Abstract
(1) Background: Parkinson’s disease (PD) is a progressively worsening neurodegenerative disorder affecting movement, mental well-being, sleep, and pain. While no cure exists, treatments like hyperbaric oxygen therapy (HBOT) offer potential relief. However, the molecular biology perspective, especially when intertwined with machine learning dynamics, remains underexplored. (2) Methods: We employed machine learning techniques to analyze single-cell RNA-seq data from human PD cell samples. This approach aimed to identify pivotal genes associated with PD and understand their relationship with HBOT. (3) Results: Our analysis indicated genes such as MAP2, CAP2, and WSB1, among others, as being crucially linked with Parkinson’s disease (PD) and showed their significant correlation with Hyperbaric oxygen therapy (HBOT) indicatively. This suggests that certain genomic factors might influence the efficacy of HBOT in PD treatment. (4) Conclusions: HBOT presents promising therapeutic potential for Parkinson’s disease, with certain genomic factors playing a pivotal role in its efficacy. Our findings emphasize the need for further machine learning-driven research harnessing diverse omics data to better understand and treat PD.
1. Introduction
The Parkinson’s disease is characterized by a spectrum of symptoms that can differ in nature and intensity, making the prediction of treatment outcomes challenging [1]. Interestingly, the therapeutic potential of hyperbaric oxygen therapy (HBOT) for Parkinson’s was sometimes identified in an unanticipated manner [2]. For example, a diabetic patient undergoing HBOT for a foot ulcer unexpectedly reported a marked alleviation in Parkinson’s symptoms. Animal studies consistently indicate that HBOT exhibits anti-inflammatory properties [3], which could be beneficial in addressing the inflammatory conditions observed in the substantia nigra region of the brain in Parkinson’s patients. Anecdotal evidence further suggests that some Parkinson’s patients, even those with advanced stages of the disease, have shown significant improvements after HBOT sessions.
The realm of hyperbaric oxygen therapy (HBOT) in treating neurodegenerative diseases [4], particularly Parkinson’s disease (PD), is burgeoning with potential. As we delve deeper into this field, a multitude of studies have emerged, shedding light on the transformative effects of HBOT on neuronal health, motor function, and overall quality of life for patients. For instance, recent research has illuminated the capacity of HBOT to target specific brain circuits, enhance neurotrophic factors, and even modulate epigenetic pathways, offering a beacon of hope for those grappling with the debilitating effects of PD.
For instance, research has demonstrated that HBOT can significantly increase the number of TH-positive neurons in MPTP-treated mice, enhancing the neurotrophic factor BDNF while reducing apoptotic signaling and attenuating inflammatory mediators in the midbrain [5]. This treatment also promotes mitochondrial biogenesis and improves locomotor activity and grip strength in these mice.
Further insights [6] highlighted the potential of HBOT in targeting specific brain circuits involved in “Kinesia Paradoxa”, including the noradrenergic system, basal ganglia, and the cerebellum circuit. This study presented evidence supporting the “Norepinephrine Hypothesis”, suggesting a role for HBOT in increasing norepinephrine levels, which could restore motor deficits in Parkinson’s disease patients. When considering the combination of treatments, the research indicates that combining donepezil with HBOT and functional rehabilitation training can significantly enhance therapeutic effectiveness in Parkinson’s disease dementia (PDD) patients. This combination not only improves cognitive function, self-care ability, and quality of life but also significantly reduces inflammatory markers like serum IL-1β and IL-6 [7].
In a broader context, the potential of HBOT as a therapeutic intervention for neurodegenerative diseases has been explored, with findings emphasizing its promising effects in conditions associated with neurodegeneration and functional impairments. A special focus has been given to the role of epigenetics in these effects [8]. Lastly, in a study focused on spinocerebellar ataxias (SCAs), HBOT was found to attenuate motor coordination and cognitive impairment in SCA17 mice, with effects persisting for about a month post-treatment. SCA17 is a rare subtype of SCAs (spinocerebellar ataxias), notable for its association with a myriad of neurological symptoms including motor coordination and cognitive impairments, often leading to a substantial reduction in the quality of life of affected individuals.
This neuroprotective effect of HBOT might be attributed to the promotion of BDNF production and the reduction of neuroinflammation [9].
Despite the promising strides made in this domain, the field is still in its infancy. The intricacies of HBOT’s impact on the human brain, especially in the context of neurodegenerative diseases, remain vast and largely uncharted. While the preliminary results are indeed encouraging, they underscore the pressing need for more comprehensive, large-scale studies. Only through rigorous research, meticulous analysis, and collaborative efforts can we truly harness the full potential of HBOT and pave the way for groundbreaking therapeutic interventions in the future.
On the other hand, machine learning (ML) methodologies have been extensively applied to enhance the understanding and management of Parkinson’s disease (PD). A comprehensive review of the literature reveals the utilization of ML models in conjunction with Internet of Things technologies, such as smart devices and various sensors, to optimize predictions and estimations regarding different aspects of PD [10]. These models are trained on data acquired via these technologies and address a myriad of PD-related problems, offering insights into the most effective algorithms and commonly addressed issues in PD management. Another study provides an extensive overview of the application of ML in categorizing PD, emphasizing the use of diverse data modalities and artificial intelligence techniques to facilitate informed and systematic clinical decision-making [11]. These studies collectively underscore the pivotal role of ML in advancing diagnostic processes and therapeutic interventions for PD, highlighting its potential in contributing to more nuanced and effective approaches in PD treatment and management.
The exploration of hyperbaric oxygen therapy (HBOT) in the context of Parkinson’s disease (PD) has predominantly been rooted in traditional research methodologies [12]. Notably absent from this landscape is the integration of modern machine learning (ML) frameworks, which have the potential to revolutionize our understanding of the disease’s intricacies [13]. While several studies have explored molecular biology to understand underlying mechanisms, many have not fully utilized advanced computational methods. Our endeavor represents a pioneering effort in this direction. By employing an ML approach, we aim to meticulously examine the behavior of key genes implicated in PD. This innovative methodology allows us to unravel the intricate relationships between these genes and the therapeutic effects of HBOT, offering a fresh perspective and potentially groundbreaking insights into the treatment of PD.
2. Materials and Methods
2.1. Dataset
In our study, we utilized single-cell RNA sequencing (scRNA-seq) data derived from the work of [14]. This dataset offers a comprehensive expression profiling of human induced pluripotent stem cell (iPSC)-derived midbrain dopaminergic neurons. These neurons were sourced from both Parkinson’s disease patients and healthy controls. Novak and her team employed scRNA-seq to delve into the expression profiles of these neurons, aiming to uncover the underlying molecular networks associated with Parkinson’s disease pathology. Their findings hint at a core molecular network linked to the disease, presenting a valuable resource for further exploration of this debilitating neurological disorder.
The scRNA-seq dataset under consideration comprises a total of 4495 cells, which are profiled for their expression across 18,098 genes. This extensive gene coverage ensures a comprehensive view of the transcriptional landscape of each individual cell, allowing for a detailed understanding of cellular heterogeneity and potential differences between the two conditions. The dataset is categorized into two distinct tags or conditions such as “Control” and “PD”. The distribution of cells across these conditions is slightly imbalanced. The “Control” group consists of 2518 cells, which constitutes approximately 56% of the total cells. On the other hand, the “PD” group has 1977 cells, making up the remaining 44% of the dataset. This discrepancy in cell numbers between the two conditions should be taken into account during downstream analyses, especially when comparing gene expression patterns or inferring statistical significance. The presence of nearly 2000 cells in the “PD” group, despite being fewer than the “Control”, still offers a substantial sample size for robust analysis. Given the depth of genes profiled, this dataset is poised to provide significant insights into the molecular differences and similarities between normal (Control) cells and those affected by Parkinson’s Disease.
2.2. Hybrid Feature Selection Methodology
To ensure a comprehensive and robust feature selection, we devised a hybrid methodology that synergizes the strengths of both traditional differential gene expression (DEG) analysis and machine learning techniques. The DEG analysis offers a foundational understanding by pinpointing genes that exhibit significant expression differences, serving as an initial filter in the identification of potential key players. On the other hand, the variable importance (VI) from machine learning provides a data-driven perspective, highlighting genes that are most influential in predictive modeling. Building on these insights, we incorporated an ensemble voting scheme to establish a more robust gene ranking. This approach not only consolidates the insights from both methodologies but also prioritizes genes that are strongly associated with PD.
2.2.1. Differential Gene Expression Analysis
We initiated our feature selection process by applying the Wilcoxon test [15]. This non-parametric statistical test was used to identify genes that were differentially expressed between the PD and healthy cell samples. The Wilcoxon test, or Mann–Whitney U test, is a non-parametric method comparing the medians of two independent samples by ranking all observations and summing the ranks separately for each group to determine statistical significance. If and are the sum of ranks for the first and second groups, respectively, and and are the sizes of the two groups, the test statistic is given by . The Wilcoxon test is particularly useful when the data does not meet the assumptions of a t-test, such as when the data is not normally distributed. The outcome of the Wilcoxon test provided us with a ranked list of genes, organized based on their significance levels. We considered genes with a p-value of less than 0.05 as statistically significant. This threshold value of p < 0.05 is a conventional criterion in statistical hypothesis testing that helps in minimizing the Type I error, ensuring that the identified genes are truly differentially expressed and not due to random chance.
2.2.2. Machine Learning-Based Feature Selection Analysis
Regarding the machine learning-based feature selection, we used the XGBoost 2.0 Algorithm [16]. It is a gradient boosting framework, to further refine our feature selection. By training the model on our dataset, we extracted the variable importance scores for each gene. This allowed us to generate a second ranked list of genes, this time based on their importance in the predictive model. code. More specifically, XGBoost offers a robust mechanism to assess the importance of features in a predictive model through its variable importance (VI) metric. The VI in XGBoost is primarily derived from the number of times a feature is used to split the data across all trees, and the improvement it brings to the model, typically measured as the gain. Mathematically, if represents a feature and denotes the gain brought by when used in splits, the importance of the feature is proportional to the sum of gains over all splits where is used: . This aggregated measure provides a ranking of features based on their contribution to the model’s predictive power, allowing for the identification of the most influential predictors in the dataset. In our implementation of the XGBoost algorithm, we used a learning rate (eta) of 0.01, a max depth of 6, a subsample of 0.8, a colsample_bytree of 0.8, and built 1000 trees as the number of estimators.
2.2.3. Hybrid Ensemble Genes Ranking
To combine the insights from both the Wilcoxon test and the XGBoost algorithm, we utilized the Borda count, a consensus-based ensemble voting scheme [17]. By taking the two ranked gene lists from the previous steps, the Borda count method allowed us to derive a more robust and consolidated ranking. This combined ranking leverages the dynamics of both statistical testing and machine learning, ensuring a comprehensive selection of features that are both statistically significant and relevant for predictive modeling. The Borda count operates on the principle of assigning points to items (in this case, genes) based on their rank. For a given gene list of genes, the top-ranked gene receives points, the next receives points, and so on, with the last-ranked gene receiving 1 point.
Let us denote the ranking from the Wilcoxon test as and from the XGBoost algorithm as . For a particular gene , its Borda count score is computed as:
where and represent the ranks of gene gigi in the Wilcoxon and XGBoost rankings, respectively. After computing the Borda count scores for all genes, we can then rank them based on these scores to derive a consolidated ranking. This ensemble approach ensures that genes which are both statistically significant (from the Wilcoxon test) and important for predictive modeling (from XGBoost) receive higher ranks, providing a more robust and comprehensive feature selection.
3. Results and Discussion
We ended up with a combined list of genes (Table S1) that are strongly related to PD and decided to explore closely at the top 100 genes since it’s been demonstrated that in scRNA-seq, typically only a few dozen to a couple of hundred genes play a pivotal role in the dataset [18]. Also, focusing on the top 100 genes facilitated a more in-depth exploration of their biological functions, interactions, and roles in the context of the study, allowing for more meaningful interpretations and conclusions.
Also, by concentrating on the top genes, we’re likely capturing the most important ones that have the biggest impact on PD. A key part of our study was to see how these genes are related to HBOT. This is important because if we know which genes are affected by this therapy, it could help doctors treat PD more effectively in the future.
Our study had three four parts. First, we examine the classification performance using the 100 key genes regarding differentiating healthy samples from Parkinson’s disease samples. Furthermore, we looked at how HBOT affects each of the top 100 genes one by one. This helped us figure out which specific genes might be good targets for treatment. Next, we checked how our chosen genes fit into bigger groups of genes and how they might be linked to other diseases or treatments. This gave us a better idea of the bigger picture and how these genes work in the body. Lastly, we used a simple visual tool zooming out of the genes to show how our top genes are connected to each other along with the associated gene ontologies.
3.1. Classification Performance of Leading Genes
We investigated the role of specific genes in understanding the difference between healthy cells and those affected by Parkinson’s disease. After we had our list of 100 genes, we used a tool called PyCaret to see how well we could separate or tell apart the healthy cells from the PD ones using only these genes (Figure 1). PyCaret offers a variety of classifiers for comparison in its classification module, such as logistic regression, K nearest neighbor (KNN), naive bayes, decision tree, random forest, gradient boosting machines, support vector machines (SVM), linear discriminant analysis (LDA), quadratic discriminant analysis (QDA), ridge classifier, extreme gradient boosting (XGBoost), light gradient boosting machine (LightGBM), catboost, adaboost, extra trees, stochastic gradient descent (SGD), and dummy classifier.
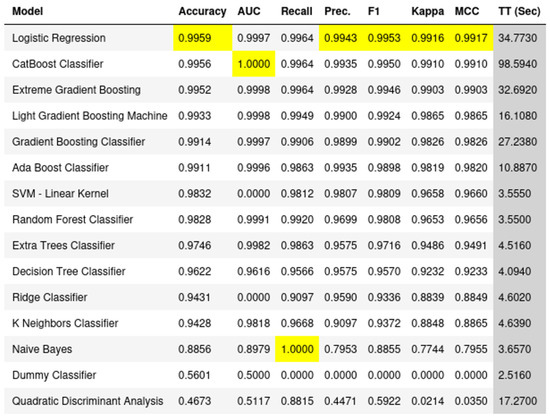
Figure 1.
Comprehensive classification performance of the top 100 genes. The figure displays the outcomes from 15 well-known classifiers, evaluated based on 7 performance measures alongside their execution time. This analysis underscores the efficacy of our selected genes in distinguishing between PD and healthy samples across diverse machine learning models.
The primary contribution of this task lies in its comprehensive examination of the performance of multiple classifiers in discerning between Parkinson’s Disease and healthy cell states using the identified key genes. By leveraging a diverse set of classifiers, ranging from logistic regression to more complex models like CatBoost and Gradient Boosting Machines, we were able to gauge the robustness and reliability of these key genes as discriminative features. This approach not only underscores the significance of the selected genes but also provides a holistic view of their potential in different machine learning paradigms. The ability of these genes to consistently separate PD from healthy samples across various classifiers reinforces their importance in the realm of Parkinson’s Disease research. This task, therefore, serves as a foundational step in understanding the potential of these genes as biomarkers and offers a blueprint for future studies aiming to harness the power of machine learning in biomedical research.
Our results were pretty clear. By using the top 100 genes we identified, the tool was able to clearly tell the difference between healthy and PD cells. This means that these genes are really important and can be key players in understanding Parkinson’s Disease. In simple words, our study shows that with the right genes, we can easily spot the difference between a healthy cell and one that is affected by PD.
3.2. Gene-Based Analysis
The 100 genes derived from our analysis were individually examined to determine their established or potential links with Parkinson’s disease and the impacts of HBOT. This targeted approach was designed to elucidate the molecular underpinnings that might be at play in the therapeutic response of Parkinson’s patients to HBOT, providing a deeper understanding of the disease mechanism and potential intervention points. Furthermore, the distribution and the associations among the top 10 genes is illustrated in Figure 2 showing their potential in our framework.
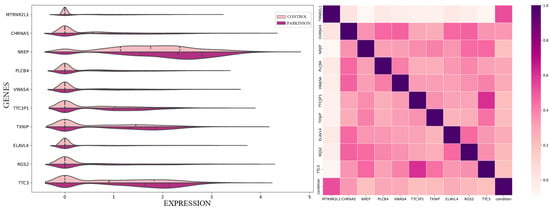
Figure 2.
Comparative analysis of the 10 dominant genes. On the left, a violin plot illustrates the distribution of expression levels for each gene, highlighting their prominence in our framework. On the right, a heatmap showcases the correlation patterns among these genes, providing insights into their interrelated dynamics.
More specifically, TXNIP (Thioredoxin Interacting Protein) plays a pivotal role in the regulation of cellular redox balance [19]. By binding to thioredoxin, a primary antioxidant protein, TXNIP inhibits its antioxidant function, potentially leading to heightened oxidative stress within cells. This interaction becomes particularly relevant in the context of hyperbaric oxygen therapy (HBOT). Given that HBOT involves the administration of oxygen at elevated pressures, there’s an inherent increase in the production of reactive oxygen species (ROS). As oxidative stress is a recognized factor in the pathogenesis of Parkinson’s Disease, the modulation of this stress, potentially influenced by TXNIP, might be crucial in determining cellular responses to HBOT and its therapeutic implications for PD.
The ELAVL4 (ELAV Like RNA Binding Protein 4) gene, a member of the ELAVL family of RNA-binding proteins, is predominantly expressed in neurons [20]. Its primary function revolves around stabilizing mRNA, a process integral to neuronal differentiation and maintenance. In the realm of HBOT, where there’s a proposed neuroprotective effect primarily through enhanced oxygen delivery to hypoxic tissues, ELAVL4’s role in neuronal maintenance becomes significant. Neurons, when exposed to increased oxygen levels during HBOT, might leverage the stabilizing influence of ELAVL4, underscoring its potential importance. Furthermore, in neurodegenerative conditions like Parkinson’s, where neuronal health is paramount, genes like ELAVL4 that bolster neuronal function could offer insights into disease progression and therapeutic responses.
Lastly, XBP1 (X-Box Binding Protein 1), a transcription factor, is activated as part of the unfolded protein response (UPR) [21]. The UPR is a cellular mechanism triggered by the accumulation of unfolded or misfolded proteins within the endoplasmic reticulum (ER). The relevance of XBP1 in HBOT stems from the therapy’s potential to induce oxidative modifications to proteins, which can lead to their misfolding. As the UPR aims to restore cellular function in the face of such protein stress, XBP1 might be a key player in this restoration process. This becomes even more pertinent in Parkinson’s Disease, where protein misfolding and aggregation are hallmark features. The potential activation of the UPR, and by extension the role of XBP1, could shed light on how Parkinsonian brain cells respond to both protein aggregation and treatments like HBOT.
3.3. Enrichment Analysis in Gene Ontologies, Disease and Pharmaceutical Terms
We conducted an enrichment analysis on the leading genes to ascertain if there was a notable overlap with predefined gene sets from established ontologies (Figure 3). We employed the EnrichR platform [22] to analyze our gene set in the context of GO cellular component processes, pathway maps, drug descriptors, and disease terms. EnrichR is a comprehensive web-based and mobile application that offers a range of gene-set libraries, a unique ranking method for enriched terms, and diverse visualization techniques for results presentation. The platform encompasses 35 gene-set libraries, accounting for a total of 31,026 gene-sets that span the entire human and mouse genome and proteome. Typically, each gene-set contains approximately 350 genes, leading to over six million interconnections between terms and genes. For the enrichment analysis, EnrichR utilizes the Fisher exact test, a standard method prevalent in many enrichment analysis tools. This test, based on a binomial distribution, evaluates the likelihood of a gene’s association with a particular set, assuming independence.
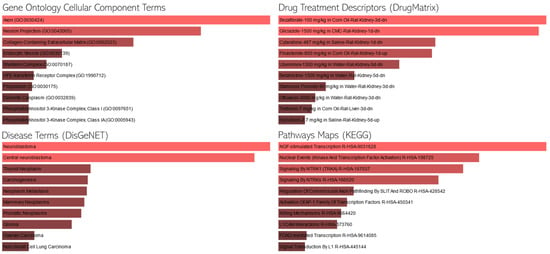
Figure 3.
Enrichment Analysis of the Top 100 Genes. This figure presents the association of our selected genes with various gene ontology (GO) terms, disease annotations, and pharmaceutical implications. Longer bars indicate greater statistical significance, highlighting the genes’ multifaceted roles and potential therapeutic significance in the broader biological and medical context.
Enrichment analysis in KEGG pathway terms reveals several intriguing pathways. Indicatively, the “NGF-stimulated Transcription R-HSA-9031628” pathway suggests a role in neurotrophic factor signaling, which is crucial for neuronal survival and has been implicated in Parkinson’s disease [23]. Neurotrophic factors could potentially be modulated by HBOT, leading to neuroprotective effects. Additionally, the “Serotonin and Melatonin Biosynthesis R-HSA-209931” pathway is noteworthy, given that serotoninergic system dysfunction is often observed in Parkinson’s disease, and HBOT might influence neurotransmitter levels or their biosynthetic pathways. Both pathways provide valuable insights into the potential mechanisms through which HBOT could exert therapeutic effects in Parkinson’s disease.
Upon examining the GO cellular component terms, several cellular structures and complexes emerge as potentially relevant. Specifically, the term “Axon (GO:0030424)” is of particular interest, as axonal degeneration is a hallmark of Parkinson’s disease, and any therapeutic intervention, including HBOT, that can influence axonal health could be beneficial. Similarly, “Neuron Projection (GO:0043005)” is another term that stands out, given that the integrity of neuronal projections is vital for proper neuronal communication, and its disruption is observed in Parkinson’s disease [24]. HBOT’s potential to modulate or protect these neuronal structures could provide a mechanistic insight into its therapeutic effects in the context of Parkinson’s disease.
Regarding the drug terms, certain drugs emerge as potentially relevant in the context of Parkinson’s disease and HBOT. Notably, epinephrine (adrenaline) plays a role in the autonomic nervous system and its dysregulation is observed in Parkinson’s disease. The potential modulation of epinephrine levels or its pathways by HBOT could provide insights into its therapeutic effects. Several disease conditions stand out in the context of Parkinson’s disease and HBOT. “Neuroblastoma” is particularly noteworthy, as it is a neural tumor that could provide insights into the neural mechanisms potentially influenced by HBOT. Additionally, “Glioma,” another type of brain tumor, is of interest, given that any therapeutic intervention, including HBOT, that can influence neural health or growth mechanisms could be beneficial in understanding its broader implications for neurological conditions like Parkinson’s disease.
These findings not only elucidate the potential biological processes influenced by the dominant genes separating PD from healthy states but also offer a preliminary understanding of how HBOT might interact with these processes. Further studies could dive deeper into these associations, paving the way for targeted therapeutic strategies in PD.
3.4. Graph-Based Analysis—Interconnectivity and Associations
In our graph-based analysis (Figure 4), we focused on understanding the relationships between genes and their associated gene ontologies, which describe their roles in molecular functions and broader biological processes. A key aspect of this was examining gene-gene interactions using protein-protein interaction (PPI) networks. These PPI networks provide a structured representation of how proteins, and by extension the genes that code for them, interact within a cell. By mapping our selected genes onto these networks, we gained insights into potential functional relationships that these genes might have with one another.
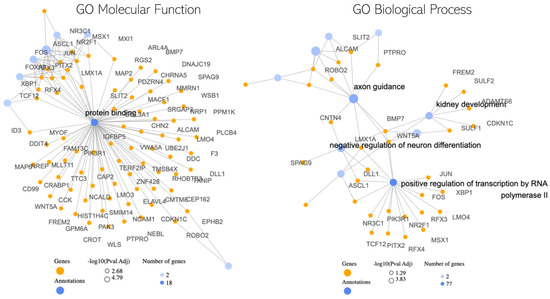
Figure 4.
Interconnectivity Graph of the Top 100 Genes. This visualization depicts the Protein–protein Interaction (PPI) associations among our significant genes, emphasizing their interconnected roles. Additionally, key gene ontologies are highlighted as hubs, demonstrating their central role in bridging multiple significant genes and underscoring their biological importance.
Alongside this, we aimed to determine how our genes fit within larger biological contexts. To do this, we conducted an enrichment analysis, which checks if certain biological categories or functions are more common among our selected genes than would be expected by chance [25]. It uses the standard hypergeometric distribution test, also known as the Fisher exact test, for this purpose, a widely accepted statistical method in gene enrichment analysis. By comparing our gene set to reference sets, this test helped us identify specific biological processes or molecular functions that our genes are likely involved in.
In our exploration of the potential molecular interplay between hyperbaric oxygen therapy (HBOT) and Parkinson’s disease (PD), the graph-based analysis has revealed several noteworthy findings. The central observation is the pronounced role of the “Protein Binding” term. Acting as a hub, this term suggests a nexus of interactions, with several genes like MAP2, CAP2, and WSB1 being pivotal [26,27,28]. The hub-like nature of “Protein Binding” implies that these genes might be central to many protein–protein interactions, potentially modulating a variety of cellular processes that could be influenced by HBOT in the context of PD [29].
Among these genes, for instance, MAP2 is known for its role in stabilizing microtubules [30], which are essential for maintaining cell structure and facilitating intracellular transport. Any modulation in its activity could impact neuronal health and function, making it a potential target of interest in PD and its response to HBOT. Furthermore, other gene ontology (GO) terms that stood out include “axon guidance” [31], “negative regulation of neuron differentiation” [32], and “positive regulation of transcription by RNA polymerase II” [33]. The presence of “axon guidance” is particularly intriguing, as it plays a crucial role in the proper formation of neural circuits. Disruptions in this process could contribute to neurodegenerative conditions like PD. The regulation of neuron differentiation and transcription further suggests that HBOT might influence the broader landscape of gene expression and neuronal development in PD.
In considering the translational potential of our findings, it is pivotal to acknowledge the prospective clinical implications. The identified genomic correlations with hyperbaric oxygen therapy (HBOT) in Parkinson’s disease (PD) suggest avenues for the development of personalized and optimized treatment strategies, potentially enhancing therapeutic outcomes and patient quality of life. These genomic insights could inform the creation of targeted therapies and predictive models, allowing for individualized treatment plans based on specific genomic profiles. However, the realization of these clinical applications necessitates rigorous validation through clinical trials, collaborative integration into clinical workflows, adherence to ethical and regulatory standards, and comprehensive educational outreach to stakeholders about the benefits and limitations of such interventions.
Our study, while offering significant insights, is subject to several limitations. The single-cell RNA-seq data utilized may not fully capture the intricate cellular heterogeneity inherent to Parkinson’s disease due to its inherent resolution and depth limitations, and the public datasets employed may harbor biases stemming from variations in sample collection, processing, and sequencing technologies across different studies. Additionally, the machine learning techniques applied in our analysis are susceptible to biases from the training data, model assumptions, and algorithmic constraints, potentially impacting the reliability of our identified gene correlations. Furthermore, the generalizability of our findings is constrained, necessitating validation in diverse and larger Parkinson’s disease populations to confirm their universal applicability and clinical relevance.
4. Conclusions
Our comprehensive study, integrating both traditional and machine learning methodologies, has shed light on the intricate molecular landscape underpinning Parkinson’s disease (PD) and its potential modulation by hyperbaric oxygen therapy (HBOT). By synergizing differential gene expression analysis with machine learning techniques, we’ve identified pivotal genes, such as MAP2, SLIT2, CAP2, DDC, WSB1, and MYOF, that play significant roles in PD and may be influenced by HBOT. The pronounced role of the “Protein Binding” term, acting as a hub in our analysis, underscores the importance of protein–protein interactions in understanding the therapeutic potential of HBOT in PD.
Furthermore, our exploration into gene ontologies and pathways, such as “axon guidance” and “negative regulation of neuron differentiation,” has provided insights into the broader biological processes that might be at play. The enrichment analysis, leveraging the Fisher exact test, has highlighted potential biological pathways and drug interactions that could be pivotal in understanding the therapeutic mechanisms of HBOT in PD. In essence, our findings emphasize the intricate interplay of genes, pathways, and cellular processes in PD and how they might be modulated by HBOT. This research not only offers a deeper understanding of the molecular mechanisms of PD but also paves the way for future studies aiming to optimize therapeutic strategies for this debilitating neurodegenerative disorder.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedinformatics4010009/s1, Table S1: List of genes that were found to be significant in the present approaches.
Author Contributions
Conceptualization, E.B. and P.V.; methodology, E.B. and A.G.V.; software, E.B.; validation, A.G.V. and M.G.K.; formal analysis, E.B. and A.G.V.; data curation, E.B. and A.G.V.; writing—original draft preparation, E.B. and A.G.V.; writing—review and editing, M.G.K. and P.V.; supervision, P.V.; funding acquisition, P.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research is funded by the European Union and Greece (Partnership Agreement for the Development Framework 2014–2020) under the Regional Operational Programme Ionian Islands 2014–2020, project title: “Study of Clinical trial protocols with biomarkers that define the evolution of non-genetic neurodegenerative diseases- NEUROTRIAL”, project number: 5016089.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
This study uses a public dataset available at Gene Expression Omnibus. The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bloem, B.R.; Okun, M.S.; Klein, C. Parkinson’s disease. Lancet 2021, 397, 2284–2303. [Google Scholar] [CrossRef] [PubMed]
- Wilmshurst, P.; Bewley, S.; Murray, P. Hyperbaric oxygen therapy for the treatment of long COVID. Clin. Med. 2023, 23, 99–100. [Google Scholar] [CrossRef] [PubMed]
- Taslipinar, M.Y.; Aydin, I.; Kaldirim, U.; Aydin, F.N.; Agilli, M.; Eyi, Y.E.; Cayci, T. Hyperbaric oxygen treatment and N-acetylcysteine ameliorate acetaminophen-induced liver injury in a rat model. Hum. Exp. Toxicol. 2013, 32, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Kidd, P.M. Multiple sclerosis, an autoimmune inflammatory disease: Prospects for its integrative management. Altern. Med. Rev. 2001, 6, 540–566. [Google Scholar] [PubMed]
- Hsu, H.T.; Yang, Y.L.; Chang, W.H.; Fang, W.Y.; Huang, S.H.; Chou, S.H.; Lo, Y.C. Hyperbaric oxygen therapy improves Parkinson’s disease by promoting mitochondrial biogenesis via the SIRT-1/PGC-1α pathway. Biomolecules 2022, 12, 661. [Google Scholar] [CrossRef]
- Banou, E. Hyperbaric oxygen therapy effect on “Kinesia Paradoxa” brain circuits. GeNeDis 2020: Genetics and Neurodegenerative Diseases. GeNeDis 2020: Genet. Neurodegener. Dis. 2021; 1339, 139–146. [Google Scholar]
- Fan, A.; Zhou, J. Effect of the combination of donepezil with hyperbaric oxygen therapy and functional rehabilitation training on parkinson’s disease dementia and the neurological function system. Int. J. Clin. Exp. Med. 2020, 13, 5867–5875. [Google Scholar]
- Mensah-Kane, P.; Sumien, N. The potential of hyperbaric oxygen as a therapy for neurodegenerative diseases. GeroScience 2023, 45, 747–756. [Google Scholar] [CrossRef]
- Shi, Q.; Luo, Q.; Gong, Q.; Wang, G. Effects of rTMS Combined with Hyperbaric Oxygen-acupuncture-rehabilitation Therapy on Motor Function, Serum CRP and Plasma Dopamine in Patients with Parkinson’s Disease. Chin. Gen. Pract. 2020, 23, 3460. [Google Scholar]
- Giannakopoulou, K.M.; Roussaki, I.; Demestichas, K. Internet of things technologies and machine learning methods for Parkinson’s disease diagnosis, monitoring and management: A systematic review. Sensors 2022, 22, 1799. [Google Scholar] [CrossRef]
- Rana, A.; Dumka, A.; Singh, R.; Panda, M.K.; Priyadarshi, N.; Twala, B. Imperative role of machine learning algorithm for detection of Parkinson’s disease: Review, challenges and recommendations. Diagnostics 2022, 12, 2003. [Google Scholar] [CrossRef]
- Atzeni, F.; Masala, I.F.; Cirillo, M.; Boccassini, L.; Sorbara, S.; Alciati, A. Hyperbaric oxygen therapy in fibromyalgia and the diseases involving the central nervous system. Clin. Exp. Rheumatol. 2020, 38, 0094–0098. [Google Scholar]
- Quazi, S. Artificial intelligence and machine learning in precision and genomic medicine. Med. Oncol. 2022, 39, 120. [Google Scholar] [CrossRef] [PubMed]
- Novak, G.; Kyriakis, D.; Grzyb, K.; Bernini, M. Single-cell transcriptomics of human iPSC differentiation dynamics reveal a core molecular network of Parkinson’s disease. Commun. Biol. 2022, 5, 49. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, X.; Peng, F.; Li, W.; Li, J.J. Wilcoxon rank-sum test still outperforms dearseq after accounting for the normalization impact in semi-synthetic RNA-seq data simulation. bioRxiv 2022, 2022-06. [Google Scholar] [CrossRef]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Zhou, T. Xgboost: Extreme gradient boosting. R Package Version 0.4-2 2015, 1, 1–4. [Google Scholar]
- Paplomatas, P.; Krokidis, M.G.; Vlamos, P.; Vrahatis, A.G. An ensemble feature selection approach for analysis and modeling of transcriptome data in alzheimer’s disease. Appl. Sci. 2023, 13, 2353. [Google Scholar] [CrossRef]
- Chatzilygeroudis, K.I.; Vrahatis, A.G.; Tasoulis, S.K.; Vrahatis, M.N. Feature Selection in single-cell RNA-seq data via a Genetic Algorithm. In Proceedings of the Learning and Intelligent Optimization: 15th International Conference, LION 15, Athens, Greece, 20–25 June 2021; Revised Selected Papers 15. Springer International Publishing: Berlin/Heidelberg, Germany, 2021; pp. 66–79. [Google Scholar]
- Tezgin, D.; Giardina, C.; Perdrizet, G.A.; Hightower, L.E. The effect of hyperbaric oxygen on mitochondrial and glycolytic energy metabolism: The caloristasis concept. Cell Stress Chaperones 2020, 25, 667–677. [Google Scholar] [CrossRef]
- Bowles, K.R.; Silva, M.C.; Whitney, K.; Bertucci, T.; Berlind, J.E.; Lai, J.D.; Temple, S. ELAVL4, splicing, and glutamatergic dysfunction precede neuron loss in MAPT mutation cerebral organoids. Cell 2021, 184, 4547–4563. [Google Scholar] [CrossRef]
- Iwakoshi, N.N.; Lee, A.H.; Glimcher, L.H. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol. Rev. 2003, 194, 29–38. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 1–14. [Google Scholar] [CrossRef]
- Zhou, L.; Too, H.P. Mitochondrial localized STAT3 is involved in NGF induced neurite outgrowth. PLoS ONE 2011, 6, e21680. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Braak, E. Pathoanatomy of Parkinson’s disease. J. Neurol. 2000, 247, II3–II10. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Moreno, A.; López-Domínguez, R.; Villatoro-García, J.A.; Ramirez-Mena, A.; Aparicio-Puerta, E.; Hackenberg, M.; Pascual-Montano, A.; Carmona-Saez, P. Functional Enrichment Analysis of Regulatory Elements. Biomedicines 2022, 10, 590. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, M.R.; Ilyin, S.; Plata-Salaman, C.R. Abnormal patterns of microtubule-associated protein-2 (MAP-2) immunolabeling in neuronal nuclei and Lewy bodies in Parkinson’s disease substantia nigra brain tissues. Neurosci. Lett. 2001, 306, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Di Maio, A.; De Rosa, A.; Pelucchi, S.; Garofalo, M.; Marciano, B.; Nuzzo, T.; Usiello, A. Analysis of mRNA and protein levels of CAP2, DLG1 and ADAM10 genes in post-mortem brain of schizophrenia, Parkinson’s and Alzheimer’s disease patients. Int. J. Mol. Sci. 2022, 23, 1539. [Google Scholar] [CrossRef] [PubMed]
- Nucifora Jr, F.C.; Nucifora, L.G.; Ng, C.H.; Arbez, N.; Guo, Y.; Roby, E.; Ross, C.A. Ubiqutination via K27 and K29 chains signals aggregation and neuronal protection of LRRK2 by WSB1. Nat. Commun. 2016, 7, 11792. [Google Scholar] [CrossRef] [PubMed]
- Ruf, W.P.; Freischmidt, A.; Grozdanov, V.; Roth, V.; Brockmann, S.J.; Mollenhauer, B.; Danzer, K.M. Protein binding partners of dysregulated miRNAs in Parkinson’s Disease Serum. Cells 2021, 10, 791. [Google Scholar] [CrossRef] [PubMed]
- Dehmelt, L.; Halpain, S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2005, 6, 1–10. [Google Scholar]
- Lesnick, T.G.; Papapetropoulos, S.; Mash, D.C.; Ffrench-Mullen, J.; Shehadeh, L.; De Andrade, M.; Maraganore, D.M. A genomic pathway approach to a complex disease: Axon guidance and Parkinson disease. PLoS Genet. 2007, 3, e98. [Google Scholar] [CrossRef]
- Pirooznia, S.K.; Wang, H.; Panicker, N.; Kumar, M.; Neifert, S.; Dar, M.A.; Dawson, T.M. Deubiquitinase CYLD acts as a negative regulator of dopamine neuron survival in Parkinson’s disease. Sci. Adv. 2022, 8, eabh1824. [Google Scholar] [CrossRef]
- Majidinia, M.; Mihanfar, A.; Rahbarghazi, R.; Nourazarian, A.; Bagca, B.; Avci, Ç.B. The roles of non-coding RNAs in Parkinson’s disease. Mol. Biol. Rep. 2016, 43, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).