholar] [CrossRef]
- Chirkov, S.; Ivanov, P.; Sheveleva, A.; Zakubanskiy, A.; Osipov, G. New highly divergent Plum pox virus isolates infecting sour cherry in Russia. Virology 2017, 502, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Ravelonandro, M.; Briard, P. Mechanisms of natural and genetically engineered resistance against viruses. In Applied Plant Virology, Advances, Detection and Antivral Strategy; Academic Press: Cambridge, MA, USA, 2020; pp. 697–704. [Google Scholar] [CrossRef]
- Kinoti, W.M.; Constable, F.E.; Nancarrow, N.; Plummer, K.; Rodoni, B. Analysis of intra-host genetic diversity of Prunus necrotic ringspot virus (PNRSV) using amplicon next generation sequencing. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [Green Version]
- Shi, W.; Yao, R.; Sunwu, R.; Huang, K.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. Incidence and Molecular Identification of Apple Necrotic Mosaic Virus (ApNMV) in Southwest China. Plants 2020, 9, 415. [Google Scholar] [CrossRef] [Green Version]
- Eichmeier, A.; Komínková, M.; Kominek, P.; Baránek, M. Comprehensive Virus Detection Using Next Generation Sequencing in Grapevine Vascular Tissues of Plants Obtained from the Wine Regions of Bohemia and Moravia (Czech Republic). PLoS ONE 2016, 11, e0167966. [Google Scholar] [CrossRef]
- Fan, X.; Dong, Y.F.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Li, Z.; Zhou, J. First Report of Grapevine Pinot gris virus in Grapevines in China. Plant Dis. 2016, 100, 540. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Golino, D.; Rowhani, A. First Report of Grapevine Pinot gris virus Infecting Grapevine in the United States. Plant Dis. 2016, 100, 1030. [Google Scholar] [CrossRef]
- Xiao, H.; Shabanian, M.; Mcfaddensmith, W.; Meng, B. First Report of Grapevine Pinot gris virus in Commercial Grapes in Canada. Plant Dis. 2016, 100, 1030. [Google Scholar] [CrossRef]
- Wu, Q.; Habili, N. The recent importation of Grapevine Pinot gris virus into Australia. Virus Genes 2017, 53, 935–938. [Google Scholar] [CrossRef] [PubMed]
- Hily, J.-M.; Poulicard, N.; Candresse, T.; Vigne, E.; Beuve, M.; Renault, L.; Velt, A.; Spilmont, A.-S.; Lemaire, O. Datamining, Genetic Diversity Analyses, and Phylogeographic Reconstructions Redefine the Worldwide Evolutionary History of Grapevine Pinot gris virus and Grapevine berry inner necrosis virus. Phytobiomes J. 2020, 4, 165–177. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Wetzel, T.; Candresse, T.; Ravelonandro, M.; Dunez, J. A polymerase chain reaction assays adapted to plum pox virus detection. J. Virol. Methods 1991, 33, 355–365. [Google Scholar] [CrossRef]
- Levy, L.; Damsteegt, V.; Welliver, R. First Report of Plum pox virus (Sharka Disease) in Prunus persica in the United States. Plant Dis. 2000, 84, 202. [Google Scholar] [CrossRef]
- Jones, S.; Baizan-Edge, A.; MacFarlane, S.; Torrance, L. Viral Diagnostics in Plants Using Next Generation Sequencing: Computational Analysis in Practice. Front. Plant Sci. 2017, 8, 1770. [Google Scholar] [CrossRef] [PubMed]
- Varsani, A.; Roumagnac, P.; Fuchs, M.; Navas-Castillo, J.; Moriones, E.; Idris, A.; Briddon, R.; Rivera-Bustamante, R.; Zerbini, F.M.; Martin, D.P. Capulavirus and Grablovirus: Two new genera in the family Geminiviridae. Arch. Virol. 2017, 162, 1819–1831. [Google Scholar] [CrossRef] [Green Version]
- Sudarshana, M.R.; Perry, K.L.; Fuchs, M.F. Grapevine red blotch-associated virus, an emerging threat to the grapevine in-dustry. Phytopathology 2015, 105, 1026–1032. [Google Scholar] [CrossRef] [Green Version]
- Al Rwahnih, M.; Alabi, O.J.; Westrick, N.M.; Golino, D.; Rowhani, A. Description of a novel monopartite geminivirus and its defective subviral genome in grapevine. Phytopathology 2017, 107, 240–251. [Google Scholar] [CrossRef] [Green Version]
- Liang, P.; Navarro, B.; Zhang, Z.; Wang, H.; Lu, M.; Xiao, H.; Wu, Q.; Zhou, X.; Di Serio, F.; Li, S. Identification and characterization of a novel geminivirus with a monopartite genome infecting apple trees. J. Gen. Virol. 2015, 96, 2411–2420. [Google Scholar] [CrossRef] [PubMed]
- Maliogka, V.I.; Minafra, A.; Saldarelli, P.; Ruiz-García, A.B.; Glasa, M.; Katis, N.; Olmos, A. Recent Advances on Detection and Characterization of Fruit Tree Viruses Using High-Throughput Sequencing Technologies. Viruses 2018, 10, 436. [Google Scholar] [CrossRef] [Green Version]
- Roossinck, M.J.; Martin, D.P.; Roumagnac, P. Plant Virus Metagenomics: Advances in Virus Discovery. Phytopathology 2015, 105, 716–727. [Google Scholar] [CrossRef] [Green Version]
- Marais, A.; Faure, C.; Bergey, B.; Candresse, T. Viral Double-Stranded RNAs (dsRNAs) from Plants: Alternative Nucleic Acid Substrates for High-Throughput Sequencing. In Viral Metagenomics, Methods in Molecular Biology; Pantaleo, V., Chiumenti, M., Eds.; Humana Press: New York, NY, USA, 2018; Volume 1746, pp. 45–53. ISBN 978-1-4939-7683-6. [Google Scholar]
- Marais, A.; Faure, C.; Mustafayev, E.; Barone, M.; Alioto, D.; Candresse, T. Characterization by Deep Sequencing of Prunus virus T, a Novel Tepovirus Infecting Prunus Species. Phytopathology 2015, 105, 135–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, D.; Phelan, J.; Jesperson, G. First Report of Prunus virus F Infecting Sweet Cherry (Prunus avium cv. Staccato) in Canada. Plant Dis. 2018, 102, 1468–1469. [Google Scholar] [CrossRef]
- Boonham, N.; Kreuze, J.; Winter, S.; van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef]
- Marais, A.; Faure, C.; Couture, C.; Bergey, B.; Gentit, P.; Candresse, T. Characterization by Deep Sequencing of Divergent Plum bark necrosis stem pitting-associated virus (PBNSPaV) Isolates and Development of a Broad-Spectrum PBNSPaV Detection Assay. Phytopathology 2014, 104, 660–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhai, Y.; Liu, W.; Zhu, D.; Pappu, H.R.; Liu, Q. Complete Genomic Characterization of Plum bark necrosis stem pitting–associated virus Infecting Sweet Cherry in China. Genome Announc. 2016, 4, e00413-16. [Google Scholar] [CrossRef] [Green Version]
- Massart, S.; Candresse, T.; Gil, J.; Lacomme, C.; Predajna, L.; Ravnikar, M.; Reynard, J.-S.; Rumbou, A.; Saldarelli, P.; Škorić, D.; et al. A Framework for the Evaluation of Biosecurity, Commercial, Regulatory, and Scientific Impacts of Plant Viruses and Viroids Identified by NGS Technologies. Front. Microbiol. 2017, 8, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrero, R.A.; Napier, K.R.; Cunnington, J.; Liefting, L.; Keenan, S.; Frampton, R.A.; Szabó, T.O.; Bulman, S.; Hunter, A.; Ward, L.; et al. An internet-based bioinformatics toolkit for plant biosecurity diagnosis and surveillance of viruses and viroids. BMC Bioinform. 2017, 18, 26. [Google Scholar] [CrossRef] [Green Version]
- Roistacher, C.N. (Ed.) Graft-transmissible diseases of citrus. In Handbook for Detection and Diagnosis; FAO: Rome, Italy, 1991; pp. 115–120. [Google Scholar]
- Martín, S.; Alioto, D.; Milne, R.G.; Guerri, J.; Moreno, P. Detection of Citrus psorosis virus in field trees by direct tissue blot immunoassay in comparison with ELISA, symptomatology, biological indexing and cross-protection tests. Plant Pathol. 2002, 51, 134–141. [Google Scholar] [CrossRef] [Green Version]
- Peña, L.; Pérez, R.M.; Cervera, M.; Juárez, J.A.; Navarro, L. Early Events in Agrobacterium-mediated Genetic Transformation of Citrus Explants. Ann. Bot. 2004, 94, 67–74. [Google Scholar] [CrossRef] [Green Version]
- Scorza, R.; Ravelonandro, M.; Callahan, A.; Zagrai, I.; Polak, J.; Malinowski, T.; Cambra, M.; Levy, L.; Damsteegt, V.; Krška, B.; et al. ‘HoneySweet’ (C5), the First Genetically Engineered Plum pox virus–resistant Plum (Prunus domestica L.) Cultivar. HortScience 2016, 51, 601–603. [Google Scholar] [CrossRef] [Green Version]
- De Francesco, A.; Costa, N.; García, M.L. Citrus psorosis virus coat protein-derived hairpin construct confers stable transgenic resistance in citrus against psorosis A and B syndromes. Transgenic Res. 2017, 26, 225–235. [Google Scholar] [CrossRef]
- Scorza, R.; Callahan, A.; Dardick, C.; Ravelonandro, M.; Polak, J.; Malinowski, T.; Zagrai, I.; Cambra, M.; Kamenova, I. Genetic engineering of Plum pox virus resistance: ‘HoneySweet’ plum—from concept to product. Plant Cell Tissue Organ Cult. 2013, 115. [Google Scholar] [CrossRef]
- Ravelonandro, M.; Scorza, R.; Michel, H.J.; Briard, P. The efficiency of RNA interference for conferring stable resistance to plum pox virus. Plant Cell Tissue Organ Cult. 2014, 118. [Google Scholar] [CrossRef]
- Malinowski, T.; Cambra, M.; Capote, N.; Zawadzka, B.; Gorris, M.T.; Scorza, R.; Ravelonandro, M. Field Trials of Plum Clones Transformed with the Plum pox virus Coat Protein (PPV-CP) Gene. Plant Dis. 2006, 90, 1012–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Kohalmi, S.E.; Svircev, A.; Wang, A.; Sanfaçon, H.; Tian, L. Silencing of the Host Factor eIF(iso)4E Gene Confers Plum Pox Virus Resistance in Plum. PLoS ONE 2013, 8, e50627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, Y.; Zhang, H.; Xu, N.; Zhang, B.; Gou, F.; Zhu, J.-K. Application of the CRISPR–Cas System for Efficient Genome Engineering in Plants. Molecular Plant. 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duroy, P.-O.; Perrier, X.; Laboureau, N.; Jacquemoud-Collet, J.-P.; Iskra-Caruana, M.-L. How endogenous plant pararetroviruses shed light on Musa evolution. Ann. Bot. 2016, 117, 625–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, J.; Ntui, V.O.; Ron, M.; Muiruri, S.K.; Britt, A.; Tripathi, L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2019, 2, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liua, J.; Gaoa, P.; Sunc, X.; Zhanga, J.; Sunb, P.; Wangb, J.; Jiaa, C.; Zhanga, J.; Hua, W.; Xua, B.; et al. Efficient regener-ation and genetic transformation platform applicable to five Musa varieties. Electron. J. Biotechnol. 2019, 25, 33–38. [Google Scholar] [CrossRef]
- Zuriaga, E.; Romero, C.; Blanca, J.M.; Badenes, M.L. Resistance to Plum Pox Virus (PPV) in apricot (Prunus armeniaca L.) is associated with down-regulation of two MATHd genes. BMC Plant Biol. 2018, 18, 25. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Zhao, Y.; Ye, J.; Cao, X.; Xu, C.; Chen, B.; An, H.; Jiao, Y.; Zhang, F.; Yang, X.; et al. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and moncot plants. Plant Biotechnol. J. 2019, 17, 1185–1187. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravelonandro, M. Reliable Methodologies and Impactful Tools to Control Fruit Tree Viruses. Crops 2021, 1, 32-41. https://doi.org/10.3390/crops1010005
Ravelonandro M. Reliable Methodologies and Impactful Tools to Control Fruit Tree Viruses. Crops. 2021; 1(1):32-41. https://doi.org/10.3390/crops1010005
Chicago/Turabian StyleRavelonandro, Michel. 2021. "Reliable Methodologies and Impactful Tools to Control Fruit Tree Viruses" Crops 1, no. 1: 32-41. https://doi.org/10.3390/crops1010005
APA StyleRavelonandro, M. (2021). Reliable Methodologies and Impactful Tools to Control Fruit Tree Viruses. Crops, 1(1), 32-41. https://doi.org/10.3390/crops1010005




