Using Geometric Approaches to the Common Transcriptomics in Acute Lymphoblastic Leukemia and Rhabdomyosarcoma: Expanding and Integrating Pathway Simulations
Abstract
1. Introduction
2. Materials and Methods
2.1. In Vitro Cell Models
2.2. Microarray Experimentation
2.3. Experimental Workflow
2.4. Creation of the Systems Biology Model
2.5. Simulation of the Systems Biology Model
2.6. The Analytical and Numerical Solutions to the System’s Simulation
2.7. Modelling the Simulated Data
3. Results
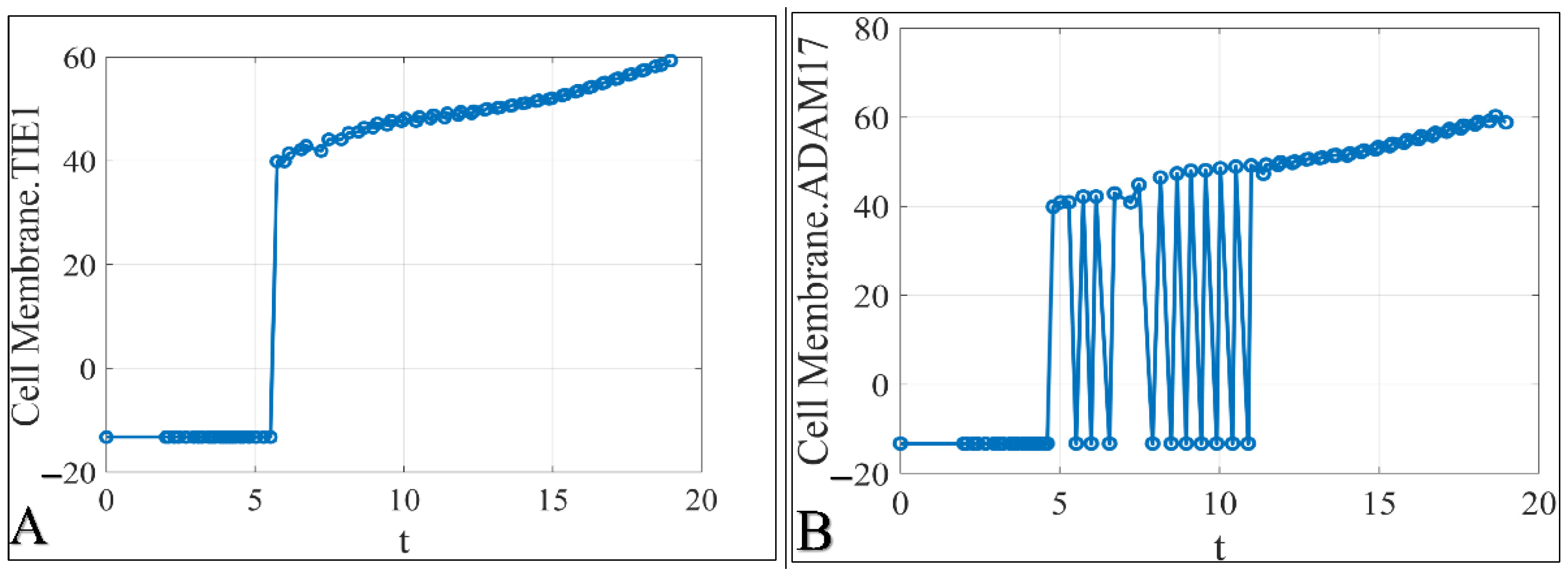
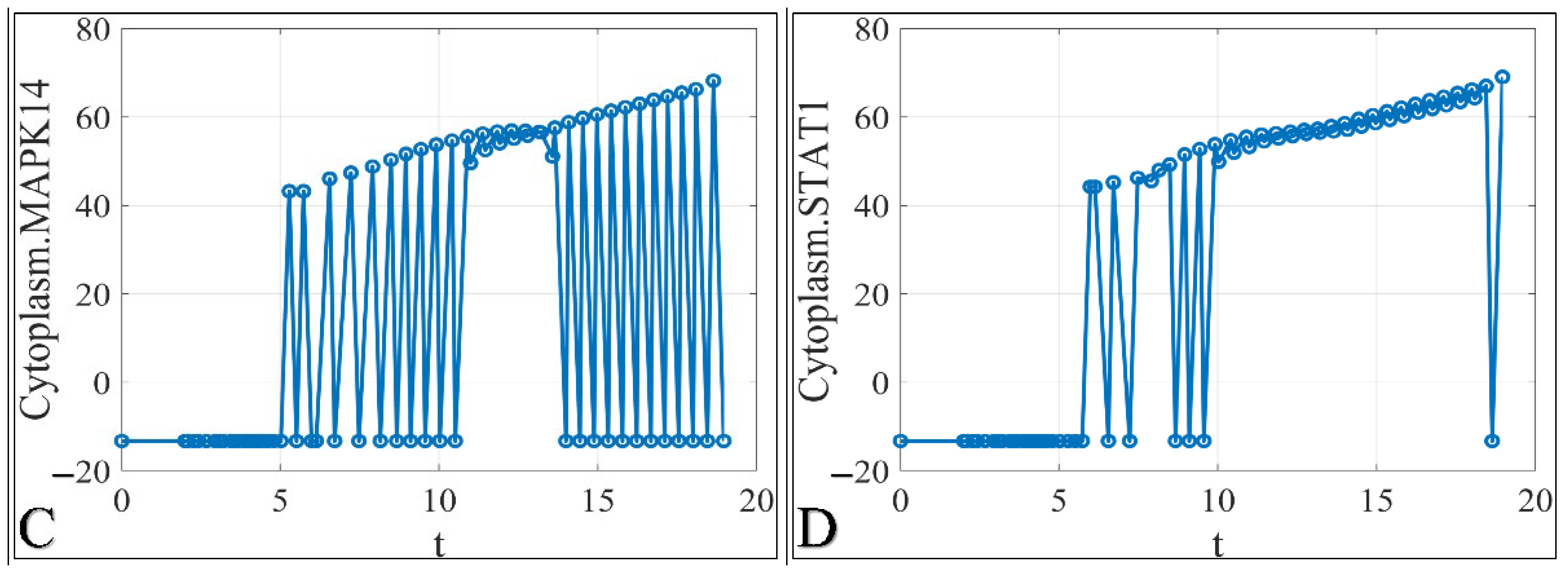
3.1. The Time-Series Iteration of the Simulated Data
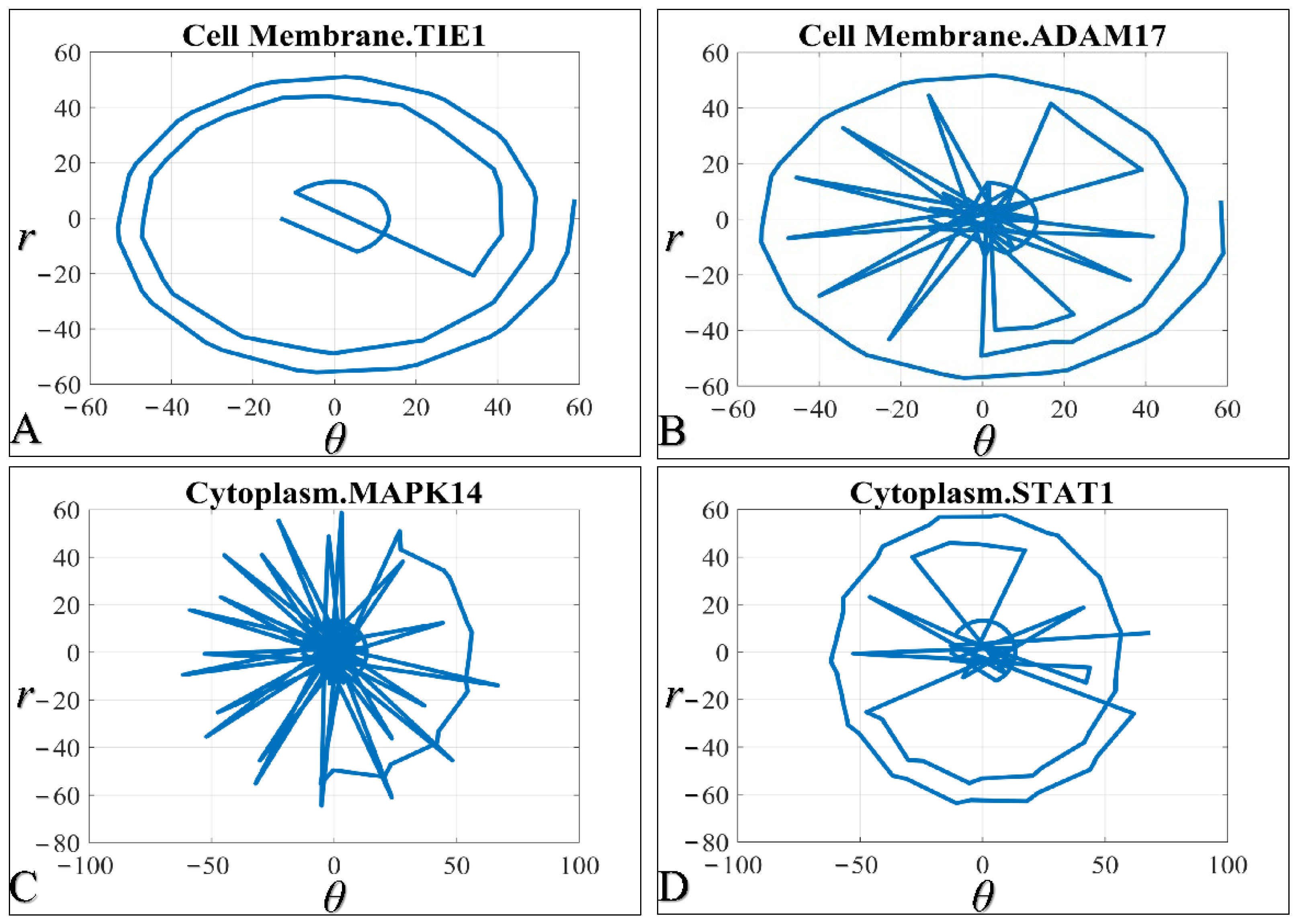
3.2. Polar Transformation of Time Series Iterations
3.3. Time-Dependent Regression of All Species
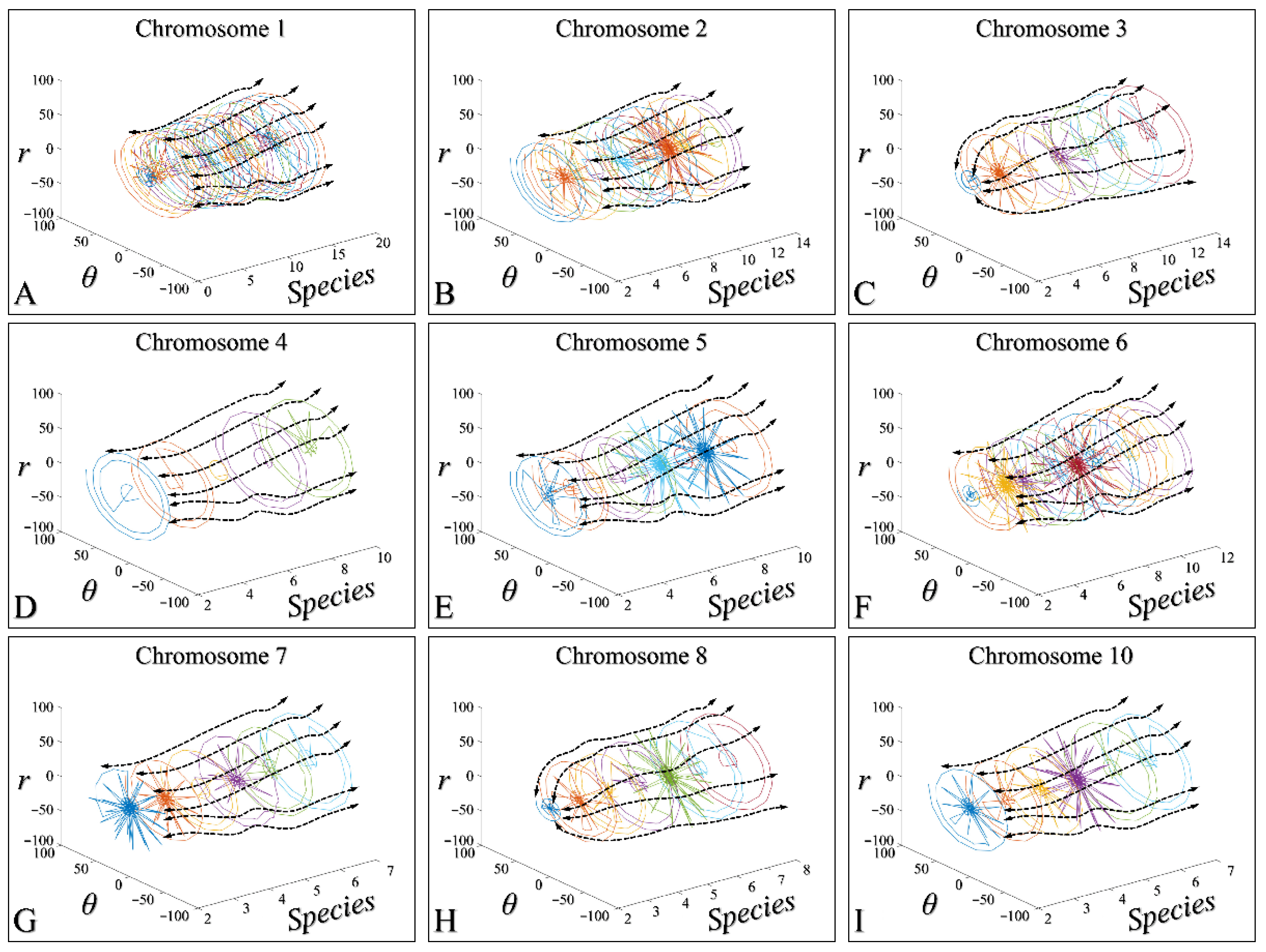
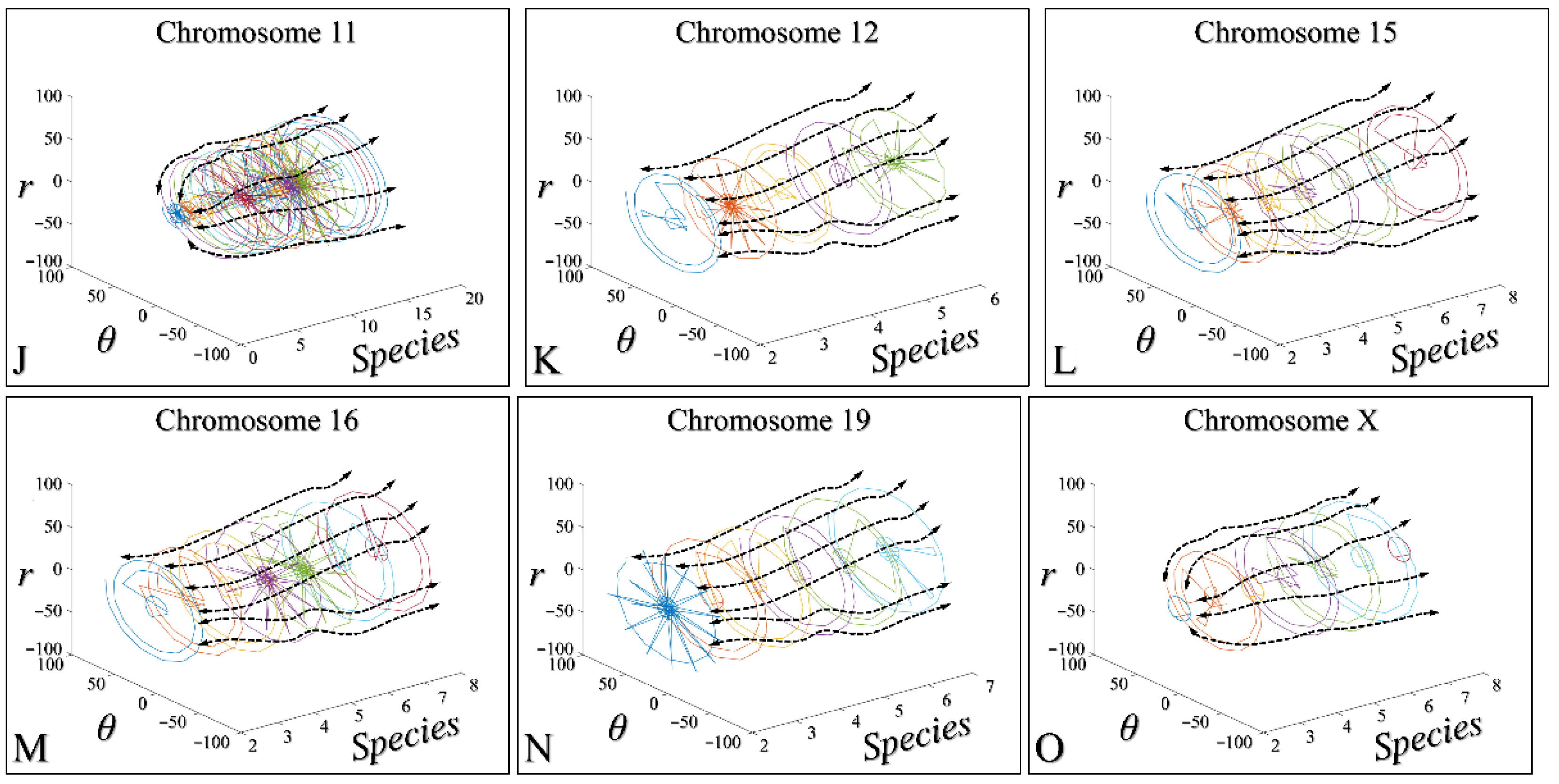
3.4. Chromosomal-Dependent Regression of All Species
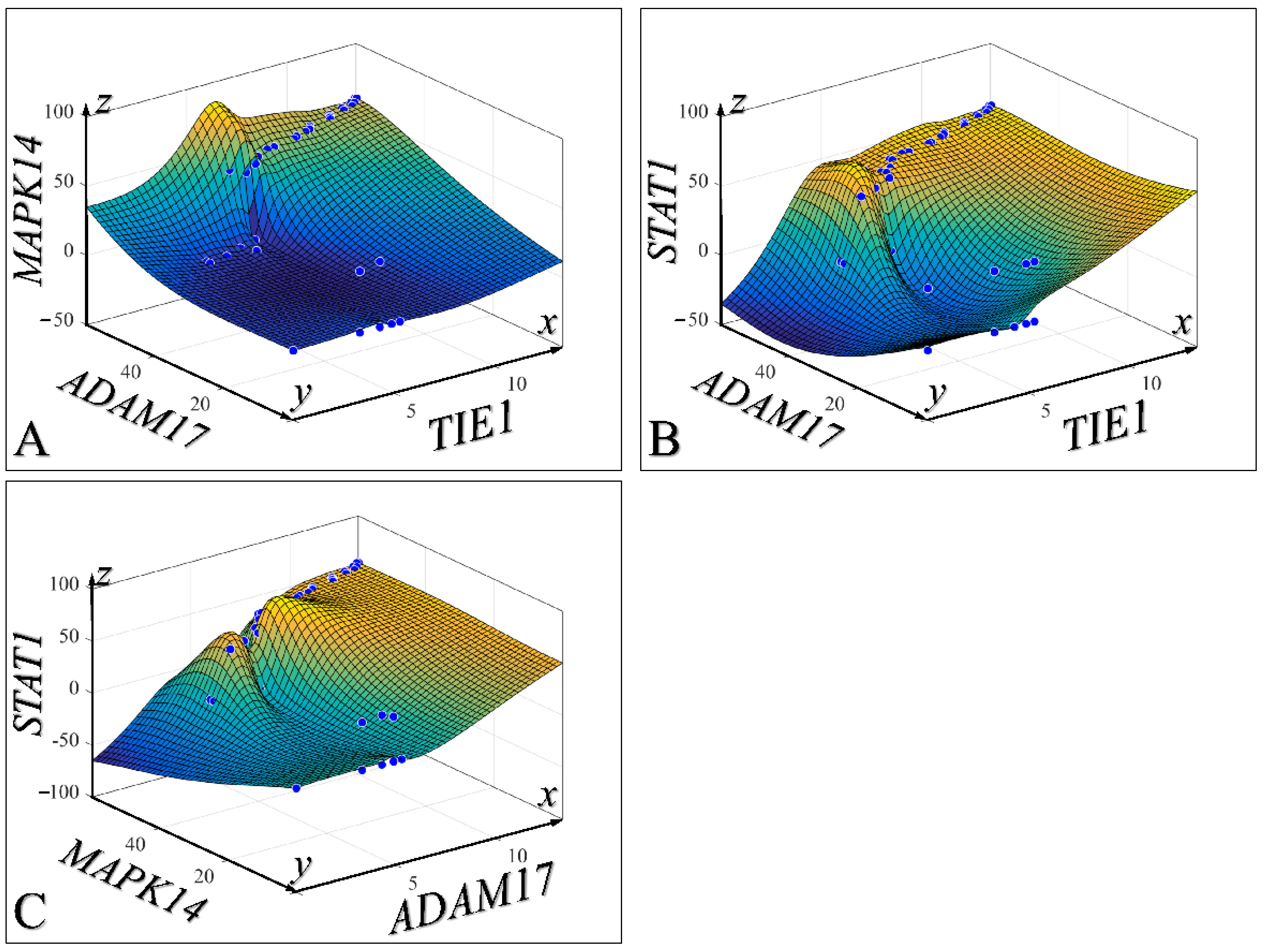
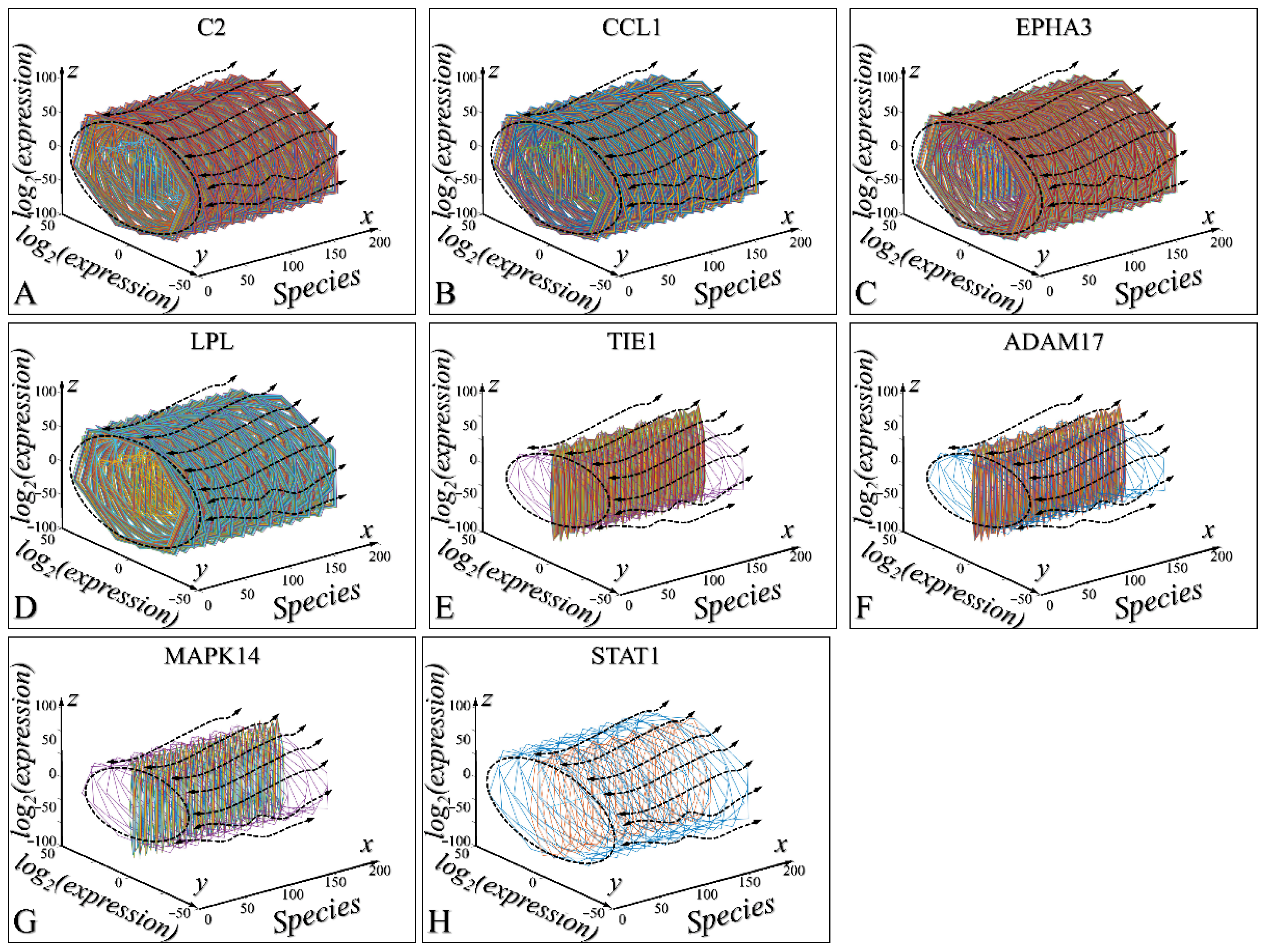
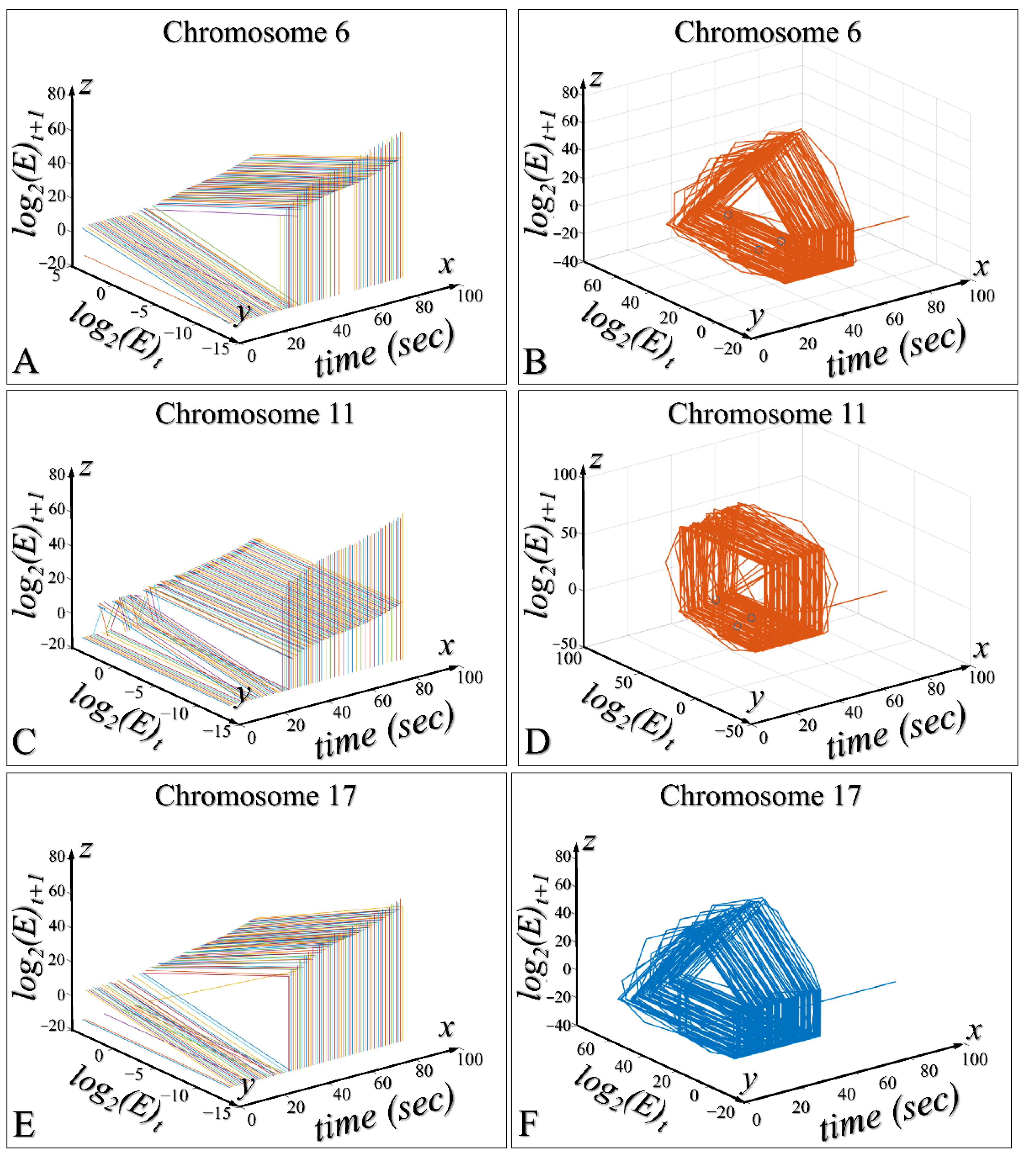
3.5. Three-Dimensional Modelling of the Time-Dependent Evolution
3.6. Time-Dependent Evolution of the System for All Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Onciu, M. Acute lymphoblastic leukemia. Hematol. Oncol. Clin. N. Am. 2009, 23, 655–674. [Google Scholar] [CrossRef] [PubMed]
- Skapek, S.X.; Ferrari, A.; Gupta, A.A.; Lupo, P.J.; Butler, E.; Shipley, J.; Barr, F.G.; Hawkins, D.S. Rhabdomyosarcoma. Nat. Rev. Dis. Primers 2019, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Dagher, R.; Helman, L. Rhabdomyosarcoma: An overview. Oncologist 1999, 4, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, R.; Fuchs, J.; Rodeberg, D. Rhabdomyosarcoma. Semin. Pediatr. Surg. 2016, 25, 276–283. [Google Scholar] [CrossRef]
- Pui, C.H.; Relling, M.V.; Downing, J.R. Acute lymphoblastic leukemia. N. Engl. J. Med. 2004, 350, 1535–1548. [Google Scholar] [CrossRef]
- Miranda, L.; Wolf, J.; Pichuantes, S.; Duke, R.; Franzusoff, A. Isolation of the human pc6 gene encoding the putative host protease for hiv-1 gp160 processing in cd4+ t lymphocytes. Proc. Natl. Acad. Sci. USA 1996, 93, 7695–7700. [Google Scholar] [CrossRef]
- Naujokat, C.; Sezer, O.; Zinke, H.; Leclere, A.; Hauptmann, S.; Possinger, K. Proteasome inhibitors induced caspase-dependent apoptosis and accumulation of p21waf1/cip1 in human immature leukemic cells. Eur. J. Haematol. 2000, 65, 221–236. [Google Scholar] [CrossRef]
- Foley, G.E.; Lazarus, H.; Farber, S.; Uzman, B.G.; Boone, B.A.; McCarthy, R.E. Continuous culture of human lymphoblasts from peripheral blood of a child with acute leukemia. Cancer 1965, 18, 522–529. [Google Scholar] [CrossRef]
- Lambrou, G.I.; Zaravinos, A.; Adamaki, M.; Spandidos, D.A.; Tzortzatou-Stathopoulou, F.; Vlachopoulos, S. Pathway simulations in common oncogenic drivers of leukemic and rhabdomyosarcoma cells: A systems biology approach. Int. J. Oncol. 2012, 40, 1365–1390. [Google Scholar] [CrossRef]
- McAllister, R.M.; Isaacs, H.; Rongey, R.; Peer, M.; Au, W.; Soukup, S.W.; Gardner, M.B. Establishment of a human medulloblastoma cell line. Int. J. Cancer 1977, 20, 206–212. [Google Scholar] [CrossRef]
- Friedman, H.S.; Bigner, S.H.; McComb, R.D.; Schold, S.C., Jr.; Pasternak, J.F.; Groothuis, D.R.; Bigner, D.D. A model for human medulloblastoma. Growth, morphology, and chromosomal analysis in vitro and in athymic mice. J. Neuropathol. Exp. Neurol. 1983, 42, 485–503. [Google Scholar] [CrossRef]
- McAllister, R.M.; Melnyk, J.; Finkelstein, J.Z.; Adams, E.C., Jr.; Gardner, M.B. Cultivation in vitro of cells derived from a human rhabdomyosarcoma. Cancer 1969, 24, 520–526. [Google Scholar] [CrossRef]
- Yeung, C.M.; An, B.S.; Cheng, C.K.; Chow, B.K.; Leung, P.C. Expression and transcriptional regulation of the gnrh receptor gene in human neuronal cells. Mol. Hum. Reprod. 2005, 11, 837–842. [Google Scholar] [CrossRef]
- Chu, E.S.; Wong, T.K.; Yow, C.M. Photodynamic effect in medulloblastoma: Downregulation of matrix metalloproteinases and human telomerase reverse transcriptase expressions. Photochem. Photobiol. Sci. 2008, 7, 76–83. [Google Scholar] [CrossRef]
- Sifakis, E.G.; Prentza, A.; Koutsouris, D.; Chatziioannou, A.A. Evaluating the effect of various background correction methods regarding noise reduction, in two-channel microarray data. Comput. Biol. Med. 2012, 42, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Tselios, C.; Zaravinos, A.; Tsartsalis, A.N.; Tagka, A.; Kotoulas, A.; Geronikolou, S.A.; Braoudaki, M.; Lambrou, G.I. Systems approaches in the common metabolomics in acute lymphoblastic leukemia and rhabdomyosarcoma cells: A computational approach. Adv. Exp. Med. Biol. 2021, 1338, 55–66. [Google Scholar]
- Elizarraras, J.M.; Liao, Y.; Shi, Z.; Zhu, Q.; Pico, A.R.; Zhang, B. Webgestalt 2024: Faster gene set analysis and new support for metabolomics and multi-omics. Nucleic Acids Res. 2024, 52, W415–W421. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. Webgestalt 2019: Gene set analysis toolkit with revamped uis and apis. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Wang, J.; Duncan, D.; Shi, Z.; Zhang, B. Web-based gene set analysis toolkit (webgestalt): Update 2013. Nucleic Acids Res. 2013, 41, W77–W83. [Google Scholar] [CrossRef]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. Webgestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef]
- Zhang, B.; Kirov, S.; Snoddy, J. Webgestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005, 33, W741–W748. [Google Scholar] [CrossRef]
- Yang, P.; Chen, N.; Jia, J.H.; Gao, X.J.; Li, S.H.; Cai, J.; Wang, Z. Tie-1: A potential target for anti-angiogenesis therapy. J. Huazhong Univ. Sci. Technol. 2015, 35, 615–622. [Google Scholar] [CrossRef]
- Teicher, B.A. Searching for molecular targets in sarcoma. Biochem. Pharmacol. 2012, 84, 1–10. [Google Scholar] [CrossRef]
- Le, Q.; Castro, S.; Tang, T.; Loeb, A.M.; Hylkema, T.; McKay, C.N.; Perkins, L.; Srivastava, S.; Call, L.; Smith, J.; et al. Therapeutic targeting of mesothelin with chimeric antigen receptor t cells in acute myeloid leukemia. Clin. Cancer Res. 2021, 27, 5718–5730. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Wei, L.; Jiang, X.; Conroy, J.; Glenn, S.; Bshara, W.; Yu, T.; Pao, A.; Tanaka, S.; Kawai, A.; et al. Analysis of mutations in primary and metastatic synovial sarcoma. Oncotarget 2018, 9, 36878–36888. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Huang, J.; Xiao, S.; Cheng, G.; Liu, Y.; Zhao, T.; Chen, C.; Yi, Y.; Peng, Y.; Cao, J. Synthesis, anti-leukemia activity, and molecular docking of novel 3,16-androstenedione derivatives. Steroids 2023, 199, 109290. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Li, X.; Yang, X.; Mou, H.; Wei, L. Indirubin, an active component of indigo naturalis, exhibits inhibitory effects on leukemia cells via targeting hsp90aa1 and pi3k/akt pathway. Anti-Cancer Agents Med. Chem. 2024, 24, 718–727. [Google Scholar] [CrossRef]
- Perdiguero, E.; Ruiz-Bonilla, V.; Serrano, A.L.; Muñoz-Cánoves, P. Genetic deficiency of p38alpha reveals its critical role in myoblast cell cycle exit: The p38alpha-jnk connection. Cell Cycle 2007, 6, 1298–1303. [Google Scholar] [CrossRef]
- Cho, H.J.; Smith, J.; Switzer, C.H.; Louka, E.; Charles, R.L.; Prysyazhna, O.; Schroder, E.; Fernandez-Caggiano, M.; de Jesus, D.S.; Eminaga, S.; et al. Sfx-01 is therapeutic against myeloproliferative disorders caused by activating mutations in shp2. EMBO Mol. Med. 2025, 17, 2115–2136. [Google Scholar] [CrossRef]
- Liu, A.; Liu, Y.; Xu, Z.; Yu, W.; Wang, H.; Li, C.; Lin, J. Novel small molecule, xzh-5, inhibits constitutive and interleukin-6-induced stat3 phosphorylation in human rhabdomyosarcoma cells. Cancer Sci. 2011, 102, 1381–1387. [Google Scholar] [CrossRef]
- Blandford, M.C.; Barr, F.G.; Lynch, J.C.; Randall, R.L.; Qualman, S.J.; Keller, C. Rhabdomyosarcomas utilize developmental, myogenic growth factors for disease advantage: A report from the children’s oncology group. Pediatr Blood Cancer 2006, 46, 329–338. [Google Scholar] [CrossRef]
- Khan, J.; Simon, R.; Bittner, M.; Chen, Y.; Leighton, S.B.; Pohida, T.; Smith, P.D.; Jiang, Y.; Gooden, G.C.; Trent, J.M.; et al. Gene expression profiling of alveolar rhabdomyosarcoma with cdna microarrays. Cancer Res. 1998, 58, 5009–5013. [Google Scholar]
- Shi, W.; Hou, X.; Li, X.; Peng, H.; Shi, M.; Jiang, Q.; Liu, X.; Ji, Y.; Yao, Y.; He, C.; et al. Differential gene expressions of the mapk signaling pathway in enterovirus 71-infected rhabdomyosarcoma cells. Braz. J. Infect. Dis. Off. Publ. Braz. Soc. Infect. Dis. 2013, 17, 410–417. [Google Scholar] [CrossRef]
- Bott, S.R.; Arya, M.; Kirby, R.S.; Williamson, M. P21waf1/cip1 gene is inactivated in metastatic prostatic cancer cell lines by promoter methylation. Prostate Cancer Prostatic Dis. 2005, 8, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; He, L.; Savell, V.H.; Jenkins, J.J.; Parham, D.M. Inhibition of the interferon-gamma/signal transducers and activators of transcription (stat) pathway by hypermethylation at a stat-binding site in the p21waf1 promoter region. Cancer Res. 2000, 60, 3290–3298. [Google Scholar] [PubMed]
- Illa, I.; Gallardo, E.; Gimeno, R.; Serrano, C.; Ferrer, I.; Juárez, C. Signal transducer and activator of transcription 1 in human muscle: Implications in inflammatory myopathies. Am. J. Pathol. 1997, 151, 81–88. [Google Scholar] [PubMed]
- Kim, J.Y.; Park, H.K.; Yoon, J.S.; Kim, S.J.; Kim, E.S.; Song, S.H.; Choi, J.H.; Kim, B.K.; Park, B.B.; Lee, Y.Y. Molecular mechanisms of cellular proliferation in acute myelogenous leukemia by leptin. Oncol. Rep. 2010, 23, 1369–1374. [Google Scholar] [CrossRef]
- Pillozzi, S.; Masselli, M.; De Lorenzo, E.; Accordi, B.; Cilia, E.; Crociani, O.; Amedei, A.; Veltroni, M.; D’Amico, M.; Basso, G.; et al. Chemotherapy resistance in acute lymphoblastic leukemia requires herg1 channels and is overcome by herg1 blockers. Blood 2011, 117, 902–914. [Google Scholar] [CrossRef]
- Wang, J.; Wakeman, T.P.; Lathia, J.D.; Hjelmeland, A.B.; Wang, X.F.; White, R.R.; Rich, J.N.; Sullenger, B.A. Notch promotes radioresistance of glioma stem cells. Stem Cells 2010, 28, 17–28. [Google Scholar] [CrossRef]
- Wysoczynski, M.; Shin, D.M.; Kucia, M.; Ratajczak, M.Z. Selective upregulation of interleukin-8 by human rhabdomyosarcomas in response to hypoxia: Therapeutic implications. Int. J. Cancer 2010, 126, 371–381. [Google Scholar] [CrossRef]
- Karp, J.E.; Kaufmann, S.H.; Adjei, A.A.; Lancet, J.E.; Wright, J.J.; End, D.W. Current status of clinical trials of farnesyltransferase inhibitors. Curr. Opin. Oncol. 2001, 13, 470–476. [Google Scholar] [CrossRef]
- Fathi, A.T.; Grant, S.; Karp, J.E. Exploiting cellular pathways to develop new treatment strategies for aml. Cancer Treat. Rev. 2010, 36, 142–150. [Google Scholar] [CrossRef]
- Suwanwela, J.; Farber, C.R.; Haung, B.L.; Song, B.; Pan, C.; Lyons, K.M.; Lusis, A.J. Systems genetics analysis of mouse chondrocyte differentiation. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2011, 26, 747–760. [Google Scholar] [CrossRef]
- Kusnezow, W.; Syagailo, Y.V.; Rüffer, S.; Baudenstiel, N.; Gauer, C.; Hoheisel, J.D.; Wild, D.; Goychuk, I. Optimal design of microarray immunoassays to compensate for kinetic limitations: Theory and experiment. Mol. Cell. Proteom. 2006, 5, 1681–1696. [Google Scholar] [CrossRef]
- Gan, X.; Liew, A.W.; Yan, H. Discovering biclusters in gene expression data based on high-dimensional linear geometries. BMC Bioinform. 2008, 9, 209. [Google Scholar] [CrossRef]
- Trink, A.; Kanter, I.; Pode-Shakked, N.; Urbach, A.; Dekel, B.; Kalisky, T. Geometry of gene expression space of wilms’ tumors from human patients. Neoplasia 2018, 20, 871–881. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tselios, C.; Vezakis, I.; Zaravinos, A.; Lambrou, G.I. Using Geometric Approaches to the Common Transcriptomics in Acute Lymphoblastic Leukemia and Rhabdomyosarcoma: Expanding and Integrating Pathway Simulations. BioMedInformatics 2025, 5, 45. https://doi.org/10.3390/biomedinformatics5030045
Tselios C, Vezakis I, Zaravinos A, Lambrou GI. Using Geometric Approaches to the Common Transcriptomics in Acute Lymphoblastic Leukemia and Rhabdomyosarcoma: Expanding and Integrating Pathway Simulations. BioMedInformatics. 2025; 5(3):45. https://doi.org/10.3390/biomedinformatics5030045
Chicago/Turabian StyleTselios, Christos, Ioannis Vezakis, Apostolos Zaravinos, and George I. Lambrou. 2025. "Using Geometric Approaches to the Common Transcriptomics in Acute Lymphoblastic Leukemia and Rhabdomyosarcoma: Expanding and Integrating Pathway Simulations" BioMedInformatics 5, no. 3: 45. https://doi.org/10.3390/biomedinformatics5030045
APA StyleTselios, C., Vezakis, I., Zaravinos, A., & Lambrou, G. I. (2025). Using Geometric Approaches to the Common Transcriptomics in Acute Lymphoblastic Leukemia and Rhabdomyosarcoma: Expanding and Integrating Pathway Simulations. BioMedInformatics, 5(3), 45. https://doi.org/10.3390/biomedinformatics5030045








