Pulmonary Nodule Detection, Segmentation and Classification Using Deep Learning: A Comprehensive Literature Review
Abstract
1. Introduction
- Bronchoscopy: A thin, flexible tube with a camera (bronchoscope) is inserted through the nose or mouth and into the airways to examine the lungs and collect tissue samples for biopsy [8].
- Needle Biopsy: A needle is used to extract a tissue sample from a suspicious lung nodule or lymph node for examination under a microscope. There are different types of needle biopsies, including transthoracic needle biopsy and endobronchial ultrasound-guided biopsy [9].
- Thoracoscopy or Video-Assisted Thoracoscopic Surgery (VATS): These minimally invasive surgical procedures involve making small incisions in the chest to access and biopsy lung tissue or remove a suspicious nodule [10].
- Mediastinoscopy: This procedure involves making a small incision in the neck and inserting a scope to examine and sample lymph nodes in the area between the lungs (mediastinum) [11].
- Chest X-rays: Historically, chest X-rays have been the primary tool for detecting lung abnormalities. They provide two-dimensional images of the chest, and can reveal the presence of lung nodules or other suspicious lesions. However, their sensitivity in detecting early-stage lung cancer is limited [12,13].
- Low-dose Computed Tomography (LDCT) Scans: Computed Tomography (CT) has become a more advanced and widely adopted method for lung cancer screening. These scans use a series of X-rays to create detailed cross-sectional images of the chest. Low-dose CT (LDCT) scans, in particular, have gained prominence in recent years due to their ability to detect smaller nodules and early-stage cancers [14,15].
- Lung Cancer Risk Assessment Models: Doctors often employ risk assessment models to identify individuals at a higher risk of developing lung cancer. These models take into account factors such as age, smoking history, and family history to stratify patients into different risk categories [16].
2. Datasets
2.1. Lung Image Database Consortium Image Collection
2.2. Lung Nodule Analysis 2016
2.3. ELCAP Public Lung Image Database
2.4. Alibaba Tianchi Competition Dataset
2.5. SPIE-AAPM Lung CT Challenge
3. Preprocessing
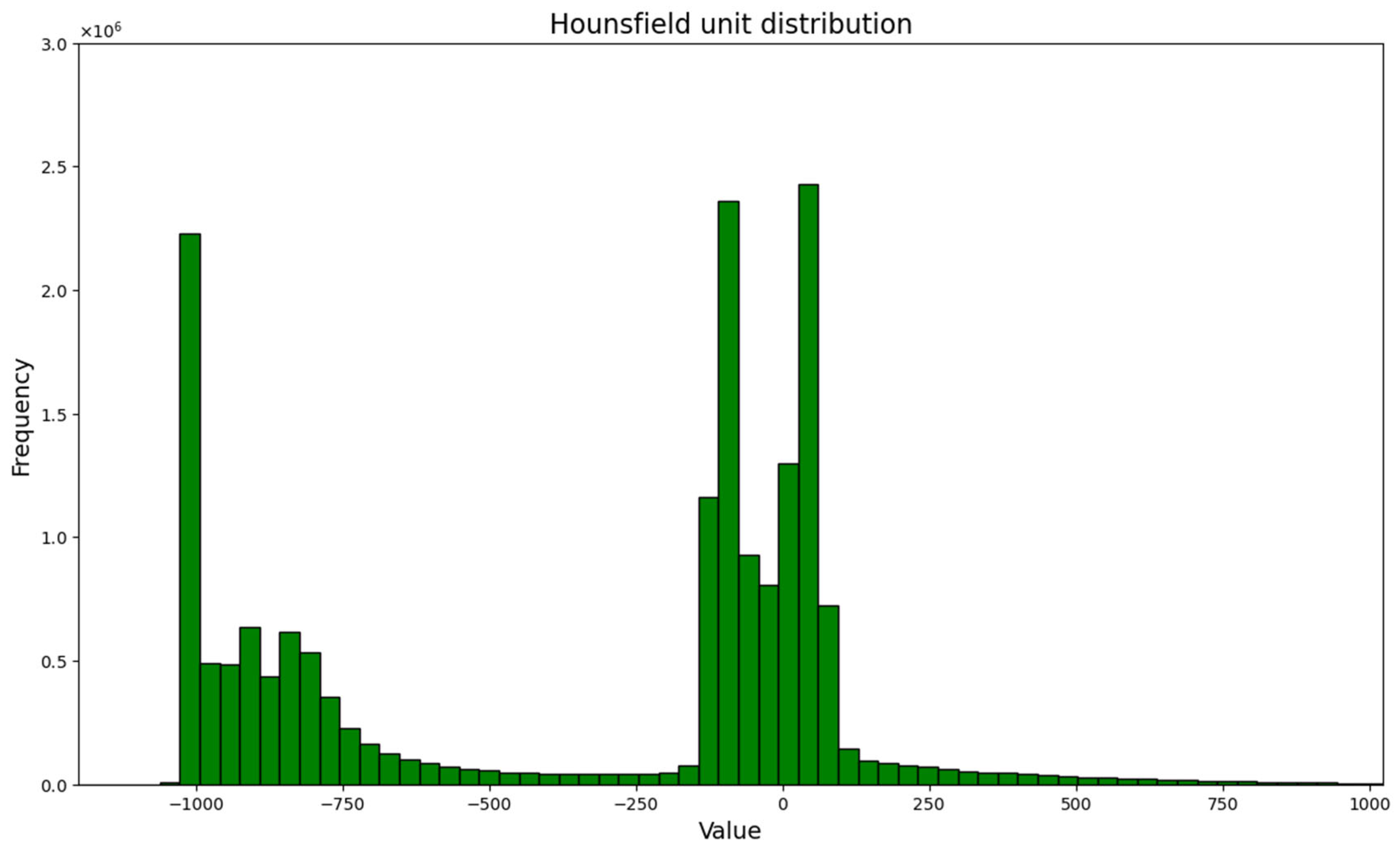
3.1. Conversion of Pixel Value to Hounsfield Units and Thresholding
3.2. Resampling for Isotropy
3.3. Lung Segmentation
3.4. Normalization
3.5. Zero Centering
3.6. Patch Extraction
3.7. Data Augmentation
4. Architectures
- Convolutional Neural Networks (CNNs)
- U-Net
- Autoencoders
- Capsule Networks
- Transformers
4.1. Nodule Detection
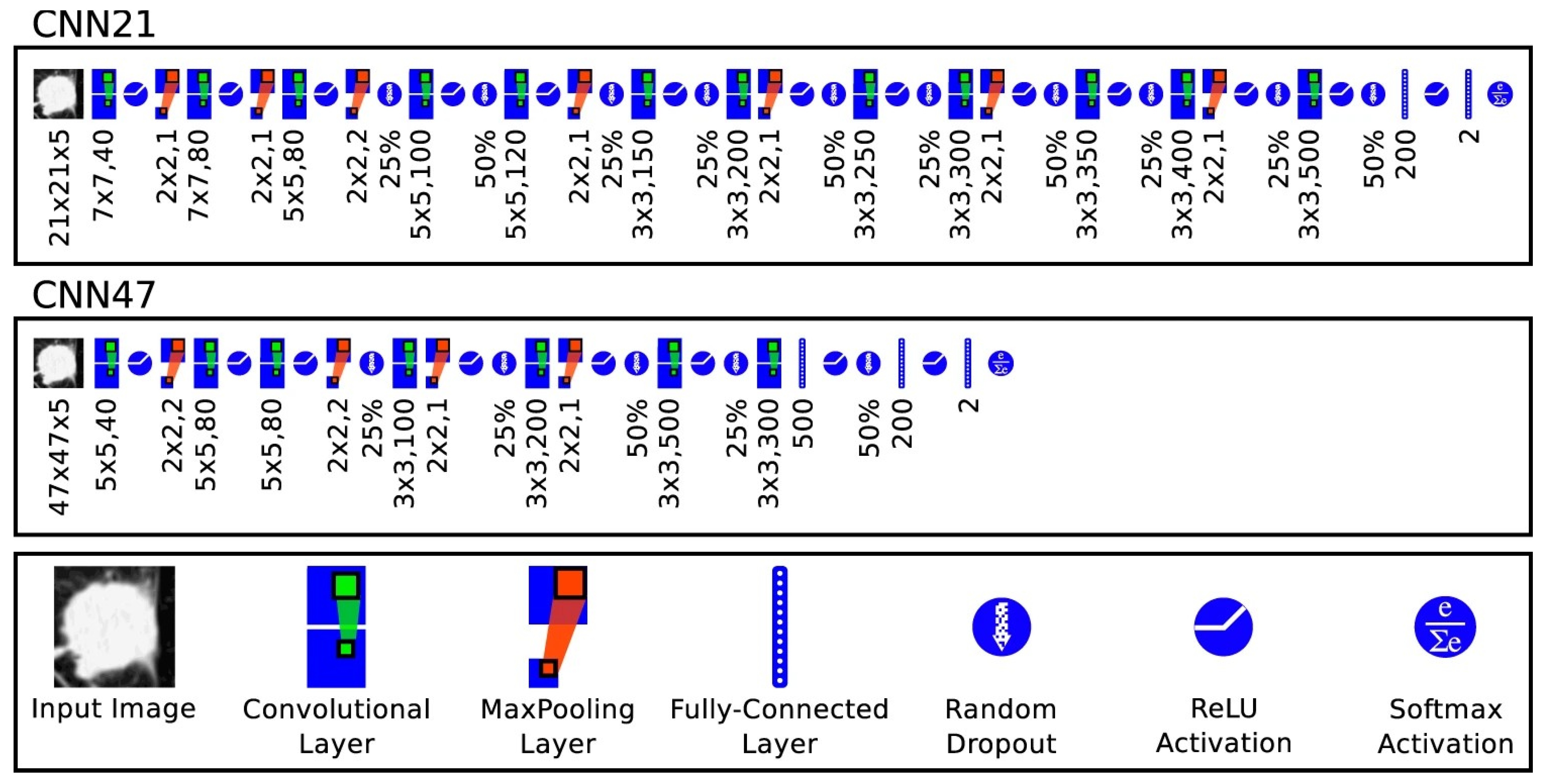
4.1.1. Two-Dimensional Convolutional Neural Networks (2D CNN)
4.1.2. Three-Dimensional Convolutional Neural Networks (3D CNN)
4.1.3. Auto Encoders
4.1.4. Transformers
4.1.5. Capsule Networks
4.1.6. Others
4.2. Nodule Segmentation
4.2.1. Two-Dimensional Convolutional Neural Networks (2D CNN)
4.2.2. Three-Dimensional Convolutional Neural Networks (3D CNN)
4.2.3. U-Net
4.2.4. Residual Networks
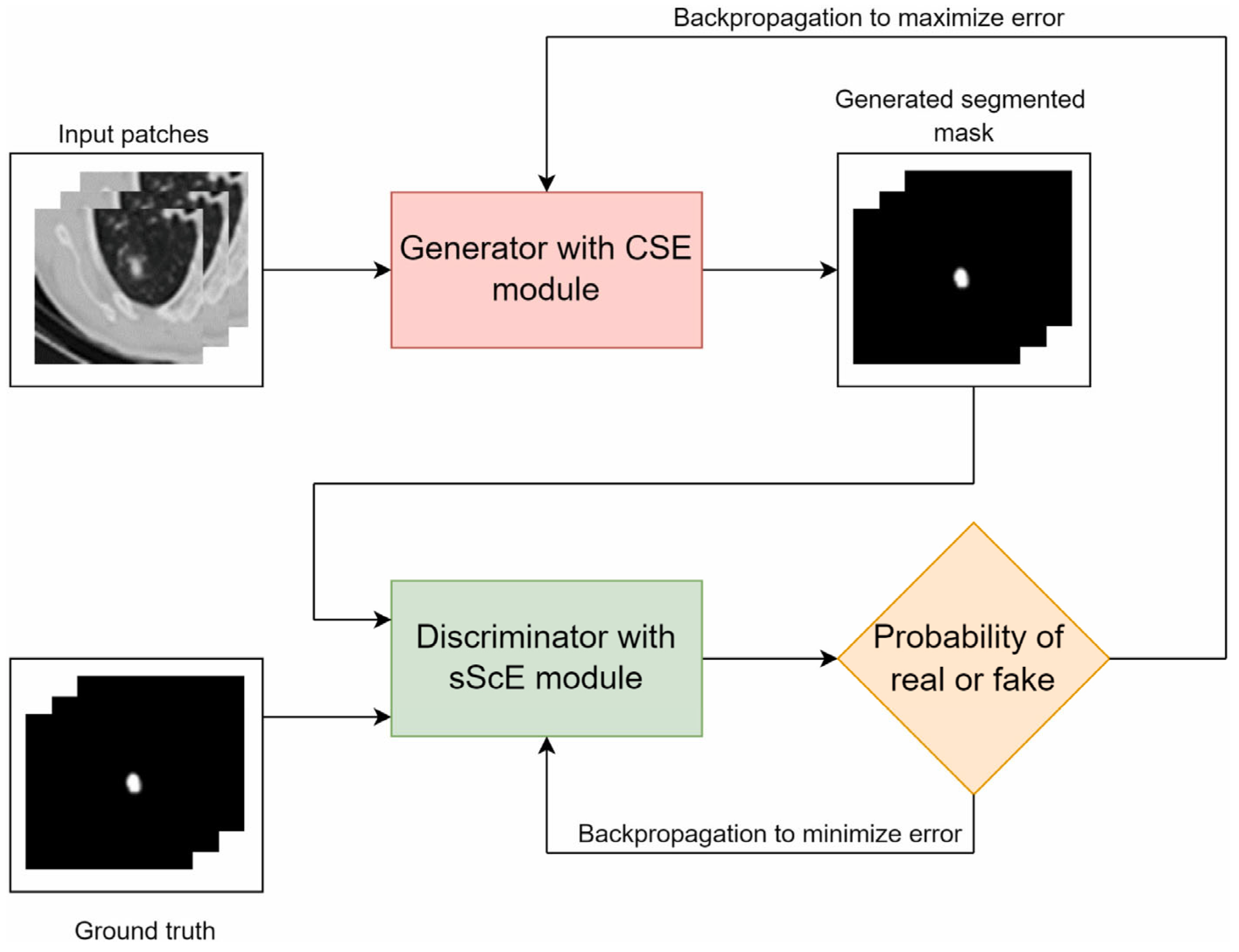
4.2.5. Generative Adversarial Networks (GANs)
4.2.6. Transformers
4.2.7. Others
4.3. Nodule Classification
4.3.1. Single-View
4.3.2. Multi-View
4.3.3. Three-Dimensional Classifiers
4.3.4. Auto Encoders
4.3.5. Multi-Task Learning
4.3.6. Transformers
4.3.7. Capsule Networks
4.3.8. Others
5. Discussion
5.1. Preprocessing
5.2. Nodule Detection
5.3. Nodule Segmentation
5.4. Nodule Classification
5.5. Radiologist vs. AI
5.6. Future Extensions and Research Directions
5.7. Practical Implications
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Reference | Year | Dataset | Preprocessing | Data Augmentation | Deep Architecture |
|---|---|---|---|---|---|
| [54] | 2017 | LUNA16 | No, TH, Cr, 3D-PA | Cr, Fl, Du | Faster R-CNN, 3D DCNN |
| [55] | 2018 | LUNA16 | No, 2D-PA | Tr, Ro, Fl | 2D CNNs, AE, DAE |
| [56] | 2018 | LUNA16 | 3D-PA, Re | Cr, Sc, 3D Fl, 3D Ro | 3D CNN |
| [35] | 2018 | LUNA16 | 3D-PA, Re, Rz, TH | Tr, Ro, Fl | Modified 3D U-Net |
| [57] | 2018 | LUNA16 | TH, Cr, 3D-PA | Ro, Fl | 3D DCNN |
| [58] | 2019 | LIDC-IDRI | LS, Re, 2D-PA (Only juxta-pleural nodules) | - | 2D CNN |
| [59] | 2019 | LUNA16 | TH, No, 2D-PA | Tr, Ro, Fl | 2D U-net, 3D CNN |
| [60] | 2019 | LIDC-IDRI | Cr, 2D-PA | Ro, Tr, SC, Re | Custom ResNet |
| [61] | 2019 | LIDC-IDRI | Re, No, Cr, 2D-PA | Tr, Ro | 3D Faster R-CNN and CMixNet with U-Net-like encoder–decoder architecture |
| [62] | 2019 | LUNA16 | 3D-PA | Ro | 3D DCNN |
| [63] | 2020 | LIDC-IDRI | No, 3D-PA | Fl | 3D U-net |
| [64] | 2020 | LUNA16, TIANCHI17 | Re, No, 3D-PA | - | CNN, TL |
| [65] | 2020 | LUNA16 | Cr, MV, 2D-PA | - | 3D multiscale DCNN, AE, TL |
| [66] | 2020 | LUNA16, private set | TH, No, Cr, 3D-PA | Ro, Fl, Tr | U-net, AE, TL |
| [67] | 2021 | LIDC-IDRI, private data set | 2D slice | - | 3D-ResNet and MKL |
| [68] | 2021 | LUNA16 | TH, Re, PSEG | Ro, Fl, Shift | 3DCNN |
| [69] | 2021 | Kaggle Data Science Bowl 2017 challenge (KDSB) and LUNA 16 | TH, No, Re, PSEG,3D-PA | Ro, Fl, Cr | U-Net |
| [70] | 2021 | LUNA16 | PSEG, No | balancing of lower target patches (thoselabeled as one) are equivalent to higher target patches (those labeled as zero) | 3DCNN |
| [71] | 2021 | LUNA16 | Re, PSEG, 3D-PA | Sampling, Ro, Tr | Multi-path 3D CNN |
| [72] | 2021 | LUNA16 | TH, No | Ro, Shift, Fl, Sc | Faster R-CNN with adaptive anchor box |
| [73] | 2021 | NLST (NLST, 2011), LHMC, Kaggle | TH, No, PSEG | Fl, Ro, Tr | 2D and 3D DNN |
| [74] | 2022 | LIDC-IDRI + Japan Chest CT Dataset | 3D-PA | n/a | 3D unet |
| [75] | 2022 | LUNA16 | Re, TH, No, 3D-pa | Ro, Fl | 3D sphere representation-based center-points matching detection network (SCPM-Net) |
| [76] | 2022 | LUNA16 | Re, No, 3D-PA | n/a | Atrous UNet+ |
| [77] | 2022 | LUNA16 | TH, No3s-PA | Ro, Fl | 3D U-shaped residual network |
| [78] | 2023 | LUNA16 | 3D-PA | Cr, Fl, Zoom | 3D CNN |
| [79] | 2023 | LUNA16 | HU, No, 3D-PA | n/a | 3D ResNet18 dual path Faster R-CNN and a federated learning algorithm |
| [80] | 2023 | LUNA16 | Re. TH, PSEG, 3D-PA | Sc, Cr, Fl | 3D ViT |
| [81] | 2023 | LUNA16 | TH, 3D-PA | n/a | 3D ViT |
| [82] | 2023 | LIDC-IDRI | TH, No, Masking gt seg | balancing classes by up-sampling cancer cases | 2D Ensemble Transformer with Attention Modules |
| [83] | 2023 | LUNA16 | TH, HU, Masking gt, No | n/a | 3Dl Multifaceted Attention Encoder–Decoder |
| [84] | 2023 | ELCAP | n/a | Ro, Re, Cr | 3D CNN-CapsNet |
| [85] | 2023 | LUNA16 | TH, No, masking gt seg, Re, Sc | Cr, Fl, Sc | A multiscale self-calibrated network (DEPMSCNet) with a dual attention mechanism |
| Reference | Dataset | Deep Architecture | Input Size | Sensitivity | Specificity | Precision | AUC | Accuracy | CPM (FROC) |
|---|---|---|---|---|---|---|---|---|---|
| [54] | LUNA16 | Faster R-CNN, 3D DCNN | 32 × 32 × 3, 36 × 36 × 20 | 92.2/1 FP, 94.4/4 FPs | - | - | - | - | 0.893 |
| [55] | LUNA16 | 2D CNNs, AE, DAE | 64 × 64 | - | - | - | - | - | 0.922 |
| [56] | LUNA16 | 3D CNN | 32 × 32 × 32 | 87.94/1 FP, 92.93/4 FPs | - | - | - | - | 0.7967 |
| [35] | LUNA16 | Modified 3D U-Net | 64 × 64 × 64 | 95.16/30.39 FPs | - | - | 93.72 | - | 0.8135 |
| [57] | LUNA16 | 3D DCNN | 32 × 32 × 32 | 94.9/1 FP | - | 0.947 | |||
| [58] | LIDC-IDRI | 2D CNN | 24 × 24 | 88/1 FP, 94.01/4 FPs | - | - | 94.3 | - | - |
| [59] | LUNA16 | 2D U-net, 3D CNN | 512 × 512 (CG), 16 × 16 × 16, 32 × 32 × 32 | 89.9/0.25 FP, 94.8/4 FPs | - | - | - | - | 0.952 |
| [60] | LIDC-IDRI | Custom ResNet | 64 × 64 | 92.8/8 FPs | - | - | - | - | - |
| [61] | LIDC-IDRI | 3D Faster R-CNN and CMixNet with U-Net-like encoder–decoder architecture | 36 × 36 × 36 | 93.97, 98.00 | 89.83, 94.35 | - | - | 88.79, 94.17 | - |
| [62] | LUNA16 | 3D DCNN | Full CT | - | 0.8727 | ||||
| [63] | LIDC-IDRI | 3D U-net | 40 × 40 × 26 | 92.4 | 94.6 | - | 94.1 | 96.8 | - |
| [64] | LUNA16, TIANCHI17 | CNN, TL | N [26, 36, 48] N × N | 97.26 | 97.38 | - | 99.54 | 97.33, 92.81 (multi-res) | 0.742 |
| [65] | LUNA16 | 3D multiscale DCNN, AE, TL | 32 × 32 × 32 | 94.2/1 FP, 96/2 FPs | - | - | - | - | 0.9403 |
| [66] | LUNA16, private set | U-net, AE, TL | 2D CT slice (512 × 512) | 98 | - | - | 95.67 | 97.96 | - |
| [67] | LIDC-IDRI, private data set | 3D-ResNet and MKL | 40 × 40 × 20 | 91.01 | 91.40 | - | - | 91.29 | |
| [68] | LUNA16 | 3DCNN | 128 × 128 × 128 | 87.2/22 FP | - | - | - | - | 0.923 |
| [69] | Kaggle Data Science Bowl 2017 challenge (KDSB) and LUNA 16 | U-Net | 128 × 128 | 89.1 | 87.4 | - | - | 87.8 | - |
| [70] | LUNA16 | 3DCNN | 64 × 64 × 64, 32 × 32 × 32, 16 × 16 × 16 | - | - | - | - | - | 0.948 |
| [71] | LUNA16 | Multi-path 3D CNN | 48 × 48 × 48 | 0.952/0.962 to 4, 8 FP/Scans | - | - | - | - | 0.881 |
| [72] | LUNA16 | Faster R-CNN with adaptive anchor box | 64 × 64 FP 512 × 512 DET | 93.8 | 97.6 | - | 95.7 | 95.7 | - |
| [73] | NLST (NLST, 2011), LHMC, Kaggle | 2D and 3D DNN | 32 × 32 × 32 | - | 86/94 | ||||
| [74] | LIDC-IDRI + Japan Chest CT Dataset | 3D unet | 64 × 96 × 96 | - | - | - | - | - | 0.947/0.833 |
| [75] | LUNA16 | 3D sphere representation-based center-points matching detection network (SCPM-Net) | 96 × 96 × 96 | 89.2/7 FP | - | - | - | - | - |
| [76] | LUNA16 | Atrous UNet+ | 3 × 64 × 64, 8 × 64 × 64, 16 × 64 × 64 | 92.8 | - | 77.2 | - | - | 0.93 |
| [77] | LUNA16 | 3D U-shaped residual network | 96 × 96 × 96 | 95 | - | - | - | - | 0.895 |
| [78] | LUNA16 | 3D CNN | 128 × 128 × 128 | 0.8808 | |||||
| [79] | LUNA16 | 3D ResNet18 dual path Faster R-CNN and a federated learning algorithm | 128 × 128 × 128 | 83.388 | - | 83.412 | 88.382 | 83.417 | - |
| [80] | LUNA16 | 3D ViT | 128 × 128 × 128 | 98.39 | - | - | - | 0.909 | |
| [81] | LUNA16 | 3D ViT | 64 × 64 × 64, 32 × 32 × 32, 16 × 16 × 16 | 97.81 | - | - | - | 0.911 | |
| [82] | LIDC-IDRI | 2D Ensemble Transformer with Attention Modules | 512 × 512 | 94.58 | 97.10 | 98.96 | 96.14 | - | |
| [83] | LUNA16 | 3Dl Multifaceted Attention Encoder–Decoder | 128 × 128 × 128 | 89.1/7 FPs | - | - | - | 0.891 | |
| [84] | ELCAP | 3D CNN-CapsNet | 32 × 32 × 8 | 92.31 | 98.08 | 95 | 95.19 | - | |
| [85] | LUNA16 | A multiscale self-calibrated network (DEPMSCNet)with a dual attention mechanism | 128 × 128 × 128 | 98.80 | - | - | - | 0.963 |
| Reference | Year | Dataset | Preprocessing | Data Augmentation | Deep Architecture |
|---|---|---|---|---|---|
| [93] | 2017 | LIDC-IDRI, private set | 3D-PA, 2D-PA, | n/a | Central Focused Convolutional Neural Networks (CF-CNN) |
| [94] | 2019 | LIDC-IDRI | 3D-PA | n/a | Cascaded Dual-Pathway Residual Network |
| [95] | 2019 | LIDC-IDRI | 2D-PA, PSEG, Re | n/a | SegNet, a deep, fully convolutional network |
| [62] | 2019 | LIDC-IDRI | 3D-PA | n/a | 3D DCNN |
| [96] | 2020 | LIDC-IDRI | 2D-PA | n/a | Deep residual deconvolutional network, TL |
| [97] | 2020 | LIDC-IDRI | 2D-PA, No | Deep Residual U-Net | |
| [98] | 2020 | LIDC-IDRI | 3D-PA | n/a | DB-ResNet, CF-CNN |
| [99] | 2020 | LIDC-IDRI | 2D-PA | Ro, Zoom, Padding | U-net |
| [100] | 2021 | LIDC-IDRI, LNDb, ILCID | TH, PSEG, Maximum intensity projection | Ro, Blur, No, Rand pixels to zero | 2D CNN |
| [101] | 2021 | LIDC-IDRI | 2D-PA, synthetic pseudo-color image | Intensive augmentations | U-Net |
| [102] | 2022 | LUNA16 | TH, No, 3D-Pa | Ro, Transpose, Affine Transform, Fl, Br, Contrast | V-net |
| [103] | 2021 | LIDC-IDRI, SHCH | TH, Contrast ench, PSEG, Lesion Localization with Region Growing | Fl, Ro, Cr, deformation | 2D–3D U-net |
| [104] | 2021 | LIDC-IDRI, LUNA16 | Cr, 2D-PA, Upscale | Ro, Fl, elastic transform | Faster R-CNN |
| [105] | 2021 | LIDC-IDRI | PSEG, 2D-PA | Ro, Fl, Sh, Zoom, Cr | U-net |
| [106] | 2021 | LIDC-IDRI | PSEG, TH, Re, 3D-PA | n/a | 3D res U-net |
| [107] | 2021 | LIDC-IDRI | 2D-PA, Re | n/a | VGG-SegNet |
| [108] | 2022 | hospital data | 3D-PA | Ro, Mirroring | 3D FCN |
| [109] | 2022 | LUNA16, ILND | TH, 3D-PA, Sampling for balance | patch-based augmentation | 3D GAN |
| [110] | 2022 | LIDC-IDRI | TH, No, 3D-PA | Ro, Sc, Fl | 3D Dual Attention Shadow Network (DAS-Net) |
| [111] | 2022 | LIDC-IDRI | 2D-PA | Ro, Tr, Fl | Transformer |
| [112] | 2023 | LIDC-IDRI | grayscale thresholding, No, Re | n/a | Dual-encoder-based CNN |
| [113] | 2023 | LIDC-IDRI, AHAMU-LC | window selection, No | Fl | RAD—U-net |
| [114] | 2023 | LIDC-IDRI, private set | 2D-PA | Cr, Sc, Br, Contrast, Sat, Random noise | SMR—U-net 2D |
| [115] | 2023 | LIDC-IDRI | 2D-PA | Ro, random luminance, random gamma rays, Gaussian noise, hue/sat | U-shaped hybrid transformer |
| [116] | 2023 | LIDC-IDRI, LUNA16 | TH, No, Re3D-PA | n/a | 3D U-net based |
| [117] | 2023 | LIDC-IDRI | mask generation | n/a | GUNet3++ |
| Reference | Dataset | Deep Architecture | Input Size | Input Shape | DSC (%) | IoU (%) | Sensitivity (%) |
| [93] | LIDC-IDRI, private set | Central Focused Convolutional Neural Networks (CF-CNN) | 572 × 572, 3 × 35 × 35 | 3D, 2D | 82.15 ± 10.76, LIDC 80.02 ± 11.09 Private set | - | - |
| [94] | LIDC-IDRI | Cascaded Dual-Pathway Residual Network | 65 × 65 × 3 | 2D, 3D (mask) | 81.58 ± 11.05 | - | - |
| [95] | LIDC-IDRI | SegNet, a deep, fully convolutional network | 128 × 128 | 2D | 93 ± 0.11 | - | - |
| [62] | LIDC-IDRI | 3D DCNN | n/a | 3D | 83.10 ± 8.85 | 71.85 ± 10.48 | - |
| [96] | LIDC-IDRI | Deep residual deconvolutional network, TL | 512 × 512 | 2D | 94.97 | 88.68 | - |
| [97] | LIDC-IDRI | Deep Residual U-Net | 128 × 128 | 3D | 87.5 ± 10.58 | - | - |
| [98] | LIDC-IDRI | DB-ResNet, CF-CNN | 3 × 35 × 35 | 3D | 82.74 ± 10.19 | - | - |
| [99] | LIDC-IDRI | U-net | 64 × 64 | 2D | - | 76.6 ± 12.3 | - |
| [100] | LIDC-IDRI, LNDb, ILCID | 2D CNN | 96 × 96 | 2D | 80 | - | |
| [101] | LIDC-IDRI | U-Net | 256 × 256 | 2D | 93.14 | - | 91.76 |
| [102] | LUNA16 | V-net | 96 × 96 × 16 | 3D | 95.01 | 83 | |
| [103] | LIDC-IDRI, SHCH | 2D–3D U-net | 2D–3D (3-slices) | 2D–3D | 83.16/81.97 | - | - |
| [104] | LIDC-IDRI, LUNA16 | Faster R-CNN | 224 × 224 | 2D | 89.79/90.35 | 82.34/83.21 | - |
| [105] | LIDC-IDRI | U-net | 32 × 32 | 2D | 86.23 | - | - |
| [106] | LIDC-IDRI | 3D res U-net | 48 × 192 × 192 | 3D | 80.5 | - | 80.5 |
| [107] | LIDC-IDRI | VGG-SegNet | 224 × 224 × 3 channels | 2D | 90.49 | 82.64 | - |
| [108] | hospital data | 3D FCN | 128 × 128 × 64 | 3D | 84.5 | 73.8 | |
| [109] | LUNA16, ILND | 3D GAN | 64 × 64 × 32 | 3D | 80.74/76.36 | - | 85.46/82.56 |
| [110] | LIDC-IDRI | 3D Dual Attention Shadow Network (DAS-Net) | 16 × 128 × 128 | 3D | 92.05 | - | 90.81 |
| [111] | LIDC-IDRI | Transformer | 64 × 64 | 2D | 89.86 | - | 90.50 |
| [112] | LIDC-IDRI | Dual-encoder-based CNN | 512 × 512 | 2D | 87.91 | - | 90.84 |
| [113] | LIDC-IDRI, AHAMU-LC | RAD—U-net | 512 × 512 | 2D | - | 87.76/88.13 | - |
| [114] | LIDC-IDRI, private set | SMR—U-net 2D | 128 × 128 | 2D | 91.87 | 86.88 | - |
| [115] | LIDC-IDRI | U-shaped hybrid transformer | 64 × 64, 96 × 96, 128 × 128 | 2D | 91.84 | 92.66 | |
| [116] | LIDC-IDRI, LUNA16 | 3D U-net based | 64 × 64 × 32 | 3D | 82.48 | 70.86 | 82.74 |
| [117] | LIDC-IDRI | GUNet3++ | n/a | 2D | 97.2 | - | 97.7 |
| Reference | Year | Dataset | Preprocessing | Data Augmentation | Deep Architecture |
|---|---|---|---|---|---|
| [125] | 2017 | LIDC-IDRI | Rs, 2D-PA, No | Ro, Sc (only in the test set) | ResNet |
| [126] | 2017 | LIDC-IDRI | 3D-PA, MV | Ro | 3D MV-CNN + SoftMax |
| [127] | 2017 | LIDC-IDRI | 2D-PA, Rs, Rz | Ro, Sh, Fl, Tr | ResNet-50, TL |
| [128] | 2018 | LIDC-IDRI | Rs, Rz, MV, Cr, 2D-Pa | Tr, Ro, Fl | MV-KBC |
| [129] | 2018 | LIDC-IDRI | 3D-PA, QIF extraction | Ro, Sc, shifted up to 30% | CNN + Random Forest |
| [130] | 2018 | LIDC-IDRI + Private set | Rs, 3D-PA, No | - | 3D DenseNet, TL |
| [131] | 2018 | LIDC-IDRI | Rs | - | CNN + PSO |
| [132] | 2018 | LUNA16 | Re, 3D-PA | Tr, Ro | 3D DCNN |
| [133] | 2018 | ELCAP | 2D-PA, Cr, Re | Ro, Cr, perturbation (brightness, saturation, hue, and contrast) | DAE |
| [134] | 2019 | LUNA16 | 2D-PA | Ro, Fl | Novel 2D CNN |
| [135] | 2019 | LIDC-IDRI | No, Cr, 2D-PA | Ro, Sc, Gaussian Blurring | Novel 2D CNN |
| [136] | 2019 | LIDC-IDRI | Re, 2D-PA | Ro, Fl | CNN, TL |
| [138] | 2020 | LIDC-IDRI | Rs, Th | yes, but no info | MAN (modified AlexNet), TL |
| [139] | 2020 | LIDC-IDRI | 2D-PA | Tr, Ro, Sc, GAN | CNN, TL |
| [58] | 2020 | LIDC-IDRI, private dataset (FAH-GMU) | Cr, 2D-PA | - | DTCNN, TL |
| [140] | 2020 | LIDC-IDRI, DeepLNDataset | No, Cr, 3D-PA | Tr, Fl | 3D CNN |
| [141] | 2020 | LIDC-IDRI, LUNGx Challenge database | Re, No, 2D-PA | Tr, Ro, Fl | 2D CNN, TL |
| [142] | 2020 | LIDC-IDRI | Cr, 3D-PA | Adjust sampling rate | MRC-DNN |
| [143] | 2020 | LIDC-IDRI | Re, No, Th, Cr, 3D-PA | Sampling different slices from the same nodule to achieve a better class balancing | CAE, TL |
| [144] | 2020 | LIDC-IDRI, LUNA16 | Re, No, Cr, 3D-PA, 2D-PA | Tr, Ro, Fl | Multi-Task CNN |
| [145] | 2020 | LUNA16 | Cr, 2D-PA | down sampling the negative samples, Ro | Fractalnet and CNN |
| [146] | 2020 | LIDC-IDRI | Re | Sc(zoom), FL, Ro | DCNN |
| [137] | 2020 | LIDC-IDRI | Full CT-PA | - | multiscale 3D-CNN, CapsNets |
| [147] | 2021 | NLST, DLCST | 3D-PA, 2D-PA (9 views) | n/a | 2D CNN 9 views, 3D CNN |
| [148] | 2021 | LUNA16/Kaggle DSB 2017 dataset | raw data to png | Ro, H-Fl, clip, blurry | Dense Convolutional Network (DenseNet) |
| [149] | 2021 | LIDC-IDRI/ELCAP | 2D-PA, 3D-PA | n/a | 2D MV-CNN 3D MV-CNN |
| [150] | 2021 | LIDC-IDRI | 3D-PA, zero-padding | n/a | Capsule networks (CapsNets) |
| [151] | 2021 | LIDC-IDRI | 3D-PA, padding, Cr | Fl | 3D NAS method, CBAM module, A-Softmax loss, and ensemble strategy to learn efficient |
| [152] | 2021 | LIDC-IDRI | 2D-PA | n/a | Deep Convolutional Generative Adversarial Network (DC-GAN)/FF-VGG19 |
| [153] | 2021 | LUNA16 | TH, 2D-PA, balancing samples | Ro, Fl | BCNN [VGG16, VGG19] combination with and without SVM |
| [154] | 2021 | LUNA16 | n/a | n/a | 3D CNN |
| [155] | 2021 | pet-ct private, LIDC-IDRI | n/a | Ro, Fl, Shift | 2d cnn |
| [156] | 2021 | LIDC-IDRI | 3D-PA, Re | Fl, Pad | 3D DPN _ attention mech |
| [157] | 2021 | LIDC-IDRI | 3D-PA | n/a | 3D CNN + biomarkers |
| [158] | 2021 | LIDC-IDRI | Re, 3D-PA, No | Ro | 3D attention |
| [159] | 2022 | LIDC-IDRI and LUNGx | Re, 3D-PA, TH, No | Ro | ProCAN |
| [160] | 2022 | LIDC-IDRI | 2D-PA | Ro, Tr | DCNN |
| [161] | 2022 | LIDC-IDRI | Re, 2D-PA | Ro, overlays on the axial, coronal, and sagittal slices | Transformers |
| [162] | 2022 | LIDC-IDRI | interpolation, TH | Fl, Ro | CNN-based MTL model that incorporates multiple attention-based learning modules |
| [163] | 2022 | LIDC-IDRI | Re, Rz, No | Ro | Transformers |
| [164] | 2022 | LUNA16 | 3D-PA, No | Fl, Gaussian noise | 3D ResNet + attention |
| [165] | 2023 | LIDC-IDRI/TC-LND Dataset/CQUCH-LND | No, 3D-PA, Sc | Ro, Fl | STLF-VA |
| [166] | 2023 | LIDC-IDRI | Re, No | n/a | Transformer |
| [167] | 2023 | LIDC-IDRI | Re, 2D-PA, Re | Fl, Brightness, contrast, Sc | F-LSTM-CNN |
| [168] | 2023 | private | TH,3D-PA | n/a | CAE |
| Reference | Dataset | Deep Architecture | Input Size | Sensitivity/Recall (%) | Specificity (%) | Precision (%) | AUC (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|---|
| [125] | LIDC-IDRI | ResNet | 64 × 64 | 91.07 | 88.64 | - | 94.59 | 89.90 |
| [126] | LIDC-IDRI | 3D MV-CNN + SoftMax | N [40, 50, 60] N × N × 6 slices | 95.60 | 93.94 | - | 99 | - |
| [127] | LIDC-IDRI | ResNet-50, TL | 200 × 200 | 91.43 | 94.09 | - | 97.78 | 93.40 |
| [128] | LIDC-IDRI | MV-KBC | 224 × 224 | 86.52 | 94 | 97.50 | 91.60 | |
| [129] | LIDC-IDRI | CNN + Random Forest | N × N × S N [47, 21, 31], S [5, 3] | 94.80 | 94.30 | - | 98.4 | 94.60 |
| [130] | LIDC-IDRI + Private set | 3D DenseNet, TL | N [50, 10] S [5, 10] N × N × S | 90.47 | 90.33 | - | 95.48 | 90.40 |
| [131] | LIDC-IDRI | CNN + PSO | 28 × 28 | 92.20 | 98.64 | - | 95.5 | 97.62 |
| [132] | LUNA16 | 3D DCNN | 64 × 64 × 64, 48 × 48 × 48 | 95.4/1 FP | - | - | 0.910 (FROC) | - |
| [133] | ELCAP | DAE | 180 × 180 | - | - | - | 0.939 (FROC) | - |
| [134] | LUNA16 | Novel 2D CNN | 64 × 64 | 96.0 | 97.3 | - | 98.2 | 97.2 |
| [135] | LIDC-IDRI | Novel 2D CNN | 32 × 32 | 92.67 | - | - | 95.14 | 92.57 |
| [136] | LIDC-IDRI | CNN, TL | 53 × 53 | 91 | - | - | 94 | 88 |
| [138] | LIDC-IDRI | MAN (modified AlexNet) + SVM, TL | 32 × 32 × 32 | - | 95.70 | 91.60 | ||
| [139] | LIDC-IDRI | CNN | 227 × 227 | 98.09 | 95.63 | - | 99.5 | 97.27 |
| [58] | LIDC-IDRI, private dataset (FAH-GMU) | DTCNN | 52 × 52 | 93.4 | 93 | - | 93.4 | 93.9 |
| [140] | LIDC-IDRI, DeepLNDataset | 3D CNN | 64 × 64 | 93.69/100 | 95.15/100 | - | 94.9 | 94.57/100 |
| [141] | LIDC-IDRI, LUNGx Challenge database | CNN, TL | N [32, 48, 64] N × N × N | 85.58 | 95.87 | - | 94 | 92.65 |
| [142] | LIDC-IDRI | MRC-DNN | 64 × 64 | 97.19 | - | - | 99.1 | 96.69 |
| [143] | LIDC-IDRI | CAE, TL | 32 × 32 × 32 | 81 | 95 | - | - | 90 |
| [144] | LIDC-IDRI, LUNA16 | Multi-Task CNN | 80 × 80 × 80 | 84.8 | - | 93.6 | - | |
| [145] | LUNA16 | Fractalnet and CNN | 64 × 64 | 87.74, 84.00 | 88.87, 96.80 | - | 0.955 (LIDC), 0.973 (LUNA) | - |
| [146] | LIDC-IDRI | DCNN | 50 × 50 | 97.52 | 86.76 | - | 98 | 94.06 |
| [147] | NLST, DLCST | 2D CNN 9 views, 3D CNN | 224 × 224 | 90.67 | 90.80 | - | - | 90.73 |
| [137] | LIDC-IDRI | multiscale 3D-CNN, CapsNets | 80 × 80 × 3 (+10 px for the next 2 scales) | 94.94 | 90 | - | 96.4 | 93.12 |
| [148] | LUNA16/Kaggle DSB 2017 dataset | Dense Convolutional Network (DenseNet) | 64 × 64, 64 × 64 × 64 | - | - | - | 93 | - |
| [149] | LIDC-IDRI/ELCAP | 2D MV-CNN 3D MV-CNN | 80 × 80, 64 × 64, 48 × 48, 32 × 32 and 16 × 16 | 98.2 | 99.45 | - | 98.83 | |
| [150] | LIDC | Capsule networks (CapsNets) | (20 × 20), (30 × 30) and (40 × 40) | 98 | 97 | - | 99 | 97 |
| [151] | LIDC-IDRI | 3D NAS method, CBAM module, A-Softmax loss, and ensemble strategy | 80 × 80 × 3 slices | 89.5 | 93.4 | - | 95.6 | 90.7 |
| [152] | LIDC-IDRI | Deep Convolutional Generative Adversarial Network (DC-GAN)/FF-VGG19 | 32 × 32 × 32 | 85.37 | 95.04 | - | - | 90.77 |
| [153] | LUNA16 | BCNN [VGG16, VGG19] combination with and without SVM | 32 × 32 | 89.3 | 94.8 | - | 92.1 | 92.1 |
| [154] | LUNA16 | 3D CNN | 50 × 50 | - | - | - | 95.9 | 91.99 |
| [155] | pet-ct private, LIDC-IDRI | 2D CNN | 64 × 64 × 240 | 94 | - | 87 | 97 | 97.17 |
| [156] | LIDC-IDRI | 3D DPN _ attention mech | 32 × 32 | 92.7 | 95.2 | - | 94 | 94 |
| [157] | LIDC-IDRI | 3D CNN + biomarkers | 32 × 32 × 32 | 91.3 (FP rate of 8.0%) | - | - | - | 91.9 |
| [158] | LIDC-IDRI | 3D attention | 32 × 32 × 16 slices | - | - | - | 86.74 | - |
| [159] | LIDC-IDRI and LUNGx | ProCAN | 32 × 32 × 32 | 92.36 | - | 92.59 | 96.17 | 92.81 |
| [160] | LIDC-IDRI | DCNN | 32 × 32 × 32 | - | - | - | 98.05 | 95.28 |
| [161] | LIDC-IDRI | Transformers | 52 × 52 | 97.1 | 97.2 | 99.56 | 97.8 | |
| [162] | LIDC-IDRI | CNN-based MTL model that incorporates multiple attention-based learning modules | 32 × 32 | - | - | - | 96.28 | 92.92 |
| [163] | LIDC-IDRI | Transformers | 64 × 64 | 96.2 | 82.9 | 97.8 | 95.9 | 94.7 |
| [164] | LUNA16 | 3D ResNet + attention | 32 × 32 | 94.4 | 95.9 | 98.5 | 96.1 | |
| [165] | LIDC-IDRI/TC-LND Dataset/CQUCH-LND | STLF-VA | 32 × 32 × 32 | 89.10 | 93.39 | 91.59 | 91.25 | 91.25 |
| [166] | LIDC-IDRI | Transformer | 64 × 64 × 32 | 91.62 | 93.08 | 92.99 | 97.17 | 92.36 |
| [167] | LIDC-IDRI | F-LSTM-CNN | 80 × 80 × 60 | 87.69 | 95.38 | - | 97.40 | 92.82 |
| [168] | private | CAE | 224 × 224 × 3 | 100 | 93.7 | - | 99.5 | 95.5 |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- The American Cancer Society Medical and Editorial Content Team. Key Statistics for Lung Cancer. Available online: https://www.cancer.org/cancer/types/lung-cancer/about/key-statistics.html (accessed on 21 September 2023).
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020. [Google Scholar]
- World Health Organization. Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 21 September 2023).
- The American Cancer Society Medical and Editorial Content Team. Lung Cancer Early Detection, Diagnosis, and Staging. Available online: https://www.cancer.org/content/dam/CRC/PDF/Public/8705.00.pdf (accessed on 21 September 2023).
- Ellis, P.M.; Vandermeer, R. Delays in the diagnosis of lung cancer. J. Thorac. Dis. 2011, 3, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Johns Hopkins Medicine. Lung Biopsy. Available online: https://www.hopkinsmedicine.org/health/treatment-tests-and-therapies/lung-biopsy (accessed on 21 September 2023).
- Mahmoud, N.; Vashisht, R.; Sanghavi, D.K.; Kalanjeri, S. Bronchoscopy. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar] [PubMed]
- Sigmon, D.F.; Fatima, S. Fine Needle Aspiration. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar] [PubMed]
- Mehrotra, M.; D’Cruz, J.R.; Bishop, M.A.; Arthur, M.E. Video-Assisted Thoracoscopy. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar] [PubMed]
- McNally, P.A.; Sharma, S.; Arthur, M.E. Mediastinoscopy. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar] [PubMed]
- Kim, J.; Kim, K.H. Role of chest radiographs in early lung cancer detection. Transl. Lung Cancer Res. 2020, 9, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Winkler, M.H.; Touw, H.R.; van de Ven, P.M.; Twisk, J.; Tuinman, P.R. Diagnostic Accuracy of Chest Radiograph, and When Concomitantly Studied Lung Ultrasound, in Critically Ill Patients with Respiratory Symptoms: A Systematic Review and Meta-Analysis. Crit. Care Med. 2018, 46, e707–e714. [Google Scholar] [CrossRef]
- Tylski, E.; Goyal, M. Low Dose CT for Lung Cancer Screening: The Background, the Guidelines, and a Tailored Approach to Patient Care. Mo. Med. 2019, 116, 414–419. [Google Scholar]
- Vonder, M.; Dorrius, M.D.; Vliegenthart, R. Latest CT technologies in lung cancer screening: Protocols and radiation dose reduction. Transl. Lung Cancer Res. 2021, 10, 1154–1164. [Google Scholar] [CrossRef]
- Rubin, K.H.; Haastrup, P.F.; Nicolaisen, A.; Möller, S.; Wehberg, S.; Rasmussen, S.; Balasubramaniam, K.; Søndergaard, J.; Jarbøl, D.E. Developing and Validating a Lung Cancer Risk Prediction Model: A Nationwide Population-Based Study. Cancers 2023, 15, 487. [Google Scholar] [CrossRef]
- Yu, K.-H.; Lee, T.-L.M.; Yen, M.-H.; Kou, S.C.; Rosen, B.; Chiang, J.-H.; Kohane, I.S. Reproducible Machine Learning Methods for Lung Cancer Detection Using Computed Tomography Images: Algorithm Development and Validation. J. Med. Internet Res. 2020, 22, e16709. [Google Scholar] [CrossRef]
- Cellina, M.; Cacioppa, L.M.; Cè, M.; Chiarpenello, V.; Costa, M.; Vincenzo, Z.; Pais, D.; Bausano, M.V.; Rossini, N.; Bruno, A.; et al. Artificial Intelligence in Lung Cancer Screening: The Future Is Now. Cancers 2023, 15, 4344. [Google Scholar] [CrossRef]
- Zhang, J.; Xia, Y.; Cui, H.; Zhang, Y. Pulmonary nodule detection in medical images: A survey. Biomed. Signal Process. Control 2018, 43, 138–147. [Google Scholar] [CrossRef]
- Gu, Y.; Chi, J.; Liu, J.; Yang, L.; Zhang, B.; Yu, D.; Zhao, Y.; Lu, X. A survey of computer-aided diagnosis of lung nodules from CT scans using deep learning. Comput. Biol. Med. 2021, 137, 104806. [Google Scholar] [CrossRef] [PubMed]
- Thanoon, M.A.; Zulkifley, M.A.; Zainuri, M.A.A.M.; Abdani, S.R. A Review of Deep Learning Techniques for Lung Cancer Screening and Diagnosis Based on CT Images. Diagnostics 2023, 13, 2617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Jiang, S.; Yang, Z.; Gong, L.; Ma, X.; Zhou, Z.; Bao, C.; Liu, Q. Automatic nodule detection for lung cancer in CT images: A review. Comput. Biol. Med. 2018, 103, 287–300. [Google Scholar] [CrossRef]
- Liu, B.; Chi, W.; Li, X.; Li, P.; Liang, W.; Liu, H.; Wang, W.; He, J. Evolving the pulmonary nodules diagnosis from classical approaches to deep learning-aided decision support: Three decades’ development course and future prospect. J. Cancer Res. Clin. Oncol. 2020, 146, 153–185. [Google Scholar] [CrossRef]
- Halder, A.; Dey, D.; Sadhu, A.K. Lung Nodule Detection from Feature Engineering to Deep Learning in Thoracic CT Images: A Comprehensive Review. J. Digit. Imaging 2020, 33, 655–677. [Google Scholar] [CrossRef]
- Gu, D.; Liu, G.; Xue, Z. On the performance of lung nodule detection, segmentation and classification. Comput. Med. Imaging Graph. 2021, 89, 101886. [Google Scholar] [CrossRef]
- Li, R.; Xiao, C.; Huang, Y.; Hassan, H.; Huang, B. Deep Learning Applications in Computed Tomography Images for Pulmonary Nodule Detection and Diagnosis: A Review. Diagnostics 2022, 12, 298. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.; Pereira, T.; Neves, I.; Morgado, J.; Freitas, C.; Malafaia, M.; Sousa, J.; Fonseca, J.; Negrão, E.; de Lima, B.F.; et al. Towards Machine Learning-Aided Lung Cancer Clinical Routines: Approaches and Open Challenges. J. Pers. Med. 2022, 12, 480. [Google Scholar] [CrossRef]
- Harzing, A.W. Publish or Perish. 2007. Available online: https://harzing.com/resources/publish-or-perish (accessed on 11 October 2023).
- Armato, S.G.; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef]
- Setio, A.A.A.; Traverso, A.; de Bel, T.; Berens, M.S.; Bogaard, C.v.D.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef]
- ELCAP and VIA Research Groups. ELCAP Public Lung Image Database. Available online: https://www.via.cornell.edu/databases/lungdb.html (accessed on 18 October 2023).
- Alibaba Tianchi Competition Organizers. Tianchi Medical AI Competition Dataset. 2017. Available online: https://tianchi.aliyun.com/competition/entrance/231601/information (accessed on 18 October 2023).
- Armato, S.G.; Hadjiisk, L.; Tourassi, G.; Drukker, K.; Giger, M.; Li, F. SPIE-AAPM-NCI Lung Nodule Classification Challenge Dataset (SPIE-AAPM Lung CT Challenge). Available online: https://wiki.cancerimagingarchive.net/display/Public/LUNGx+SPIE-AAPM-NCI+Lung+Nodule+Classification+Challenge (accessed on 18 October 2023).
- Wikipedia Contributors. Hounsfield Scale. Available online: https://en.wikipedia.org/w/index.php?title=Hounsfield_scale&oldid=1167604704 (accessed on 18 October 2023).
- Gruetzemacher, R.; Gupta, A.; Paradice, D. 3D deep learning for detecting pulmonary nodules in CT scans. J. Am. Med. Inform. Assoc. 2018, 25, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.; Lee, D.; Kang, S.; Lee, S. Performance evaluation of mask R-CNN for lung segmentation using computed tomographic images. J. Korean Phys. Soc. 2022, 81, 346–353. [Google Scholar] [CrossRef]
- Moragheb, M.A.; Badie, A.; Noshad, A. An Effective Approach for Automated Lung Node Detection using CT Scans. J. Biomed. Phys. Eng. 2022, 12, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.-M.; Fan, Y. Zero-Shot and Few-Shot Learning for Lung Cancer Multi-Label Classification using Vision Transformer. arXiv 2022, arXiv:2205.15290. [Google Scholar]
- Zhang, H.; Gu, X.; Zhang, M.; Yu, W.; Chen, L.; Wang, Z.; Yao, F.; Gu, Y.; Yang, G.Z. Re-thinking and Re-labeling LIDC-IDRI for Robust Pulmonary Cancer Prediction. In Workshop on Medical Image Learning with Limited and Noisy Data; Springer: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. In Proceedings of the Advances in Neural Information Processing Systems 27, Montreal, QC, Canada, 8–13 December 2014. [Google Scholar]
- O’Shea, K.; Nash, R. An Introduction to Convolutional Neural Networks. arXiv 2015, arXiv:1511.08458. [Google Scholar]
- Karampidis, K.; Kavallieratou, E.; Papadourakis, G. A review of image steganalysis techniques for digital forensics. J. Inf. Secur. Appl. 2018, 40, 217–235. [Google Scholar] [CrossRef]
- Karampidis, K.; Kavallieratou, E.; Papadourakis, G. A Dilated Convolutional Neural Network as Feature Selector for Spatial Image Steganalysis—A Hybrid Classification Scheme. Pattern Recognit. Image Anal. 2020, 30, 342–358. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Li, D.; Wang, H.; Huang, X.; Song, L. DSDCLA: Driving style detection via hybrid CNN-LSTM with multi-level attention fusion. Appl. Intell. 2023, 53, 19237–19254. [Google Scholar] [CrossRef]
- Varshitha, K.S.; Kumari, C.G.; Hasvitha, M.; Fiza, S.; Amarendra, K.; Rachapudi, V. Natural Language Processing using Convolutional Neural Network. In Proceedings of the 2023 7th International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 23–25 February 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 362–367. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Chen, S.; Guo, W. Auto-Encoders in Deep Learning—A Review with New Perspectives. Mathematics 2023, 11, 1777. [Google Scholar] [CrossRef]
- Girin, L.; Leglaive, S.; Bie, X.; Diard, J.; Hueber, T.; Alameda-Pineda, X. Dynamical Variational Autoencoders: A Comprehensive Review. Found. Trends® Mach. Learn. 2021, 15, 1–175. [Google Scholar] [CrossRef]
- Pawan, S.J.; Rajan, J. Capsule networks for image classification: A review. Neurocomputing 2022, 509, 102–120. [Google Scholar] [CrossRef]
- Islam, S.; Elmekki, H.; Elsebai, A.; Bentahar, J.; Drawel, N.; Rjoub, G.; Pedrycz, W. A comprehensive survey on applications of transformers for deep learning tasks. Expert. Syst. Appl. 2024, 241, 122666. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Yenduri, G.; Ramalingam, M.; Selvi, G.C.; Supriya, Y.; Srivastava, G.; Maddikunta, P.K.R.; Raj, G.D.; Jhaveri, R.H.; Prabadevi, B.; Wang, W.; et al. GPT (Generative Pre-Trained Transformer)—A Comprehensive Review on Enabling Technologies, Potential Applications, Emerging Challenges, and Future Directions. IEEE Access 2024, 12, 54608–54649. [Google Scholar] [CrossRef]
- Park, N.; Kim, S. How Do Vision Transformers Work? arXiv 2022, arXiv:2202.06709. [Google Scholar]
- Ding, J.; Li, A.; Hu, Z.; Wang, L. Accurate Pulmonary Nodule Detection in Computed Tomography Images Using Deep Convolutional Neural Networks. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2017, Quebec City, QC, Canada, 11–13 September 2017; pp. 559–567. [Google Scholar] [CrossRef]
- Eun, H.; Kim, D.; Jung, C.; Kim, C. Single-view 2D CNNs with fully automatic non-nodule categorization for false positive reduction in pulmonary nodule detection. Comput. Methods Programs Biomed. 2018, 165, 215–224. [Google Scholar] [CrossRef]
- Gu, Y.; Lu, X.; Yang, L.; Zhang, B.; Yu, D.; Zhao, Y.; Gao, L.; Wu, L.; Zhou, T. Automatic lung nodule detection using a 3D deep convolutional neural network combined with a multi-scale prediction strategy in chest CTs. Comput. Biol. Med. 2018, 103, 220–231. [Google Scholar] [CrossRef]
- Zhang, J.; Xia, Y.; Zeng, H.; Zhang, Y. NODULe: Combining constrained multi-scale LoG filters with densely dilated 3D deep convolutional neural network for pulmonary nodule detection. Neurocomputing 2018, 317, 159–167. [Google Scholar] [CrossRef]
- Tan, J.; Huo, Y.; Liang, Z.; Li, L. Expert knowledge-infused deep learning for automatic lung nodule detection. J. Xray Sci. Technol. 2019, 27, 17–35. [Google Scholar] [CrossRef]
- Zheng, S.; Guo, J.; Cui, X.; Veldhuis, R.N.J.; Oudkerk, M.; van Ooijen, P.M.A. Automatic Pulmonary Nodule Detection in CT Scans Using Convolutional Neural Networks Based on Maximum Intensity Projection. IEEE Trans Med. Imaging 2020, 39, 797–805. [Google Scholar] [CrossRef]
- Wang, Q.; Shen, F.; Shen, L.; Huang, J.; Sheng, W. Lung Nodule Detection in CT Images Using a Raw Patch-Based Convolutional Neural Network. J. Digit. Imaging 2019, 32, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Nasrullah, N.; Sang, J.; Alam, M.S.; Mateen, M.; Cai, B.; Hu, H. Automated Lung Nodule Detection and Classification Using Deep Learning Combined with Multiple Strategies. Sensors 2019, 19, 3722. [Google Scholar] [CrossRef]
- Tang, H.; Zhang, C.; Xie, X. NoduleNet: Decoupled False Positive Reduction for Pulmonary Nodule Detection and Segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019, Shenzhen, China, 13–17 October 2019; pp. 266–274. [Google Scholar] [CrossRef]
- Tang, S.; Yang, M.; Bai, J. Detection of pulmonary nodules based on a multiscale feature 3D U-Net convolutional neural network of transfer learning. PLoS ONE 2020, 15, e0235672. [Google Scholar] [CrossRef]
- Zuo, W.; Zhou, F.; Li, Z.; Wang, L. Multi-Resolution CNN and Knowledge Transfer for Candidate Classification in Lung Nodule Detection. IEEE Access 2019, 7, 32510–32521. [Google Scholar] [CrossRef]
- Zheng, S.; Cornelissen, L.J.; Cui, X.; Jing, X.; Veldhuis, R.N.J.; Oudkerk, M.; van Ooijen, P.M.A. Deep convolutional neural networks for multiplanar lung nodule detection: Improvement in small nodule identification. Med. Phys. 2021, 48, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Shrey, S.B.; Hakim, L.; Kavitha, M.; Kim, H.W.; Kurita, T. Transfer Learning by Cascaded Network to Identify and Classify Lung Nodules for Cancer Detection. In Proceedings of the Frontiers of Computer Vision—IW-FCV 2020, Ibusuki, Japan, 20–22 February 2020; pp. 262–273. [Google Scholar] [CrossRef]
- Tong, C.; Liang, B.; Su, Q.; Yu, M.; Hu, J.; Bashir, A.K.; Zheng, Z. Pulmonary Nodule Classification Based on Heterogeneous Features Learning. IEEE J. Sel. Areas Commun. 2021, 39, 574–581. [Google Scholar] [CrossRef]
- Peng, H.; Sun, H.; Guo, Y. 3D multi-scale deep convolutional neural networks for pulmonary nodule detection. PLoS ONE 2021, 16, e0244406. [Google Scholar] [CrossRef]
- Sori, W.J.; Feng, J.; Godana, A.W.; Liu, S.; Gelmecha, D.J. DFD-Net: Lung cancer detection from denoised CT scan image using deep learning. Front. Comput. Sci. 2021, 15, 152701. [Google Scholar] [CrossRef]
- Mittapalli, P.S.; Thanikaiselvan, V. Multiscale CNN with compound fusions for false positive reduction in lung nodule detection. Artif. Intell. Med. 2021, 113, 102017. [Google Scholar] [CrossRef]
- Yuan, H.; Fan, Z.; Wu, Y.; Cheng, J. An efficient multi-path 3D convolutional neural network for false-positive reduction of pulmonary nodule detection. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 2269–2277. [Google Scholar] [CrossRef]
- Nguyen, C.C.; Tran, G.S.; Nguyen, V.T.; Burie, J.-C.; Nghiem, T.P. Pulmonary Nodule Detection Based on Faster R-CNN With Adaptive Anchor Box. IEEE Access 2021, 9, 154740–154751. [Google Scholar] [CrossRef]
- Trajanovski, S.; Mavroeidis, D.; Swisher, C.L.; Gebre, B.G.; Veeling, B.S.; Wiemker, R.; Klinder, T.; Tahmasebi, A.; Regis, S.M.; Wald, C.; et al. Towards radiologist-level cancer risk assessment in CT lung screening using deep learning. Comput. Med. Imaging Graph. 2021, 90, 101883. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Otsuka, Y.; Nomura, Y.; Kumamaru, K.K.; Kuwatsuru, R.; Aoki, S. Development and Validation of a Modified Three-Dimensional U-Net Deep-Learning Model for Automated Detection of Lung Nodules on Chest CT Images From the Lung Image Database Consortium and Japanese Datasets. Acad. Radiol. 2022, 29, S11–S17. [Google Scholar] [CrossRef]
- Luo, X.; Song, T.; Wang, G.; Chen, J.; Chen, Y.; Li, K.; Metaxas, D.N.; Zhang, S. SCPM-Net: An anchor-free 3D lung nodule detection network using sphere representation and center points matching. Med. Image Anal. 2022, 75, 102287. [Google Scholar] [CrossRef]
- Agnes, S.A.; Anitha, J.; Solomon, A.A. Two-stage lung nodule detection framework using enhanced UNet and convolutional LSTM networks in CT images. Comput. Biol. Med. 2022, 149, 106059. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, X.; Shi, Y.; Ren, S.; Wang, W. Channel-Wise Attention Mechanism in the 3D Convolutional Network for Lung Nodule Detection. Electronics 2022, 11, 1600. [Google Scholar] [CrossRef]
- Jian, M.; Zhang, L.; Jin, H.; Li, X. 3DAGNet: 3D Deep Attention and Global Search Network for Pulmonary Nodule Detection. Electronics 2023, 12, 2333. [Google Scholar] [CrossRef]
- Liu, L.; Fan, K.; Yang, M. Federated learning: A deep learning model based on resnet18 dual path for lung nodule detection. Multimed. Tools Appl. 2023, 82, 17437–17450. [Google Scholar] [CrossRef]
- Mkindu, H.; Wu, L.; Zhao, Y. Lung nodule detection in chest CT images based on vision transformer network with Bayesian optimization. Biomed. Signal Process. Control 2023, 85, 104866. [Google Scholar] [CrossRef]
- Mkindu, H.; Wu, L.; Zhao, Y. 3D multi-scale vision transformer for lung nodule detection in chest CT images. Signal Image Video Process 2023, 17, 2473–2480. [Google Scholar] [CrossRef]
- Zhang, J.; Xia, K.; Huang, Z.; Wang, S.; Akindele, R.G. ETAM: Ensemble transformer with attention modules for detection of small objects. Expert. Syst. Appl. 2023, 224, 119997. [Google Scholar] [CrossRef]
- Cao, K.; Tao, H.; Wang, Z. Three-Dimensional Multifaceted Attention Encoder–Decoder Networks for Pulmonary Nodule Detection. Appl. Sci. 2023, 13, 10822. [Google Scholar] [CrossRef]
- Song, L.; Zhang, M.; Wu, L. Detection of low-dose computed tomography pulmonary nodules based on 3D CNN-CapsNet. Electron. Lett. 2023, 59, e12952. [Google Scholar] [CrossRef]
- Zhu, Y.; Xu, L.; Liu, Y.; Guo, P.; Zhang, J. Multiscale self-calibrated pulmonary nodule detection network fusing dual attention mechanism. Phys. Med. Biol. 2023, 68, 165007. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans. Med. Imaging 2017, 35, 1299–1312. [Google Scholar] [CrossRef]
- Wu, H.; Flierl, M. Vector Quantization-Based Regularization for Autoencoders. Proc. AAAI Conf. Artif. Intell. 2020, 34, 6380–6387. [Google Scholar] [CrossRef]
- Bechar, A.; Elmir, Y.; Medjoudj, R.; Himeur, Y.; Amira, A. Harnessing Transformers: A Leap Forward in Lung Cancer Image Detection. In Proceedings of the 2023 6th International Conference on Signal Processing and Information Security (ICSPIS), Dubai, United Arab Emirates, 8–9 November 2023. [Google Scholar]
- Shao, H.; Lu, J.; Wang, M.; Wang, Z. An Efficient Training Accelerator for Transformers With Hardware-Algorithm Co-Optimization. IEEE Trans. Very Large Scale Integr. VLSI Syst. 2023, 31, 1788–1801. [Google Scholar] [CrossRef]
- Choudhary, S.; Saurav, S.; Saini, R.; Singh, S. Capsule networks for computer vision applications: A comprehensive review. Appl. Intell. 2023, 53, 21799–21826. [Google Scholar] [CrossRef]
- Marchisio, A.; De Marco, A.; Colucci, A.; Martina, M.; Shafique, M. RobCaps: Evaluating the Robustness of Capsule Networks against Affine Transformations and Adversarial Attacks. In Proceedings of the 2023 International Joint Conference on Neural Networks (IJCNN), Gold Coast, Australia, 18–23 June 2023. [Google Scholar]
- Renzulli, R.; Grangetto, M. Towards Efficient Capsule Networks. In Proceedings of the 2022 IEEE International Conference on Image Processing (ICIP), Bordeaux, France, 16–19 October 2022. [Google Scholar]
- Wang, S.; Zhou, M.; Liu, Z.; Liu, Z.; Gu, D.; Zang, Y.; Dong, D.; Gevaert, O.; Tian, J. Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation. Med. Image Anal. 2017, 40, 172–183. [Google Scholar] [CrossRef]
- Liu, H.; Cao, H.; Song, E.; Ma, G.; Xu, X.; Jin, R.; Jin, Y.; Hung, C.-C. A cascaded dual-pathway residual network for lung nodule segmentation in CT images. Phys. Med. 2019, 63, 112–121. [Google Scholar] [CrossRef]
- Roy, R.; Chakraborti, T.; Chowdhury, A.S. A deep learning-shape driven level set synergism for pulmonary nodule segmentation. Pattern Recognit. Lett. 2019, 123, 31–38. [Google Scholar] [CrossRef]
- Singadkar, G.; Mahajan, A.; Thakur, M.; Talbar, S. Deep Deconvolutional Residual Network Based Automatic Lung Nodule Segmentation. J. Digit. Imaging 2020, 33, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Usman, M.; Lee, B.-D.; Byon, S.-S.; Kim, S.-H.; Lee, B.; Shin, Y.-G. Volumetric lung nodule segmentation using adaptive ROI with multi-view residual learning. Sci. Rep. 2020, 10, 12839. [Google Scholar] [CrossRef]
- Cao, H.; Liu, H.; Song, E.; Hung, C.-C.; Ma, G.; Xu, X.; Jin, R.; Lu, J. Dual-branch residual network for lung nodule segmentation. Appl. Soft Comput. 2020, 86, 105934. [Google Scholar] [CrossRef]
- Pezzano, G.; Ripoll, V.R.; Radeva, P. CoLe-CNN: Context-learning convolutional neural network with adaptive loss function for lung nodule segmentation. Comput. Methods Programs Biomed. 2021, 198, 105792. [Google Scholar] [CrossRef]
- Dutande, P.; Baid, U.; Talbar, S. LNCDS: A 2D-3D cascaded CNN approach for lung nodule classification, detection and segmentation. Biomed. Signal Process. Control 2021, 67, 102527. [Google Scholar] [CrossRef]
- Hesamian, M.H.; Jia, W.; He, X.; Wang, Q.; Kennedy, P.J. Synthetic CT images for semi-sequential detection and segmentation of lung nodules. Appl. Intell. 2021, 51, 1616–1628. [Google Scholar] [CrossRef]
- Dodia, S.; Basava, A.; Anand, M.P. A novel receptive field-regularized V-net and nodule classification network for lung nodule detection. Int. J. Imaging Syst. Technol. 2022, 32, 88–101. [Google Scholar] [CrossRef]
- Wu, Z.; Zhou, Q.; Wang, F. Coarse-to-Fine Lung Nodule Segmentation in CT Images with Image Enhancement and Dual-Branch Network. IEEE Access 2021, 9, 7255–7262. [Google Scholar] [CrossRef]
- Banu, S.F.; Sarker, M.M.K.; Abdel-Nasser, M.; Puig, D.; Raswan, H.A. AWEU-Net: An Attention-Aware Weight Excitation U-Net for Lung Nodule Segmentation. Appl. Sci. 2021, 11, 10132. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Zhang, B.; Dong, J.; Zhao, S.; Li, S. Accurate segmentation for different types of lung nodules on CT images using improved U-Net convolutional network. Medicine 2021, 100, e27491. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Li, J.; Zhang, L.; Cao, Y.; Yu, X.; Sun, J. Design of lung nodules segmentation and recognition algorithm based on deep learning. BMC Bioinform. 2021, 22, 314. [Google Scholar] [CrossRef]
- Khan, M.A.; Rajinikanth, V.; Satapathy, S.C.; Taniar, D.; Mohanty, J.R.; Tariq, U.; Damaševičius, R. VGG19 Network Assisted Joint Segmentation and Classification of Lung Nodules in CT Images. Diagnostics 2021, 11, 2208. [Google Scholar] [CrossRef]
- Kido, S.; Kidera, S.; Hirano, Y.; Mabu, S.; Kamiya, T.; Tanaka, N.; Suzuki, Y.; Yanagawa, M.; Tomiyama, N. Segmentation of Lung Nodules on CT Images Using a Nested Three-Dimensional Fully Connected Convolutional Network. Front. Artif. Intell. 2022, 5, 782225. [Google Scholar] [CrossRef]
- Tyagi, S.; Talbar, S.N. CSE-GAN: A 3D conditional generative adversarial network with concurrent squeeze-and-excitation blocks for lung nodule segmentation. Comput. Biol. Med. 2022, 147, 105781. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Zhang, J.; Xiao, N.; Qiang, Y.; Li, K.; Zhao, J.; Meng, L.; Song, P. DAS-Net: A lung nodule segmentation method based on adaptive dual-branch attention and shadow mapping. Appl. Intell. 2022, 52, 15617–15631. [Google Scholar] [CrossRef]
- Wang, S.; Jiang, A.; Li, X.; Qiu, Y.; Li, M.; Li, F. DPBET: A dual-path lung nodules segmentation model based on boundary enhancement and hybrid transformer. Comput. Biol. Med. 2022, 151, 106330. [Google Scholar] [CrossRef] [PubMed]
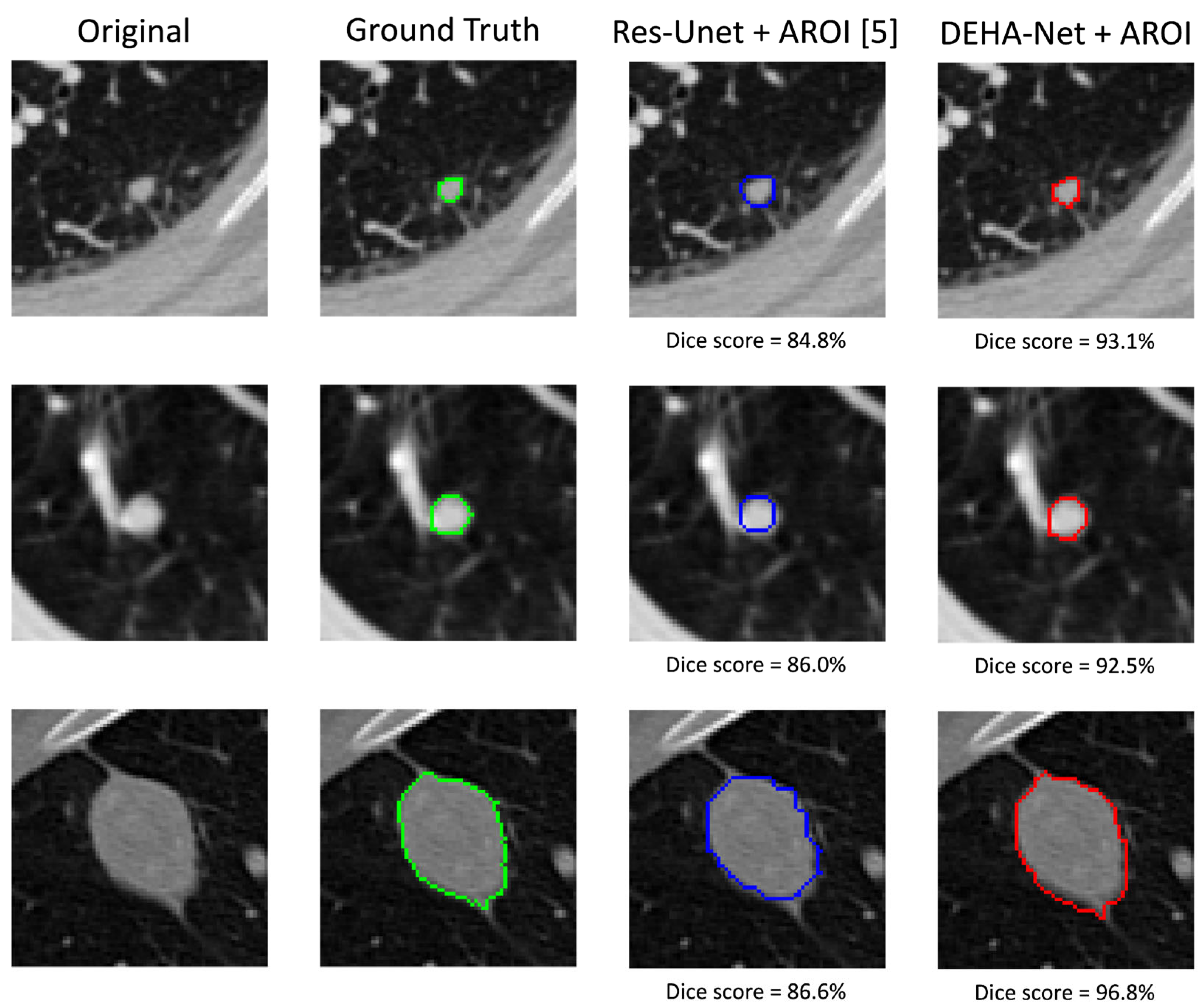
- Usman, M.; Shin, Y.-G. DEHA-Net: A Dual-Encoder-Based Hard Attention Network with an Adaptive ROI Mechanism for Lung Nodule Segmentation. Sensors 2023, 23, 1989. [Google Scholar] [CrossRef]
- Wu, Z.; Li, X.; Zuo, J. RAD-UNet: Research on an improved lung nodule semantic segmentation algorithm based on deep learning. Front. Oncol. 2023, 13, 1084096. [Google Scholar] [CrossRef]
- Hou, J.; Yan, C.; Li, R.; Huang, Q.; Fan, X.; Lin, F. Lung Nodule Segmentation Algorithm With SMR-UNet. IEEE Access 2023, 11, 34319–34331. [Google Scholar] [CrossRef]
- Li, X.; Jiang, A.; Qiu, Y.; Li, M.; Zhang, X.; Yan, S. TPFR-Net: U-shaped model for lung nodule segmentation based on transformer pooling and dual-attention feature reorganization. Med. Biol. Eng. Comput. 2023, 61, 1929–1946. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Li, B.; Liao, R.; Mo, H.; Tian, L. A dual-task region-boundary aware neural network for accurate pulmonary nodule segmentation. J. Vis. Commun. Image Represent. 2023, 96, 103909. [Google Scholar] [CrossRef]
- Ardimento, P.; Aversano, L.; Bernardi, M.L.; Cimitile, M.; Iammarino, M.; Verdone, C. Evo-GUNet3++: Using evolutionary algorithms to train UNet-based architectures for efficient 3D lung cancer detection. Appl. Soft Comput. 2023, 144, 110465. [Google Scholar] [CrossRef]
- Crespi, L.; Loiacono, D.; Sartori, P. Are 3D better than 2D Convolutional Neural Networks for Medical Imaging Semantic Segmentation? In Proceedings of the 2022 International Joint Conference on Neural Networks (IJCNN), Padua, Italy, 18–23 July 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–8. [Google Scholar] [CrossRef]
- Kolmogorov–Smirnov Test. In Encyclopedia of Research Design; SAGE Publications, Inc.: Thousand Oaks, CA, USA, 2010. [CrossRef]
- Gao, W.; McDonnell, M.D. Analysis of Gradient Degradation and Feature Map Quality in Deep All-Convolutional Neural Networks Compared to Deep Residual Networks. In Proceedings of the Neural Information Processing: 24th International Conference, ICONIP 2017, Guangzhou, China, 14–18 November 2017; pp. 612–621. [Google Scholar] [CrossRef]
- AbdulRazek, M.; Khoriba, G.; Belal, M. GAN-GA: A Generative Model based on Genetic Algorithm for Medical Image Generation. In Proceedings of the 27th Conference on Medical Image Understanding and Analysis, Aberdeen, UK, 19–21 July 2023. [Google Scholar] [CrossRef]
- Karampidis, K.; Linardos, E.; Kavallieratou, E. StegoPass—Utilization of steganography to produce a novel unbreakable biometric based password authentication scheme. In Proceedings of the 14th International Conference on Computational Intelligence in Security for Information Systems and 12th International Conference on European Transnational Educational (CISIS 2021 and ICEUTE 2021), Bilbao, Spain, 21 September 2021; Springer International Publishing: Berlin/Heidelberg, Germany, 2021; pp. 146–155. [Google Scholar] [CrossRef]
- Crespi, L.; Camnasio, S.; Dei, D.; Lambri, N.; Mancosu, P.; Scorsetti, M.; Loiacono, D. Leveraging Multimodal CycleGAN for the Generation of Anatomically Accurate Synthetic CT Scans from MRIs. arXiv 2024, arXiv:2407.10888. [Google Scholar]
- Saad, M.M.; O’Reilly, R.; Rehmani, M.H. A survey on training challenges in generative adversarial networks for biomedical image analysis. Artif. Intell. Rev. 2024, 57, 19. [Google Scholar] [CrossRef]
- Nibali, A.; He, Z.; Wollersheim, D. Pulmonary nodule classification with deep residual networks. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.; Liu, K.; Hou, B.; Zhang, N. 3D multi-view convolutional neural networks for lung nodule classification. PLoS ONE 2017, 12, e0188290. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Xia, Y.; Zhang, J.; Feng, D.D.; Fulham, M.; Cai, W. Transferable Multi-model Ensemble for Benign-Malignant Lung Nodule Classification on Chest CT. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2017, Quebec City, QC, Canada, 11–13 September 2017; pp. 656–664. [Google Scholar] [CrossRef]
- Xie, Y.; Xia, Y.; Zhang, J.; Song, Y.; Feng, D.; Fulham, M.; Cai, W. Knowledge-based Collaborative Deep Learning for Benign-Malignant Lung Nodule Classification on Chest CT. IEEE Trans. Med. Imaging 2019, 38, 991–1004. [Google Scholar] [CrossRef]
- Causey, J.L.; Zhang, J.; Ma, S.; Jiang, B.; Qualls, J.A.; Politte, D.G.; Prior, F.; Zhang, S.; Huang, X. Highly accurate model for prediction of lung nodule malignancy with CT scans. Sci. Rep. 2018, 8, 9286. [Google Scholar] [CrossRef]
- Dey, R.; Lu, Z.; Hong, Y. Diagnostic classification of lung nodules using 3D neural networks. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 774–778. [Google Scholar] [CrossRef]
- da Silva, G.L.F.; Valente, T.L.A.; Silva, A.C.; de Paiva, A.C.; Gattass, M. Convolutional neural network-based PSO for lung nodule false positive reduction on CT images. Comput. Methods Programs Biomed. 2018, 162, 109–118. [Google Scholar] [CrossRef]
- Jung, H.; Kim, B.; Lee, I.; Lee, J.; Kang, J. Classification of lung nodules in CT scans using three-dimensional deep convolutional neural networks with a checkpoint ensemble method. BMC Med. Imaging 2018, 18, 48. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Tang, R.; Wang, X.; Zhang, W.; Wu, H. Feature Representation Using Deep Autoencoder for Lung Nodule Image Classification. Complexity 2018, 2018, 3078374. [Google Scholar] [CrossRef]
- Tran, G.S.; Nghiem, T.P.; Nguyen, V.T.; Luong, C.M.; Burie, J.-C. Improving Accuracy of Lung Nodule Classification Using Deep Learning with Focal Loss. J. Healthc. Eng. 2019, 2019, 5156416. [Google Scholar] [CrossRef] [PubMed]
- Al-Shabi, M.; Lee, H.K.; Tan, M. Gated-Dilated Networks for Lung Nodule Classification in CT Scans. IEEE Access 2019, 7, 178827–178838. [Google Scholar] [CrossRef]
- Zhao, X.; Qi, S.; Zhang, B.; Ma, H.; Qian, W.; Yao, Y.; Sun, J. Deep CNN models for pulmonary nodule classification: Model modification, model integration, and transfer learning. J. Xray Sci. Technol. 2019, 27, 615–629. [Google Scholar] [CrossRef]
- Afshar, P.; Oikonomou, A.; Naderkhani, F.; Tyrrell, P.N.; Plataniotis, K.N.; Farahani, K.; Mohammadi, A. 3D-MCN: A 3D Multi-scale Capsule Network for Lung Nodule Malignancy Prediction. Sci. Rep. 2020, 10, 7948. [Google Scholar] [CrossRef]
- Bhandary, A.; Prabhu, G.A.; Rajinikanth, V.; Thanaraj, K.P.; Satapathy, S.C.; Robbins, D.E.; Shasky, C.; Zhang, Y.-D.; Tavares, J.M.R.; Raja, N.S.M. Deep-learning framework to detect lung abnormality—A study with chest X-Ray and lung CT scan images. Pattern Recognit. Lett. 2020, 129, 271–278. [Google Scholar] [CrossRef]
- Suresh, S.; Mohan, S. ROI-based feature learning for efficient true positive prediction using convolutional neural network for lung cancer diagnosis. Neural Comput. Appl. 2020, 32, 15989–16009. [Google Scholar] [CrossRef]
- Xu, X.; Wang, C.; Guo, J.; Gan, Y.; Wang, J.; Bai, H.; Zhang, L.; Li, W.; Yi, Z. MSCS-DeepLN: Evaluating lung nodule malignancy using multi-scale cost-sensitive neural networks. Med. Image Anal. 2020, 65, 101772. [Google Scholar] [CrossRef]
- Ali, I.; Muzammil, M.; Haq, I.U.; Khaliq, A.A.; Abdullah, S. Efficient Lung Nodule Classification Using Transferable Texture Convolutional Neural Network. IEEE Access 2020, 8, 175859–175870. [Google Scholar] [CrossRef]
- Ren, Y.; Tsai, M.-Y.; Chen, L.; Wang, J.; Li, S.; Liu, Y.; Jia, X.; Shen, C. A manifold learning regularization approach to enhance 3D CT image-based lung nodule classification. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.; Pereira, T.; Frade, J.; Mendes, J.; Freitas, C.; Hespanhol, V.; Costa, J.L.; Cunha, A.; Oliveira, H.P. Pre-Training Autoencoder for Lung Nodule Malignancy Assessment Using CT Images. Appl. Sci. 2020, 10, 7837. [Google Scholar] [CrossRef]
- Zhai, P.; Tao, Y.; Chen, H.; Cai, T.; Li, J. Multi-Task Learning for Lung Nodule Classification on Chest CT. IEEE Access 2020, 8, 180317–180327. [Google Scholar] [CrossRef]
- Naik, A.; Edla, D.R.; Kuppili, V. A combination of FractalNet and CNN for Lung Nodule Classification. In Proceedings of the 2020 11th International Conference on Computing, Communication and Networking Technologies (ICCCNT), Kharagpur, India, 1–3 July 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–7. [Google Scholar] [CrossRef]
- Zia, M.B.; Juan, Z.J.; Xiao, N.; Wang, J.; Khan, A.; Zhou, X. Classification of malignant and benign lung nodule and prediction of image label class using multi-deep model. Int. J. Adv. Comput. Sci. Appl. 2020, 11, 35–41. [Google Scholar] [CrossRef]
- Venkadesh, K.V.; Setio, A.A.A.; Schreuder, A.; Scholten, E.T.; Chung, K.; Wille, M.M.W.; Saghir, Z.; van Ginneken, B.; Prokop, M.; Jacobs, C. Deep Learning for Malignancy Risk Estimation of Pulmonary Nodules Detected at Low-Dose Screening CT. Radiology 2021, 300, 438–447. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Hu, F.; Feng, L.; Zhou, T.; Zheng, C. LDNNET: Towards Robust Classification of Lung Nodule and Cancer Using Lung Dense Neural Network. IEEE Access 2021, 9, 50301–50320. [Google Scholar] [CrossRef]
- Abid, M.M.N.; Zia, T.; Ghafoor, M.; Windridge, D. Multi-view Convolutional Recurrent Neural Networks for Lung Cancer Nodule Identification. Neurocomputing 2021, 453, 299–311. [Google Scholar] [CrossRef]
- Afshar, P.; Naderkhani, F.; Oikonomou, A.; Rafiee, M.J.; Mohammadi, A.; Plataniotis, K.N. MIXCAPS: A capsule network-based mixture of experts for lung nodule malignancy prediction. Pattern Recognit. 2021, 116, 107942. [Google Scholar] [CrossRef]
- Jiang, H.; Shen, F.; Gao, F.; Han, W. Learning efficient, explainable and discriminative representations for pulmonary nodules classification. Pattern Recognit. 2021, 113, 107825. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Papathanasiou, N.D.; Panayiotakis, G.S. Classification of lung nodule malignancy in computed tomography imaging utilising generative adversarial networks and semi-supervised transfer learning. Biocybern. Biomed. Eng. 2021, 41, 1243–1257. [Google Scholar] [CrossRef]
- Mastouri, R.; Khlifa, N.; Neji, H.; Hantous-Zannad, S. A bilinear convolutional neural network for lung nodules classification on CT images. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Joshua, E.S.N.; Bhattacharyya, D.; Chakkravarthy, M.; Byun, Y.-C. 3D CNN with Visual Insights for Early Detection of Lung Cancer Using Gradient-Weighted Class Activation. J. Healthc. Eng. 2021, 2021, 6695518. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Pintelas, E.G.; Livieris, I.E.; Apostolopoulos, D.J.; Papathanasiou, N.D.; Pintelas, P.E.; Panayiotakis, G.S. Automatic classification of solitary pulmonary nodules in PET/CT imaging employing transfer learning techniques. Med. Biol. Eng. Comput. 2021, 59, 1299–1310. [Google Scholar] [CrossRef]
- Xia, K.; Chi, J.; Gao, Y.; Jiang, Y.; Wu, C. Adaptive Aggregated Attention Network for Pulmonary Nodule Classification. Applied Sciences 2021, 11, 610. [Google Scholar] [CrossRef]
- Mehta, K.; Jain, A.; Mangalagiri, J.; Menon, S.; Nguyen, P.; Chapman, D.R. Lung Nodule Classification Using Biomarkers, Volumetric Radiomics, and 3D CNNs. J. Digit. Imaging 2021, 34, 647–666. [Google Scholar] [CrossRef] [PubMed]
- Al-Shabi, M.; Shak, K.; Tan, M. 3D axial-attention for lung nodule classification. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 1319–1324. [Google Scholar] [CrossRef]
- Al-Shabi, M.; Shak, K.; Tan, M. ProCAN: Progressive growing channel attentive non-local network for lung nodule classification. Pattern Recognit. 2022, 122, 108309. [Google Scholar] [CrossRef]
- Suresh, S.; Mohan, S. NROI based feature learning for automated tumor stage classification of pulmonary lung nodules using deep convolutional neural networks. J. King Saud. Univ. Comput. Inf. Sci. 2022, 34, 1706–1717. [Google Scholar] [CrossRef]
- Liu, D.; Liu, F.; Tie, Y.; Qi, L.; Wang, F. Res-trans networks for lung nodule classification. Int. J. Comput. Assist. Radiol. Surg. 2022, 17, 1059–1068. [Google Scholar] [CrossRef]
- Fu, X.; Bi, L.; Kumar, A.; Fulham, M.; Kim, J. An attention-enhanced cross-task network to analyse lung nodule attributes in CT images. Pattern Recognit. 2022, 126, 108576. [Google Scholar] [CrossRef]
- Wu, K.; Peng, B.; Zhai, D. Multi-Granularity Dilated Transformer for Lung Nodule Classification via Local Focus Scheme. Appl. Sci. 2022, 13, 377. [Google Scholar] [CrossRef]
- Zhu, Q.; Wang, Y.; Chu, X.; Yang, X.; Zhong, W. Multi-View Coupled Self-Attention Network for Pulmonary Nodules Classification. 2022. Available online: https://github.com/ahukui/MVCS (accessed on 21 September 2023).
- Wu, R.; Liang, C.; Li, Y.; Shi, X.; Zhang, J.; Huang, H. Self-supervised transfer learning framework driven by visual attention for benign–malignant lung nodule classification on chest CT. Expert. Syst. Appl. 2023, 215, 119339. [Google Scholar] [CrossRef]
- Dai, D.; Sun, Y.; Dong, C.; Yan, Q.; Li, Z.; Xu, S. Effectively fusing clinical knowledge and AI knowledge for reliable lung nodule diagnosis. Expert. Syst. Appl. 2023, 230, 120634. [Google Scholar] [CrossRef]
- Qiao, J.; Fan, Y.; Zhang, M.; Fang, K.; Li, D.; Wang, Z. Ensemble framework based on attributes and deep features for benign-malignant classification of lung nodule. Biomed. Signal Process. Control 2023, 79, 104217. [Google Scholar] [CrossRef]
- Nemoto, M.; Ushifusa, K.; Kimura, Y.; Nagaoka, T.; Yamada, T.; Yoshikawa, T. Unsupervised Feature Extraction for Various Computer-Aided Diagnosis Using Multiple Convolutional Autoencoders and 2.5-Dimensional Local Image Analysis. Appl. Sci. 2023, 13, 8330. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, F.; Wang, N.; Zhang, C.; Yu, Q.; Zhang, M.; Babyn, P.; Zhong, H. Computer-Aided Diagnosis (CAD) of Pulmonary Nodule of Thoracic CT Image Using Transfer Learning. J. Digit. Imaging 2019, 32, 995–1007. [Google Scholar] [CrossRef]
- Huang, X.; Lei, Q.; Xie, T.; Zhang, Y.; Hu, Z.; Zhou, Q. Deep Transfer Convolutional Neural Network and Extreme Learning Machine for lung nodule diagnosis on CT images. Knowl. Based Syst. 2020, 204, 106230. [Google Scholar] [CrossRef]
- Rheey, J.; Choi, D.; Park, H. Adaptive Loss Function Design Algorithm for Input Data Distribution in Autoencoder. In Proceedings of the 2022 13th International Conference on Information and Communication Technology Convergence (ICTC), Jeju Island, Republic of Korea, 19–21 October 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 489–491. [Google Scholar] [CrossRef]

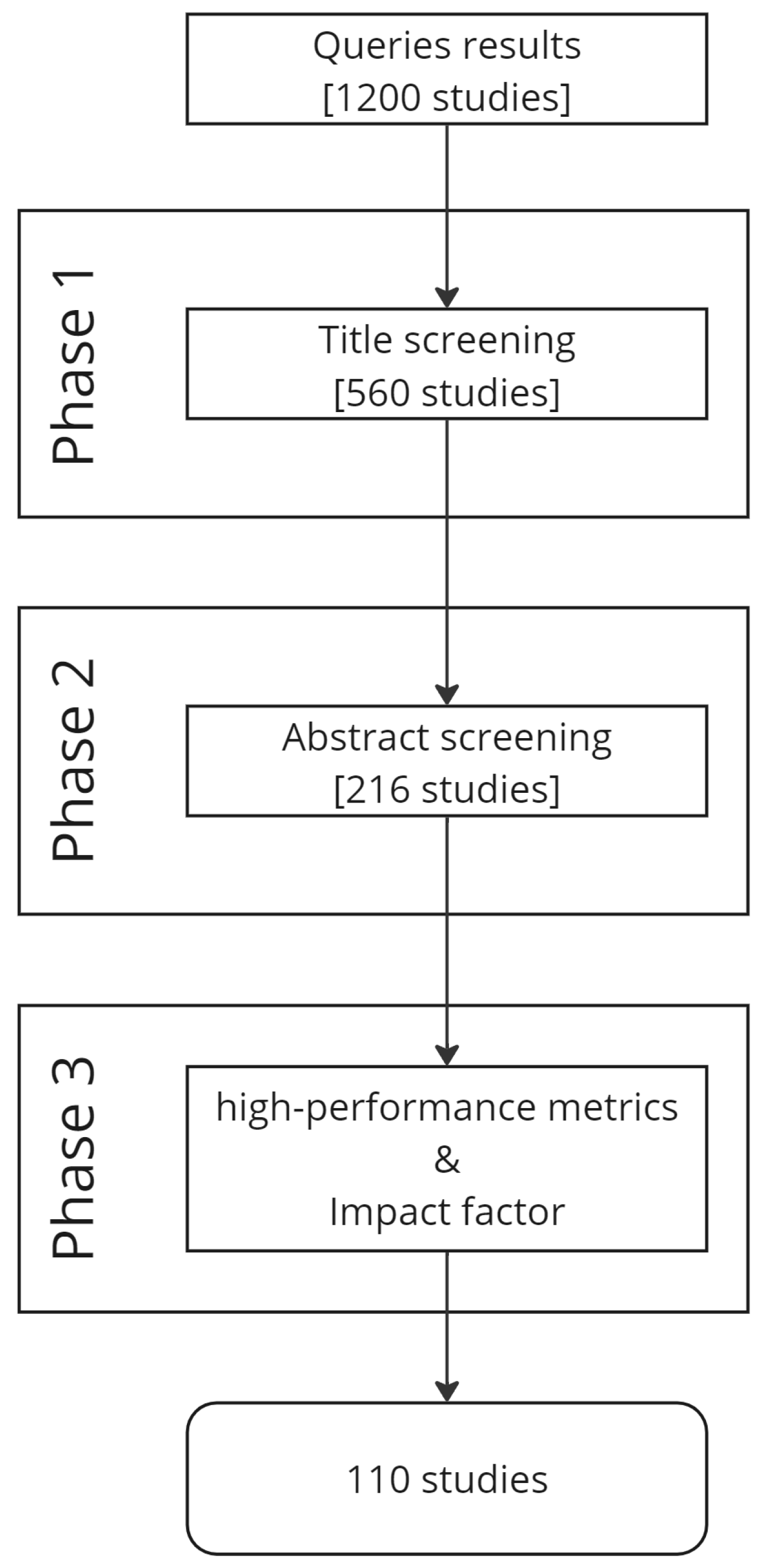
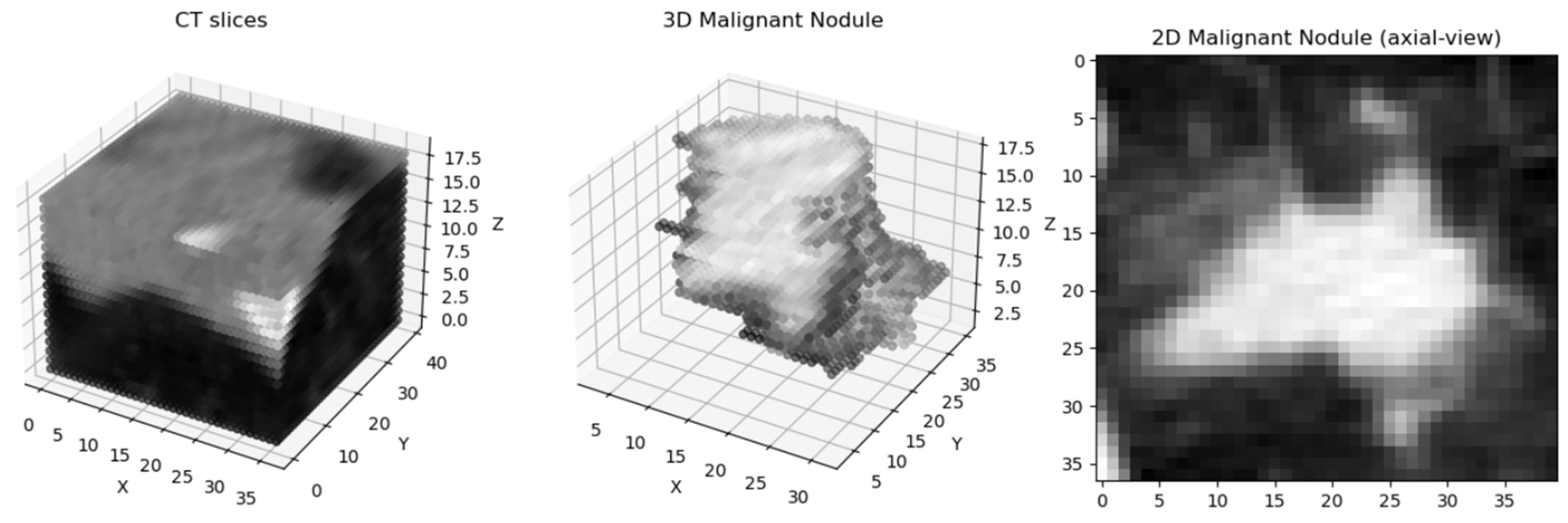
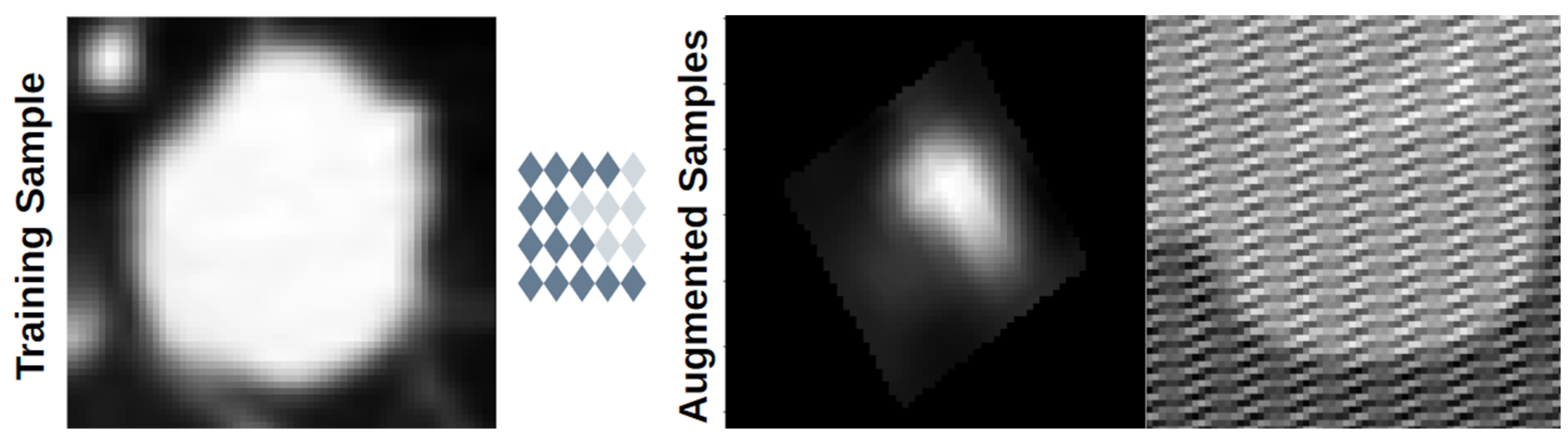
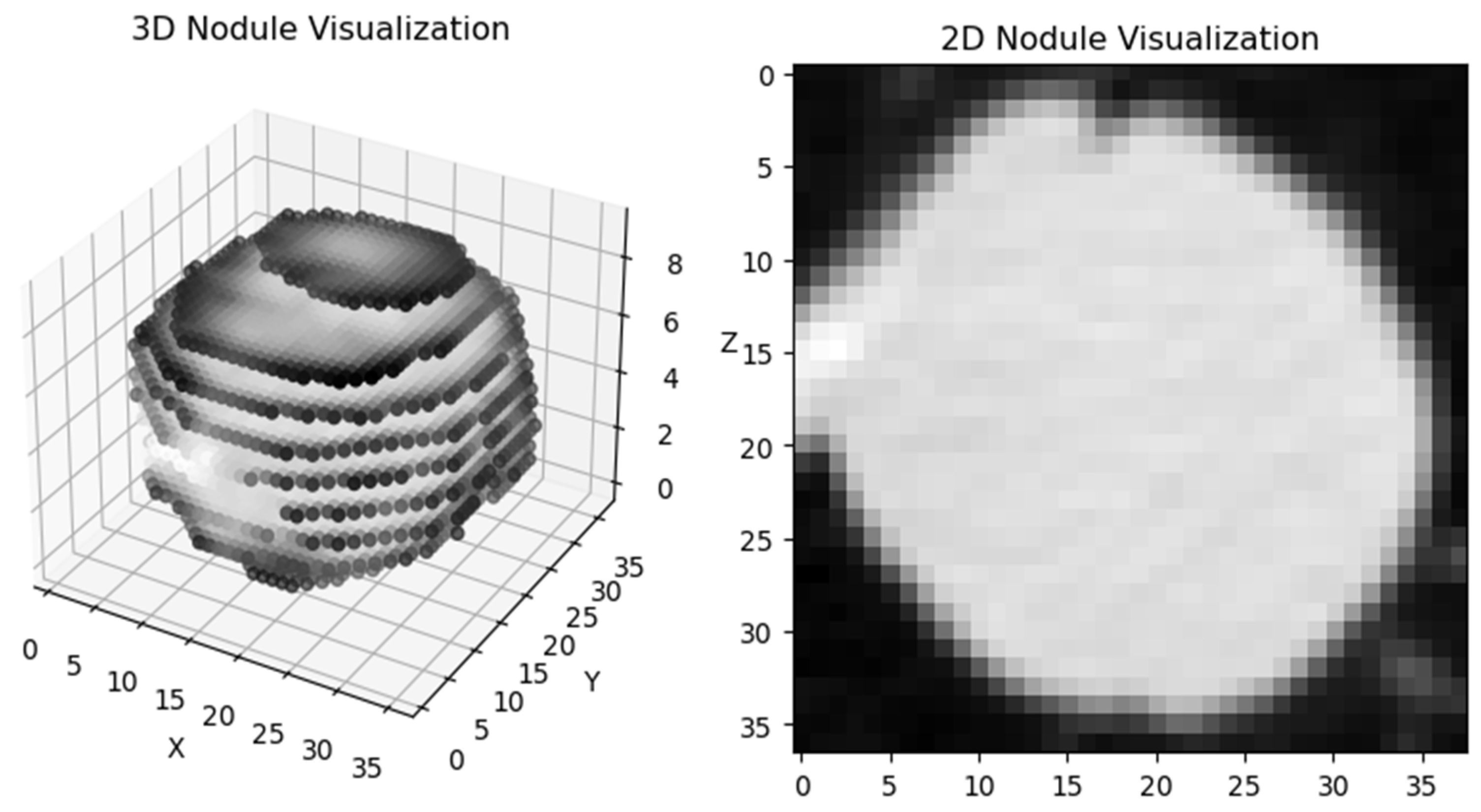

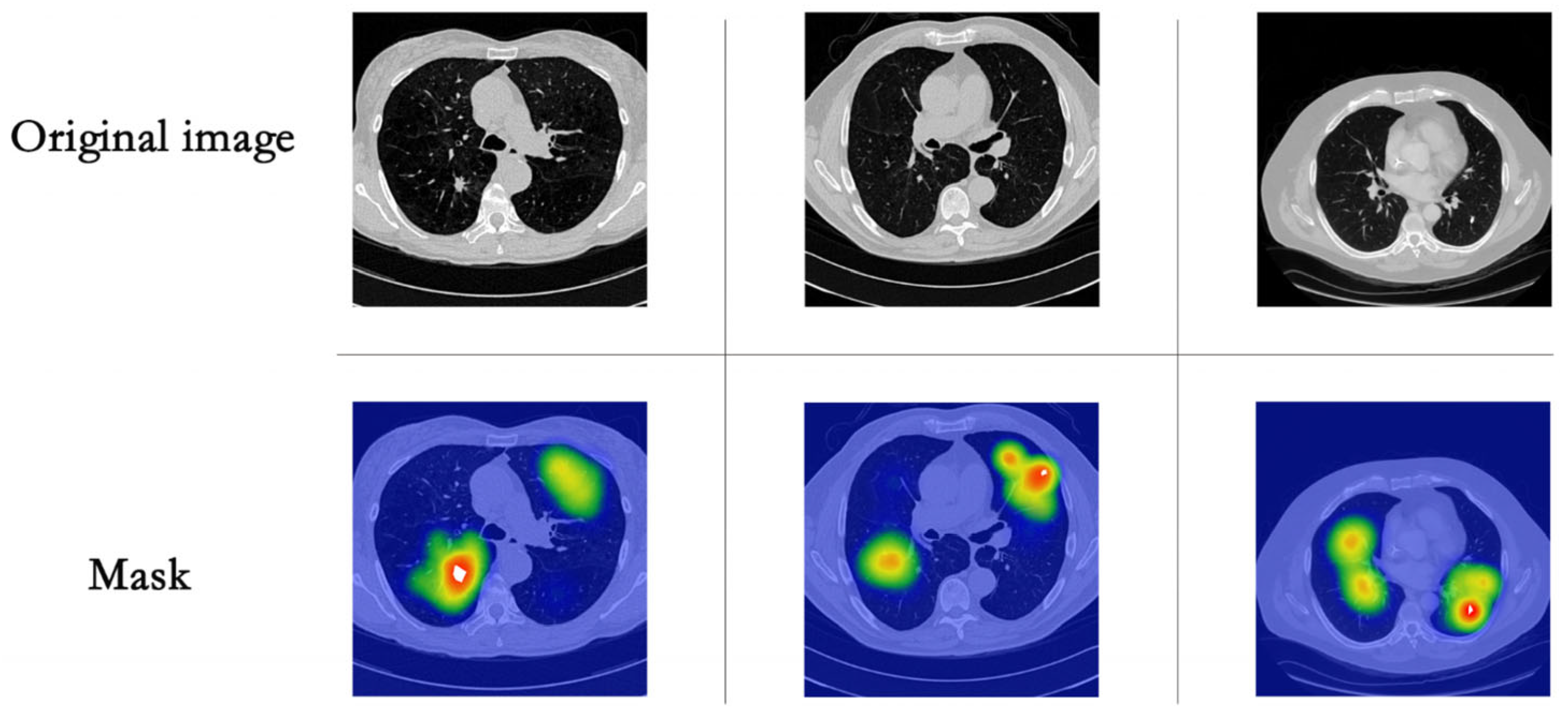

| Review | Publication Date | Date Range | Datasets | Preprocessing | Methods |
|---|---|---|---|---|---|
| [22] | 2018 | 2009–2018 | yes | yes | Traditional ML, DL |
| [19] | 2018 | 2006–2017 | few (2) | briefly | Traditional ML, DL |
| [23] | 2019 | 1990–2020 | no | no | Traditional ML, DL |
| [24] | 2020 | 2009–2018 | yes | yes | Traditional ML, DL |
| [25] | 2021 | 2005–2020 | yes | no | Non-DL, DL |
| [20] | 2021 | 2015–2020 | yes | yes | DL, GAN |
| [26] | 2022 | 2015–2021 | yes | briefly | CNN |
| [27] | 2022 | 2020–2021 | no | no | Traditional ML, DL |
| [21] | 2023 | 2018–2023 | yes | no | DL |
| Ours | 2024 | 2015–2023 | yes | yes | CNN, DL, RNN, AE, GAN, Transformers |
| Query Keywords | Description |
|---|---|
| Lung nodule detection deep learning | Research on lung nodule detection using deep learning techniques. |
| Lung nodule convolutional neural networks | Studies involving convolutional neural networks (CNNs) for lung nodule detection. |
| Lung nodule segmentation | Research on the segmentation of lung nodules is often a critical step in detection. |
| Lung nodule transfer learning | Investigations into the use of transfer learning for lung nodule detection. |
| Lung nodule Generative Adversarial Networks synthetic data | Utilizing GANs to generate synthetic data for lung nodule detection. |
| Lung nodule convolutional autoencoders | Studies involving convolutional autoencoders for lung nodule analysis. |
| Reference | Dataset Name | Modalities | #Patients | Annotations | Image Format |
|---|---|---|---|---|---|
| [29] | LIDC-IDRI | CT *, DX *, CR * | 1018 | pixel-based, patient info | DICOM |
| [30] | LUNA16 | CT, DX, CR | 888 | pixel-based, candidate nodules | MetaImage |
| [31] | ELCAP | CT | 50 | pixel-based | DICOM |
| [32] | TIANCHI17 | CT | 1000 | pixel-based | MetaImage |
| [33] | SPIE-AAPM-NCI LungX | CT | 70 | pixel-based | DICOM |
| Substance | HU |
|---|---|
| Air | −1000 |
| Lung | −500 |
| Fat | −100 to −50 |
| Water | 0 |
| Blood | +30 to +70 |
| Muscle | +10 to +40 |
| Liver | +40 to +60 |
| Bone | +700 (cancellous bone) to +3000 (cortical bone) |
| Reference | Year | Dataset | Deep Architecture | Sensitivity |
|---|---|---|---|---|
| [54] | 2017 | LUNA16 | Faster R-CNN, 3D DCNN | 92.2/1 FP, 94.4/4 FPs |
| [55] | 2018 | LUNA16 | 2D CNNs, AE, DAE | - |
| [56] | 2018 | LUNA16 | 3D CNN | 87.94/1 FP, 92.93/4 FPs |
| [35] | 2018 | LUNA16 | Modified 3D U-Net | 95.16/30.39 FPs |
| [57] | 2018 | LUNA16 | 3D DCNN | 94.9/1 FP |
| [58] | 2019 | LIDC-IDRI | 2D CNN | 88/1 FP, 94.01/4 FPs |
| [59] | 2019 | LUNA16 | 2D U-net, 3D CNN | 89.9/0.25 FP, 94.8/4 FPs |
| [60] | 2019 | LIDC-IDRI | Custom ResNet | 92.8/8 FPs |
| [61] | 2019 | LIDC-IDRI | 3D Faster R-CNN and CMixNet with U-Net-like encoder–decoder architecture | 93.97, 98.00 |
| [62] | 2019 | LUNA16 | 3D DCNN | - |
| [63] | 2020 | LIDC-IDRI | 3D U-net | 92.4 |
| [64] | 2020 | LUNA16, TIANCHI17 | CNN, TL | 97.26 |
| [65] | 2020 | LUNA16 | 3D multiscale DCNN, AE, TL | 94.2/1 FP, 96/2 FPs |
| [66] | 2020 | LUNA16, private set | U-net, AE, TL | 98 |
| [67] | 2021 | LIDC-IDRI, private data set | 3D-ResNet and MKL | 91.01 |
| [68] | 2021 | LUNA16 | 3DCNN | 87.2/22 FP |
| [69] | 2021 | Kaggle Data Science Bowl 2017 challenge (KDSB) and LUNA 16 | U-Net | 0.891 |
| [70] | 2021 | LUNA16 | 3DCNN | - |
| [71] | 2021 | LUNA16 | Multi-path 3D CNN | 0.952/0.962 to 4, 8 FP/Scans. |
| [72] | 2021 | LUNA16 | Faster R-CNN with adaptive anchor box | 93.8 |
| [73] | 2021 | NLST (NLST, 2011), LHMC, Kaggle | 2D and 3D DNN | - |
| [74] | 2022 | LIDC-IDRI + Japan Chest CT Dataset | 3D unet | - |
| [75] | 2022 | LUNA16 | 3D sphere representation-based center-points matching detection network (SCPM-Net) | 89.2/7 FP |
| [76] | 2022 | LUNA16 | Atrous UNet+ | 92.8 |
| [77] | 2022 | LUNA16 | 3D U-shaped residual network | 95 |
| [78] | 2023 | LUNA16 | 3D CNN | |
| [79] | 2023 | LUNA16 | 3D ResNet18 dual path Faster R-CNN and a federated learning algorithm | 83.388 |
| [80] | 2023 | LUNA16 | 3D ViT | 98.39 |
| [81] | 2023 | LUNA16 | 3D ViT | 97.81 |
| [82] | 2023 | LIDC-IDRI | 2D Ensemble Transformer with Attention Modules | 94.58 |
| [83] | 2023 | LUNA16 | 3Dl Multifaceted Attention Encoder–Decoder | 89.1/7 FPs |
| [84] | 2023 | ELCAP | 3D CNN-CapsNet | 92.31 |
| [85] | 2023 | LUNA16 | A multiscale self-calibrated network (DEPMSCNet)with a dual attention mechanism | 98.80 |
| Reference | Year | Dataset | Deep Architecture | DSC (%) |
|---|---|---|---|---|
| [93] | 2017 | LIDC-IDRI, private set | Central Focused Convolutional Neural Networks (CF-CNN) | 82.15 ± 10.76, LIDC 80.02 ± 11.09 Private set |
| [94] | 2019 | LIDC-IDRI | Cascaded Dual-Pathway Residual Network | 81.58 ± 11.05 |
| [95] | 2019 | LIDC-IDRI | SegNet, a deep, fully convolutional network | 93 ± 0.11 |
| [62] | 2019 | LIDC-IDRI | 3D DCNN | 83.10 ± 8.85 |
| [96] | 2020 | LIDC-IDRI | Deep residual deconvolutional network, TL | 94.97 |
| [97] | 2020 | LIDC-IDRI | Deep Residual U-Net | 87.5 ± 10.58 |
| [98] | 2020 | LIDC-IDRI | DB-ResNet, CF-CNN | 82.74 ± 10.19 |
| [99] | 2020 | LIDC-IDRI | U-net | - |
| [100] | 2021 | LIDC-IDRI, LNDb, ILCID | 2D CNN | 80 |
| [101] | 2021 | LIDC-IDRI | U-Net | 93.14 |
| [102] | 2022 | LUNA16 | V-net | 95.01 |
| [103] | 2021 | LIDC-IDRI, SHCH | 2D–3D U-net | 83.16/81.97 |
| [104] | 2021 | LIDC-IDRI, LUNA16 | Faster R-CNN | 89.79/90.35 |
| [105] | 2021 | LIDC-IDRI | U-net | 86.23 |
| [106] | 2021 | LIDC-IDRI | 3D res U-net | 80.5 |
| [107] | 2021 | LIDC-IDRI | VGG-SegNet | 90.49 |
| [108] | 2022 | hospital data | 3D FCN | 84.5 |
| [109] | 2022 | LUNA16, ILND | 3D GAN | 80.74/76.36 |
| [110] | 2022 | LIDC-IDRI | 3D Dual Attention Shadow Network (DAS-Net) | 92.05 |
| [111] | 2022 | LIDC-IDRI | Transformer | 89.86 |
| [112] | 2023 | LIDC-IDRI | Dual-encoder-based CNN | 87.91 |
| [113] | 2023 | LIDC-IDRI, AHAMU-LC | RAD—U-net | - |
| [114] | 2023 | LIDC-IDRI, private set | SMR—U-net 2D | 91.87 |
| [115] | 2023 | LIDC-IDRI | U-shaped hybrid transformer | 91.84 |
| [116] | 2023 | LIDC-IDRI, LUNA16 | 3D U-net based | 82.48 |
| [117] | 2023 | LIDC-IDRI | GUNet3++ | 97.2 |
| Reference | Year | Dataset | Deep Architecture | Accuracy (%) |
|---|---|---|---|---|
| [125] | 2017 | LIDC-IDRI | ResNet | 89.90 |
| [126] | 2017 | LIDC-IDRI | 3D MV-CNN + SoftMax | - |
| [127] | 2017 | LIDC-IDRI | ResNet-50, TL | 93.40 |
| [128] | 2018 | LIDC-IDRI | MV-KBC | 91.60 |
| [129] | 2018 | LIDC-IDRI | CNN + Random Forest | 94.60 |
| [130] | 2018 | LIDC-IDRI + Private set | 3D DenseNet, TL | 90.40 |
| [131] | 2018 | LIDC-IDRI | CNN + PSO | 97.62 |
| [132] | 2018 | LUNA16 | 3D DCNN | - |
| [133] | 2018 | ELCAP | DAE | - |
| [134] | 2019 | LUNA16 | Novel 2D CNN | 97.2 |
| [135] | 2019 | LIDC-IDRI | Novel 2D CNN | 92.57 |
| [136] | 2019 | LIDC-IDRI | CNN, TL | 88 |
| [137] | 2020 | LIDC-IDRI | multiscale 3D-CNN, CapsNets | 94.94 |
| [138] | 2020 | LIDC-IDRI | MAN (modified AlexNet), TL | 91.60 |
| [139] | 2020 | LIDC-IDRI | CNN, TL | 97.27 |
| [58] | 2020 | LIDC-IDRI, private dataset (FAH-GMU) | DTCNN, TL | 93.9 |
| [140] | 2020 | LIDC-IDRI, DeepLNDataset | 3D CNN | 94.57/100 |
| [141] | 2020 | LIDC-IDRI, LUNGx Challenge database | 2D CNN, TL | 92.65 |
| [142] | 2020 | LIDC-IDRI | MRC-DNN | 96.69 |
| [143] | 2020 | LIDC-IDRI | CAE, TL | 90 |
| [144] | 2020 | LIDC-IDRI, LUNA16 | Multi-Task CNN | - |
| [145] | 2020 | LUNA16 | Fractalnet and CNN | - |
| [146] | 2020 | LIDC-IDRI | DCNN | 94.06 |
| [147] | 2021 | NLST, DLCST | 2D CNN 9 views, 3D CNN | 90.73 |
| [137] | 2020 | LIDC-IDRI | multiscale 3D-CNN, CapsNets | 93.12 |
| [148] | 2021 | LUNA16/Kaggle DSB 2017 dataset | Dense Convolutional Network (DenseNet) | - |
| [149] | 2021 | LIDC-IDRI/ELCAP | 2D MV-CNN 3D MV-CNN | 98.83 |
| [150] | 2021 | LIDC | Capsule networks (CapsNets) | Accuracy (%) |
| [151] | 2021 | LIDC-IDRI | 3D NAS method, CBAM module, A-Softmax loss, and ensemble strategy to learn efficient, | 89.90 |
| [152] | 2021 | LIDC-IDRI | Deep Convolutional Generative Adversarial Network (DC-GAN)/FF-VGG19 | - |
| [153] | 2021 | LUNA16 | BCNN [VGG16, VGG19] combination with and without SVM | 93.40 |
| [154] | 2021 | LUNA16 | 3D CNN | 91.60 |
| [155] | 2021 | pet-ct private, LIDC-IDRI | 2d cnn | 94.60 |
| [156] | 2021 | LIDC-IDRI | 3D DPN _ attention mech | 90.40 |
| [157] | 2021 | LIDC-IDRI | 3D CNN + biomarkers | 97.62 |
| [158] | 2021 | LIDC-IDRI | 3D attention | - |
| [159] | 2022 | LIDC-IDRI and LUNGx | ProCAN | 97.2 |
| [160] | 2022 | LIDC-IDRI | DCNN | 92.57 |
| [161] | 2022 | LIDC-IDRI | Transformers | 88 |
| [162] | 2022 | LIDC-IDRI | CNN-based MTL model that incorporates multiple attention-based learning modules | 91.60 |
| [163] | 2022 | LIDC-IDRI | Transformers | 97.27 |
| [164] | 2022 | LUNA16 | 3D ResNet + attention | 93.9 |
| [165] | 2023 | LIDC-IDRI/TC-LND Dataset/CQUCH-LND | STLF-VA | 94.57/100 |
| [166] | 2023 | LIDC-IDRI | Transformer | 92.65 |
| [167] | 2023 | LIDC-IDRI | F-LSTM-CNN | 96.69 |
| [168] | 2023 | private | CAE | 90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marinakis, I.; Karampidis, K.; Papadourakis, G. Pulmonary Nodule Detection, Segmentation and Classification Using Deep Learning: A Comprehensive Literature Review. BioMedInformatics 2024, 4, 2043-2106. https://doi.org/10.3390/biomedinformatics4030111
Marinakis I, Karampidis K, Papadourakis G. Pulmonary Nodule Detection, Segmentation and Classification Using Deep Learning: A Comprehensive Literature Review. BioMedInformatics. 2024; 4(3):2043-2106. https://doi.org/10.3390/biomedinformatics4030111
Chicago/Turabian StyleMarinakis, Ioannis, Konstantinos Karampidis, and Giorgos Papadourakis. 2024. "Pulmonary Nodule Detection, Segmentation and Classification Using Deep Learning: A Comprehensive Literature Review" BioMedInformatics 4, no. 3: 2043-2106. https://doi.org/10.3390/biomedinformatics4030111
APA StyleMarinakis, I., Karampidis, K., & Papadourakis, G. (2024). Pulmonary Nodule Detection, Segmentation and Classification Using Deep Learning: A Comprehensive Literature Review. BioMedInformatics, 4(3), 2043-2106. https://doi.org/10.3390/biomedinformatics4030111






