Overall Survival Time Estimation for Epithelioid Peritoneal Mesothelioma Patients from Whole-Slide Images
Abstract
1. Introduction
2. Dataset and Methods
2.1. Ethical Considerations
2.2. Population Characteristics
2.3. Dataset
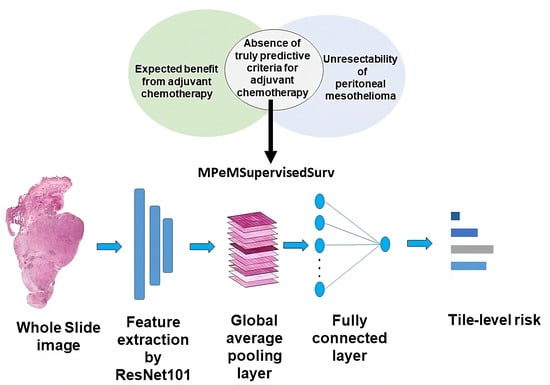
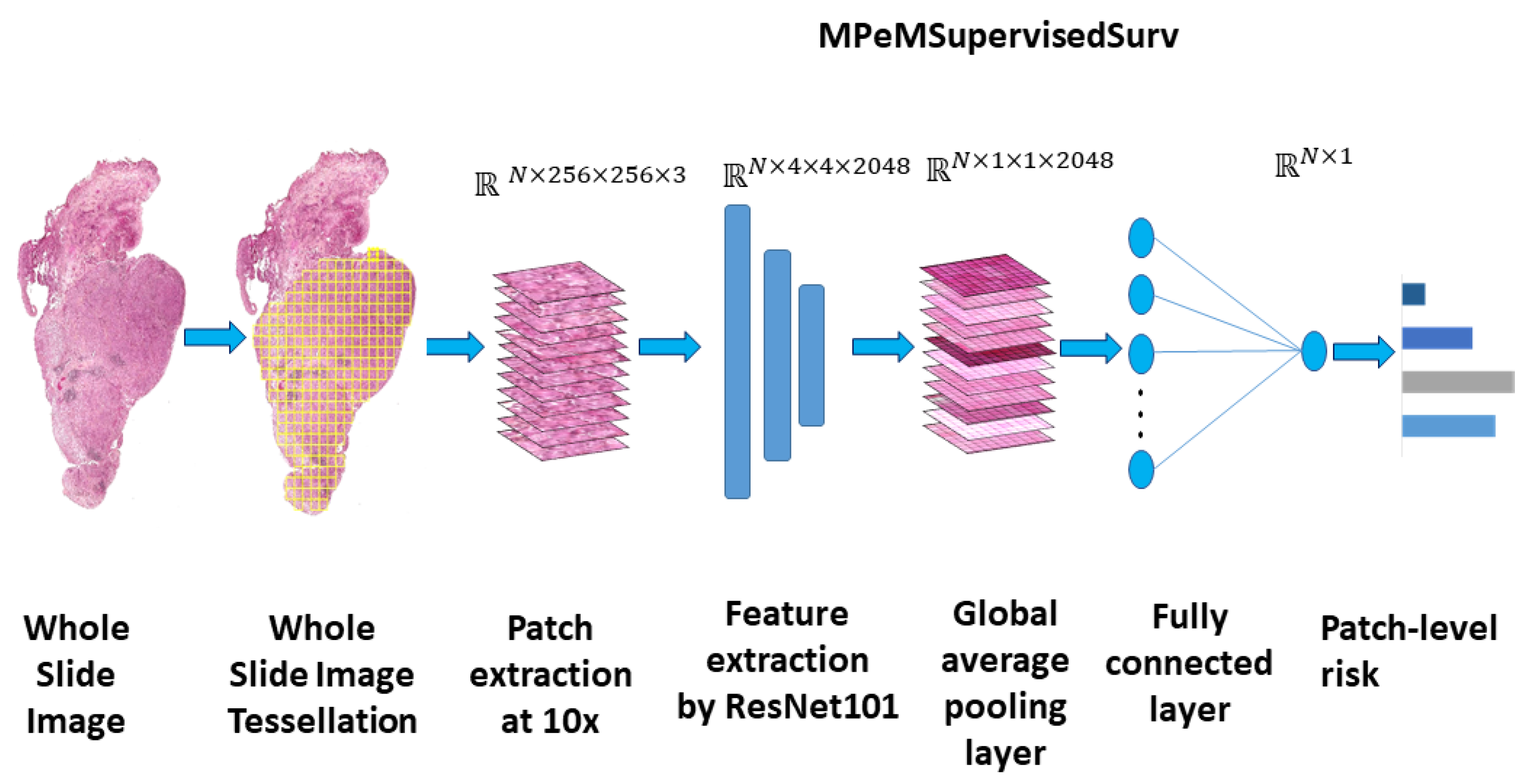
2.4. Methods
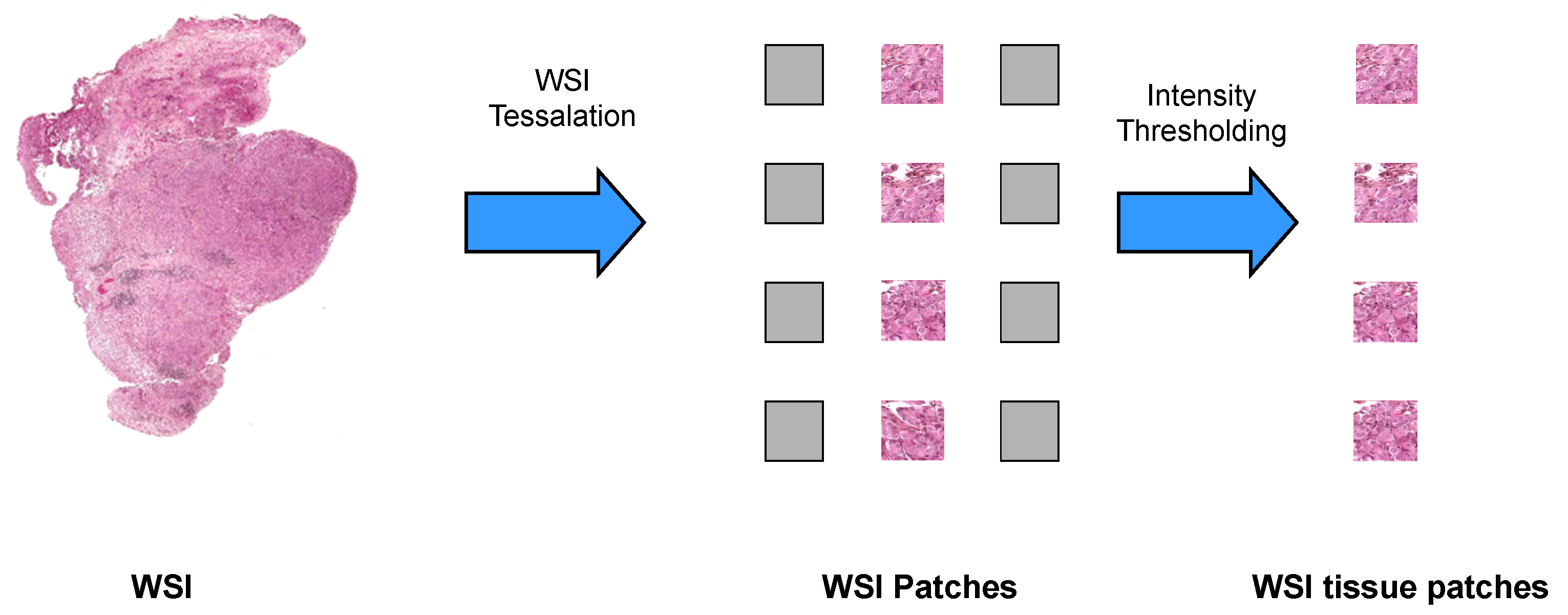
2.4.1. WSI Preprocessing
2.4.2. Model Architecture
2.4.3. Training Procedure
2.4.4. Loss Function and Model Evaluation
3. Results
3.1. Model Accuracy Results
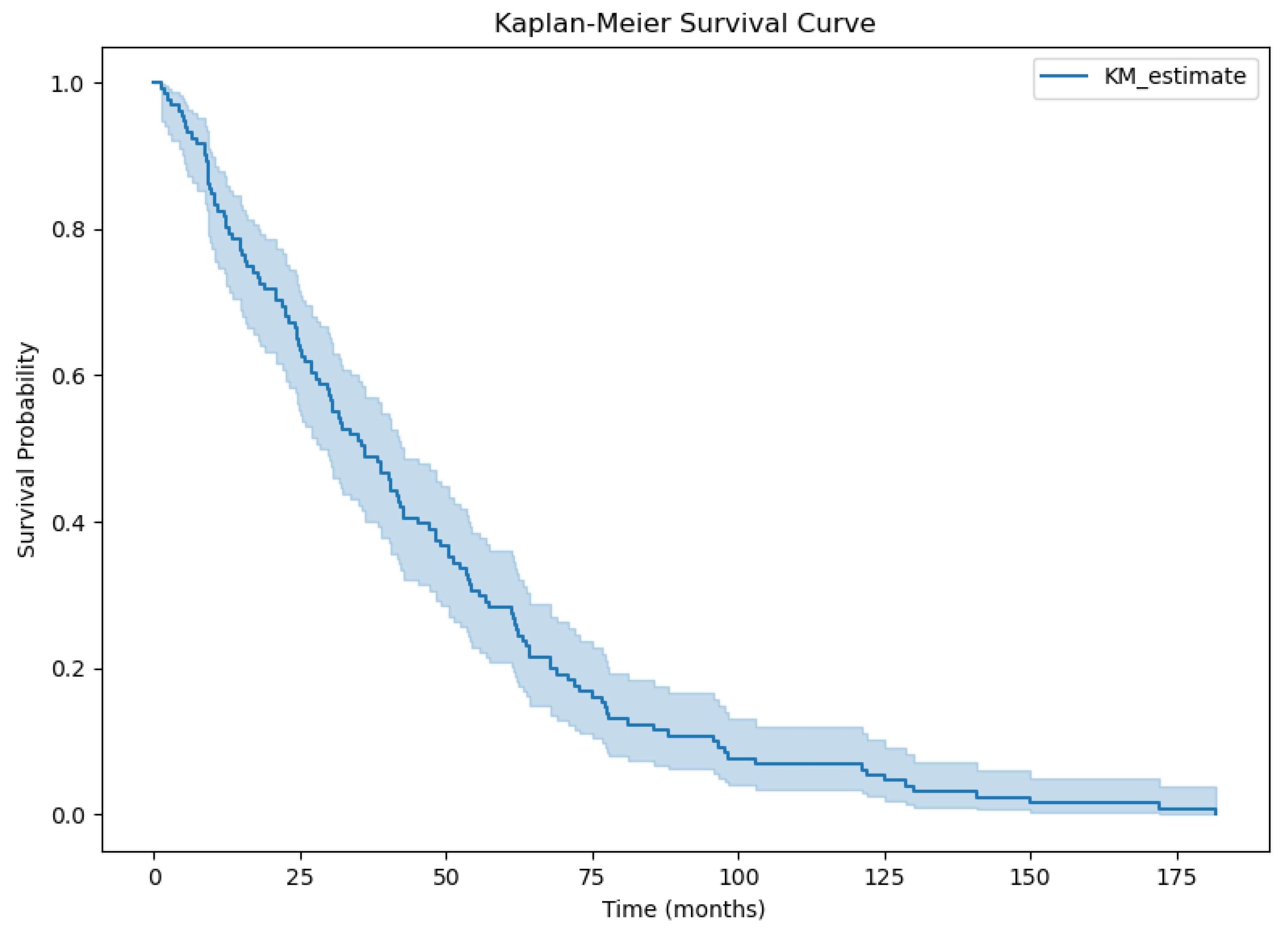
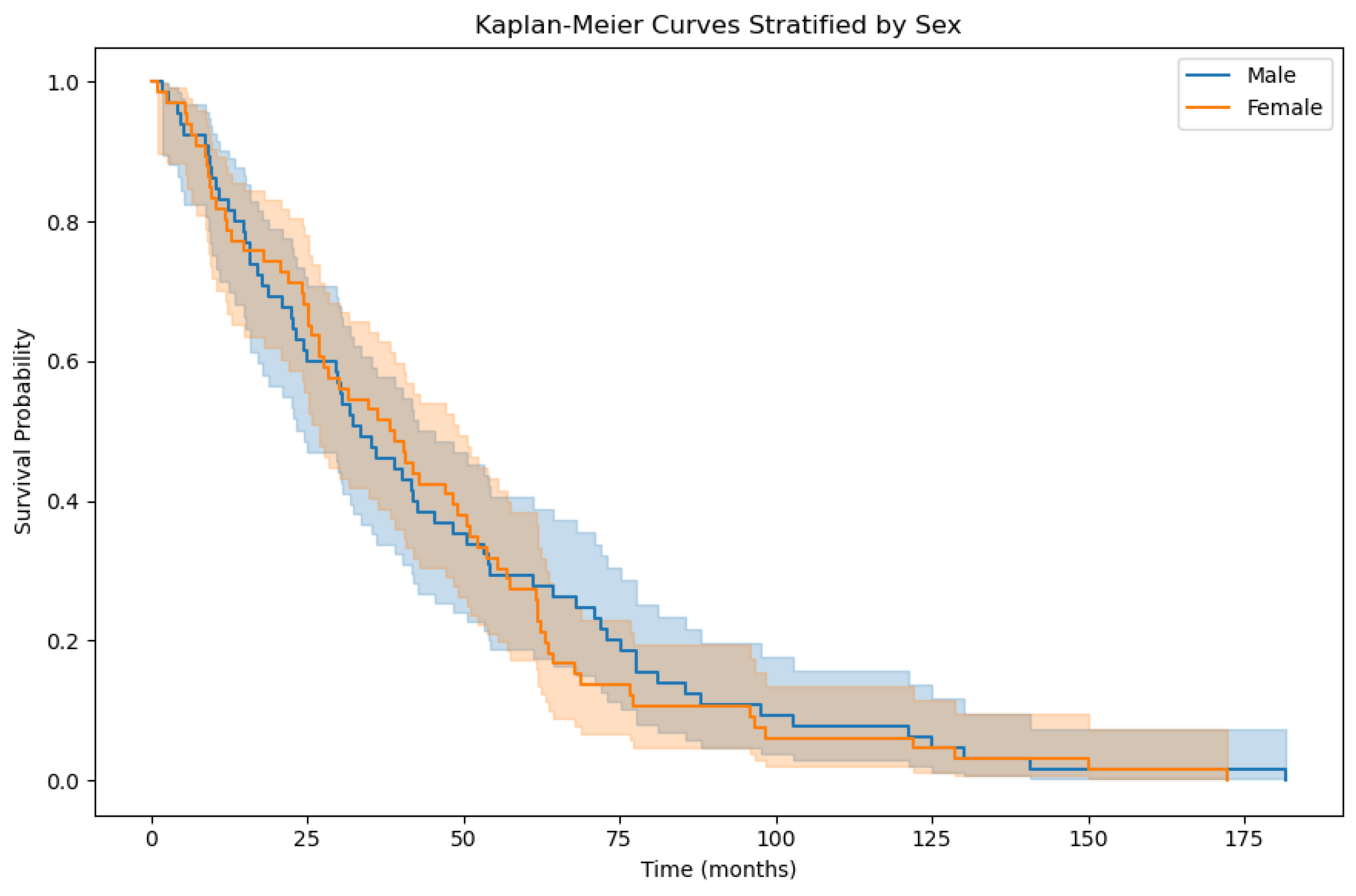
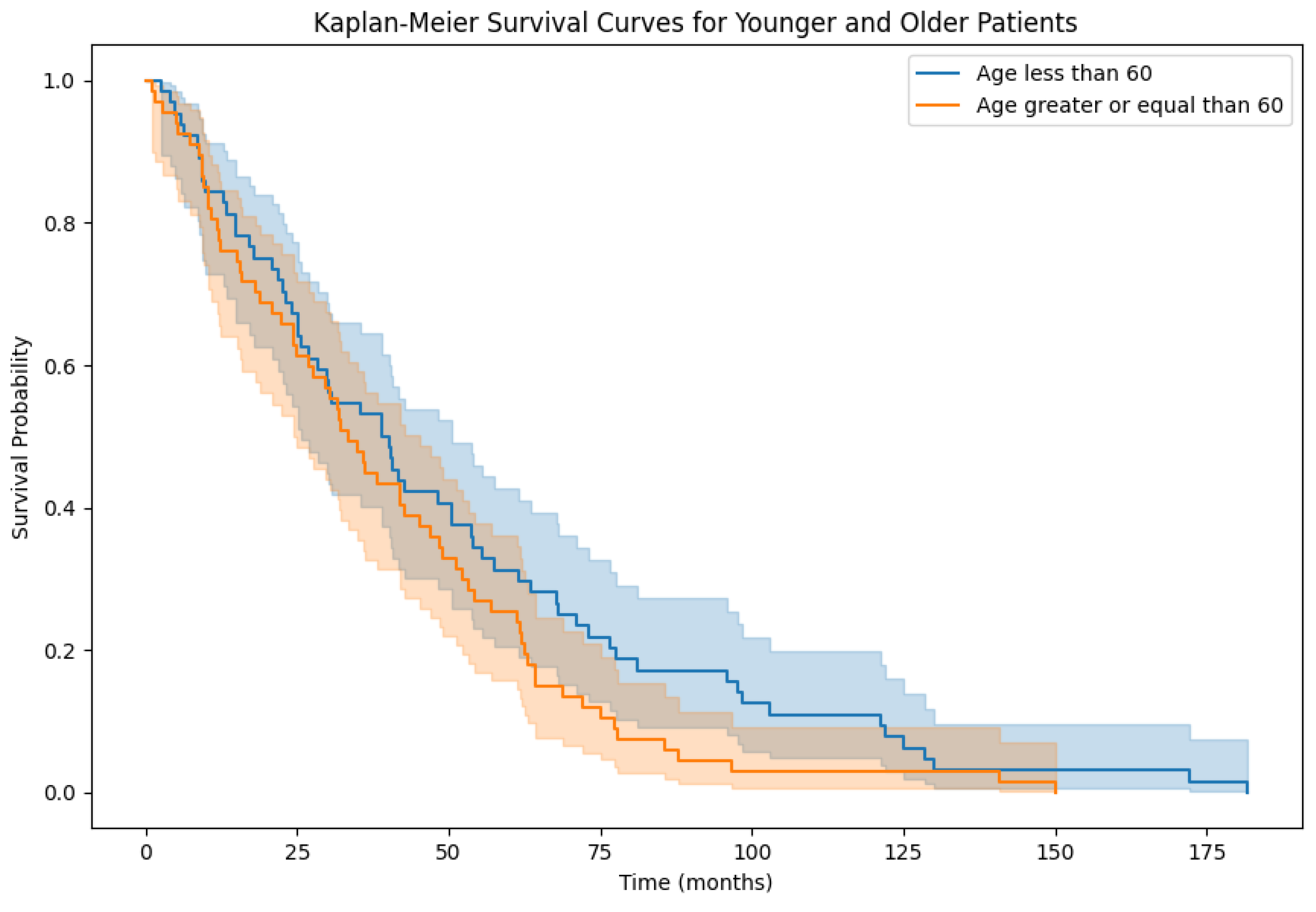
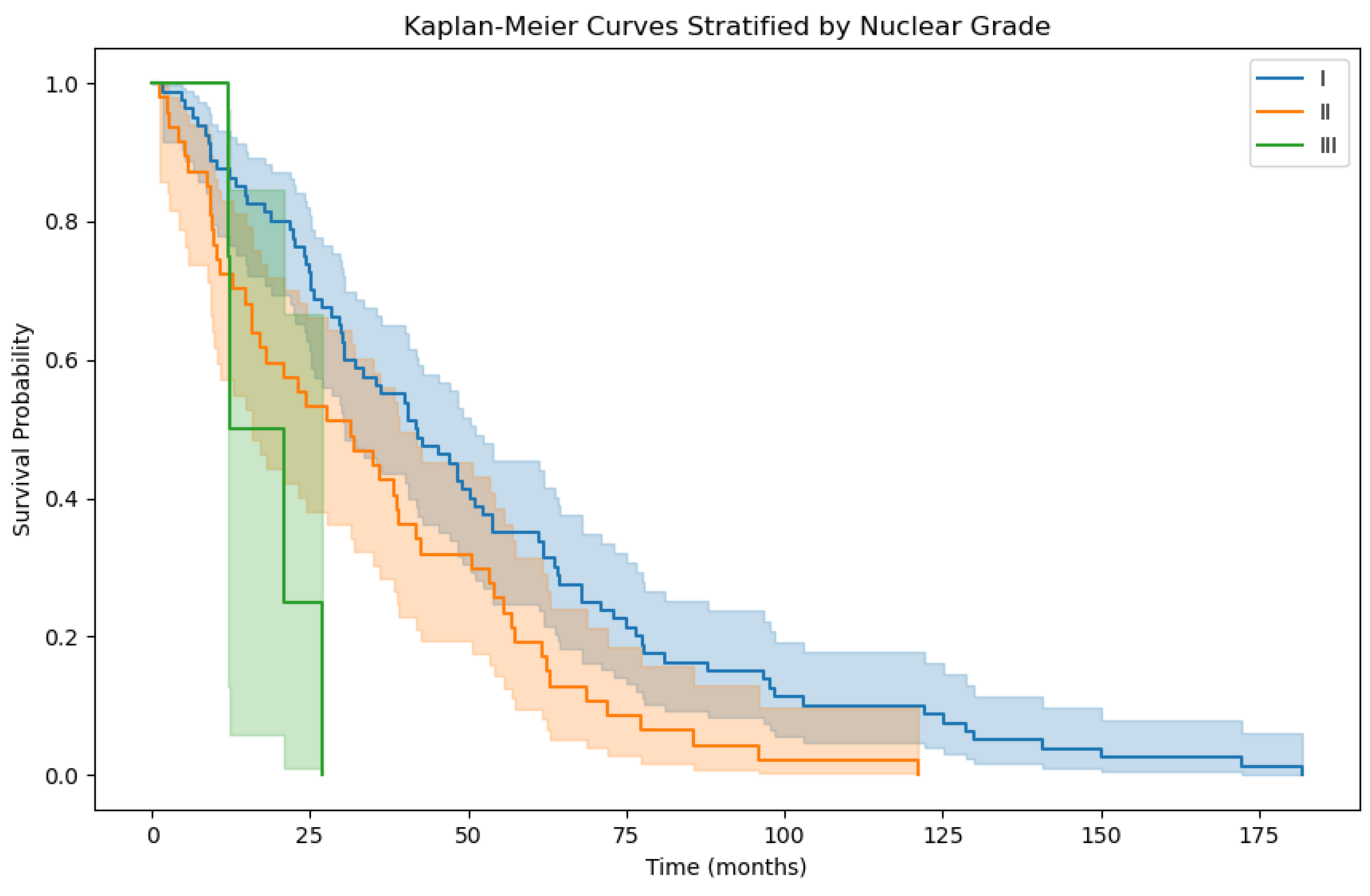
3.2. Overall Survival Factor Analysis Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| OS | Overall Survival |
| EMPeM | Epithelioid Malignant Peritoneal Mesothelioma |
| CNN | Convolutional Neural Network |
| WSI | Whole-Slide Image |
| PCI | Peritoneal Cancer Index |
| ResNet | Residual Network |
| HIPEC | Hyperthermic Intraperitoneal Chemotherapy |
| SVM | Support Vector Machine |
| RSF | Random Survival Forest |
Appendix A
References
- Boffetta, P. Epidemiology of peritoneal mesothelioma: A review. Ann. Oncol. 2007, 18, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Bhagwandin, S.; Labow, D.M. Malignant peritoneal mesothelioma: A review. Ann. Transl. Med. 2017, 5, 236. [Google Scholar] [CrossRef] [PubMed]
- Van der Laak, J.; Litjens, G.; Ciompi, F. Deep learning in histopathology: The path to the clinic. Nat. Med. 2021, 27, 775–784. [Google Scholar] [CrossRef]
- Qaiser, T.; Lee, C.Y.; Vandenberghe, M.; Yeh, J.; Gavrielides, M.; Hipp, J.; Scott, M.; Reischl, J. Usability of deep learning and HE images predict disease outcome-emerging tool to optimize clinical trials. NPJ Precis. Oncol. 2022, 6, 37. [Google Scholar] [CrossRef]
- Hess, L.M.; Brnabic, A.; Mason, O.; Lee, P.; Barker, S. Relationship between Progression-free Survival and Overall Survival in Randomized Clinical Trials of Targeted and Biologic Agents in Oncology. J. Cancer 2019, 10, 3717–3727. [Google Scholar] [CrossRef] [PubMed]
- Ishwaran, H.; Kogalur, U.B.; Blackstone, E.H.; Lauer, M.S. Random survival forests. Ann. Appl. Stat. 2008, 2, 841–860. [Google Scholar] [CrossRef]
- Ding, Z. The application of support vector machine in survival analysis. In Proceedings of the 2011 2nd International Conference on Artificial Intelligence, Management Science and Electronic Commerce (AIMSEC), Zhengzhou, China, 8–10 August 2011; pp. 6816–6819. [Google Scholar]
- Zhu, X.; Yao, J.; Huang, J. Deep convolutional neural network for survival analysis with pathological images. In Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, 15–18 December 2016; pp. 544–547. [Google Scholar]
- Zhu, X.; Yao, J.; Zhu, F.; Huang, J. WSISA: Making Survival Prediction From Whole Slide Histopathological Images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6855–6863. [Google Scholar]
- Laleh, N.G.; Echle, A.; Muti, H.S.; Hewitt, K.J.; Volkmar, S.; Kather, J.N. Deep Learning for interpretable end-to-end survival (E-ESurv) prediction in gastrointestinal cancer histopathology. In Proceedings of the MICCAI Workshop on Computational Pathology, Online, 27 September–1 October 2021; Volume 156, pp. 81–93. [Google Scholar]
- Courtiol, P.; Maussion, C.; Moarii, M.; Pronier, E.; Pilcer, S.; Sefta, M.; Manceron, P.; Toldo, S.; Zaslavskiy, M.; Le Stang, N.; et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat. Med. 2019, 25, 1519–1525. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Liu, L.; Tang, Z.; Chen, Z.; Ma, G.; Dong, J.; Zhang, X.; Yang, L.; Zheng, Q. Explainable Survival Analysis with Convolution-Involved Vision Transformer. In Proceedings of the AAAI Conference on Artificial Intelligence, Singapore, China, 17–20 June 2022; Volume 36, pp. 2207–2215. [Google Scholar]
- Steck, H.; Krishnapuram, B.; Dehing-oberije, C.; Lambin, P.; Raykar, V.C. On Ranking in Survival Analysis: Bounds on the Concordance Index. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 3–6 December 2007; Volume 20, pp. 1209–1216. [Google Scholar]
- Wulczyn, E.; Steiner, D.F.; Xu, Z.; Sadhwani, A.; Wang, H.; Flament-Auvigne, I.; Mermel, C.H.; Chen, P.H.C.; Liu, Y.; Stumpe, M.C. Deep learning-based survival prediction for multiple cancer types using histopathology images. PLOS ONE 2020, 15, e0233678. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gao, Q.; Yi, X.; Zhang, X.; Zhang, Y.; Zhang, D.; Liò, P.; Bain, C.; Bassed, R.; Li, S.; et al. Surformer: An interpretable pattern-perceptive survival transformer for cancer survival prediction from histopathology whole slide images. Comput. Methods Programs Biomed. 2023, 241, 107733. [Google Scholar] [CrossRef] [PubMed]
- French National Registry of Rare Peritoneal Surface Malignancies (RENAPE). Available online: https://clinicaltrials.gov/study/NCT02834169 (accessed on 10 September 2023).
- Echle, A.; Rindtorff, N.T.; Brinker, T.J.; Luedde, T.; Pearson, A.T.; Kather, J.N. Deep learning in cancer pathology: A new generation of clinical biomarkers. Br. J. Cancer 2021, 124, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Wsi-Tile-Cleanup. Available online: https://github.com/lucasrla/wsi-tile-cleanup (accessed on 23 September 2023).
- Jiang, J.; Trundle, P.; Ren, J. Medical image analysis with artificial neural networks. Comput. Med. Imaging Graph. 2010, 34, 617–631. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Toğaçar, M.; Ergen, B.; Tümen, V. Use of dominant activations obtained by processing OCT images with the CNNs and slime mold method in retinal disease detection. Biocybern. Biomed. Eng. 2022, 42, 646–666. [Google Scholar] [CrossRef]
- Mormont, R.; Geurts, P.; Marée, R. Multi-Task Pre-Training of Deep Neural Networks for Digital Pathology. IEEE J. Biomed. Health Inform. 2021, 25, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Scikit-Survival. Available online: https://scikit-survival.readthedocs.io/en/stable/ (accessed on 13 December 2023).
- Hosna, A.; Merry, E.; Gyalmo, J.; Alom, Z.; Aung, Z.; Azim, M.A. Transfer learning: A friendly introduction. J. Big Data 2022, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Naffouje, S.A.; Tulla, K.A.; Salti, G.I. The impact of chemotherapy and its timing on survival in malignant peritoneal mesothelioma treated with complete debulking. Med. Oncol. 2018, 35, 69. [Google Scholar] [CrossRef] [PubMed]
- Salo, S.; Lantto, E.; Robinson, E.; Myllarniemi, M.; Laaksonen, S.; Salo, J.; Rantanen, T.; Ilonen, I. Prognostic role of radiological peritoneal cancer index in malignant peritoneal mesothelioma: National cohort study. Sci. Rep. 2020, 10, 13257. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Asao, T.; Tsutsumi, S.; Fujii, T.; Yamauchi, H.; Kigure, W.; Morita, H.; Yajima, R.; Suto, T.; Kuwano, H. Effectiveness of Intraperitoneal Hyperthermo-Chemotherapy for Malignant Peritoneal Mesothelioma and Estimation of its Effect by Repeated FDG-PET: A Case Report. Hepato-Gastroentorology 2011, 58, 861–864. [Google Scholar]
| Hyperparameter | Value |
|---|---|
| Number of patches per WSI | 250 |
| Batch size | 128 |
| Learning rate | 0.0001 |
| Dataset | Method | C-Index |
|---|---|---|
| EMPeM WSIs | SurvivalSVM [7] | 0.48 |
| EMPeM WSIs | Random Survival Forests [6] | 0.50 |
| EMPeM WSIs | DeepConvSurv [8] | 0.60 |
| EMPeM WSIs | EE-Surv [10] | 0.65 |
| EMPeM WSIs | MPeMSupervisedSurv | 0.66 |
| Dataset | Cohort | C-Index |
|---|---|---|
| EMPeM WSIs | Male patients | 0.60 |
| EMPeM WSIs | Female patients | 0.59 |
| Dataset | Cohort | C-Index |
|---|---|---|
| EMPeM WSIs | Patients with age ≥ 60 | 0.59 |
| EMPeM WSIs | Patients with age < 60 | 0.60 |
| Dataset | Cohort | C-Index |
|---|---|---|
| EMPeM WSIs | Patients with Neoadjuvant Chemotherapy | 0.61 |
| EMPeM WSIs | Patients without Neoadjuvant Chemotherapy | 0.59 |
| Dataset | Cohort | C-Index |
|---|---|---|
| EMPeM WSIs | Patients with Adjuvant Chemotherapy | 0.73 |
| EMPeM WSIs | Patients without Adjuvant Chemotherapy | 0.59 |
| Dataset | Cohort | C-Index |
|---|---|---|
| EMPeM WSIs | Patients with HIPEC | 0.57 |
| EMPeM WSIs | Patients without HIPEC | 0.59 |
| Dataset | Cohort | C-Index |
|---|---|---|
| EMPeM WSIs | Patients with | 0.64 |
| EMPeM WSIs | Patients with | 0.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadopoulos, K.M.; Barmpoutis, P.; Stathaki, T.; Kepenekian, V.; Dartigues, P.; Valmary-Degano, S.; Illac-Vauquelin, C.; Avérous, G.; Chevallier, A.; Laverriere, M.-H.; et al. Overall Survival Time Estimation for Epithelioid Peritoneal Mesothelioma Patients from Whole-Slide Images. BioMedInformatics 2024, 4, 823-836. https://doi.org/10.3390/biomedinformatics4010046
Papadopoulos KM, Barmpoutis P, Stathaki T, Kepenekian V, Dartigues P, Valmary-Degano S, Illac-Vauquelin C, Avérous G, Chevallier A, Laverriere M-H, et al. Overall Survival Time Estimation for Epithelioid Peritoneal Mesothelioma Patients from Whole-Slide Images. BioMedInformatics. 2024; 4(1):823-836. https://doi.org/10.3390/biomedinformatics4010046
Chicago/Turabian StylePapadopoulos, Kleanthis Marios, Panagiotis Barmpoutis, Tania Stathaki, Vahan Kepenekian, Peggy Dartigues, Séverine Valmary-Degano, Claire Illac-Vauquelin, Gerlinde Avérous, Anne Chevallier, Marie-Hélène Laverriere, and et al. 2024. "Overall Survival Time Estimation for Epithelioid Peritoneal Mesothelioma Patients from Whole-Slide Images" BioMedInformatics 4, no. 1: 823-836. https://doi.org/10.3390/biomedinformatics4010046
APA StylePapadopoulos, K. M., Barmpoutis, P., Stathaki, T., Kepenekian, V., Dartigues, P., Valmary-Degano, S., Illac-Vauquelin, C., Avérous, G., Chevallier, A., Laverriere, M.-H., Villeneuve, L., Glehen, O., Isaac, S., Hommell-Fontaine, J., Ng Kee Kwong, F., & Benzerdjeb, N. (2024). Overall Survival Time Estimation for Epithelioid Peritoneal Mesothelioma Patients from Whole-Slide Images. BioMedInformatics, 4(1), 823-836. https://doi.org/10.3390/biomedinformatics4010046