From Code to Cure: The Impact of Artificial Intelligence in Biomedical Applications
1. Introduction
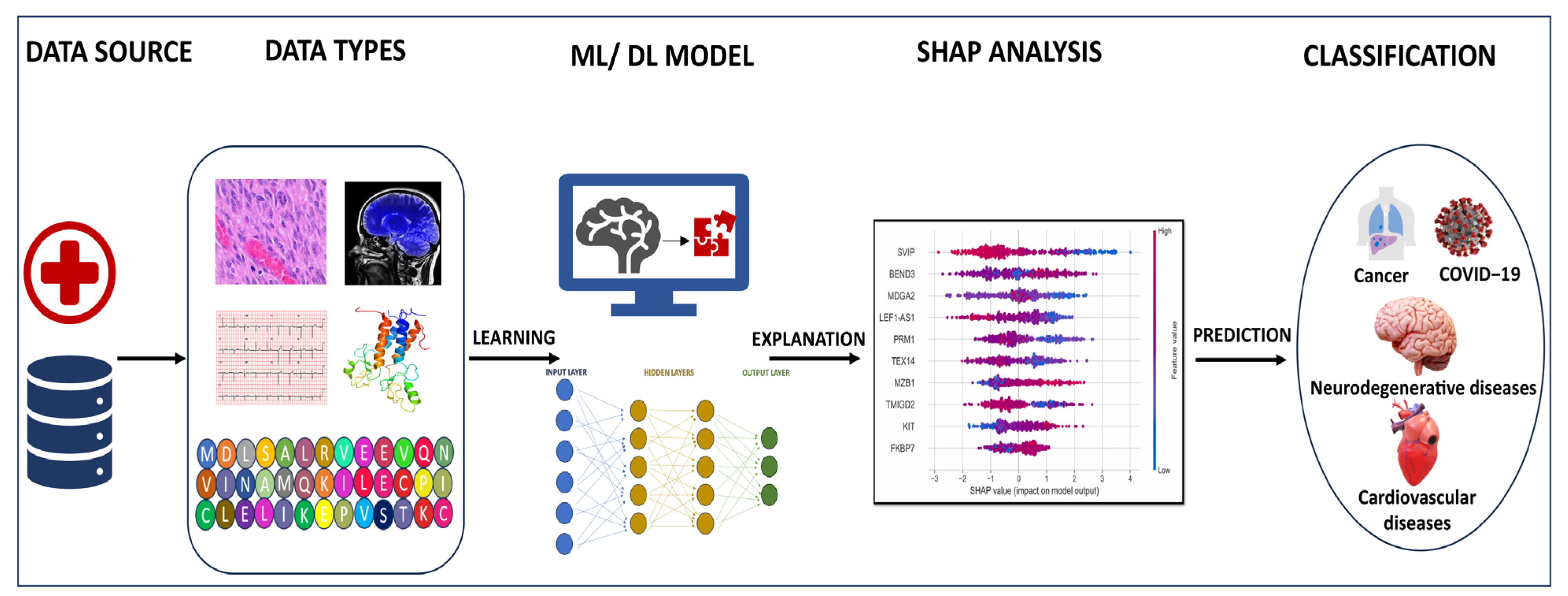
2. Applications of Machine Learning and Deep Learning
2.1. ML and DL in Cancer
2.2. Application in COVID-19 and Neurodegenerative Diseases
2.3. Applications in Omics
3. Challenges in ML and DL
4. Explainable AI
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Athanasopoulou, K.; Daneva, G.N.; Adamopoulos, P.G.; Scorilas, A. Artificial Intelligence: The Milestone in Modern Biomedical Research. BioMedInformatics 2022, 2, 727–744. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Novati, G.; Pan, J.; Bycroft, C.; Zemgulyte, A.; Applebaum, T.; Pritzel, A.; Wong, L.H.; Zielinski, M.; Avsec, Z. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 2023, 381, eadg7492. [Google Scholar] [CrossRef] [PubMed]
- Rajapaksha, L.T.; Vidanagamachchi, S.M.; Gunawardena, S.; Thambawita, V. An Open-Access Dataset of Hospitalized Cardiac-Arrest Patients: Machine-Learning-Based Predictions Using Clinical Documentation. BioMedInformatics 2024, 4, 34–49. [Google Scholar] [CrossRef]
- Cheng, E.R.; Steinhardt, R.; Ben Miled, Z. Predicting Childhood Obesity Using Machine Learning: Practical Considerations. BioMedInformatics 2022, 2, 184–203. [Google Scholar] [CrossRef]
- Bai, Q.; Tan, S.; Xu, T.; Liu, H.; Huang, J.; Yao, X. MolAICal: A soft tool for 3D drug design of protein targets by artificial intelligence and classical algorithm. Brief. Bioinform. 2021, 22(3), bbaa161. [Google Scholar] [CrossRef]
- Heid, E.; Greenman, K.P.; Chung, Y.; Li, S.; Graff, D.; Vermeire, F.H.; Wu, H.; Green, W.H.; McGill, C.J. Chemprop: A Machine Learning Package for Chemical Property Prediction. J. Chem. Inf. Model. 2023, 64, 9–17. [Google Scholar] [CrossRef]
- Gentile, F.; Yaacoub, J.C.; Gleave, J.; Fernandez, M.; Ton, A.; Ban, F.; Stern, A.; Cherkasov, A. Artificial intelligence–enabled virtual screening of ultra-large chemical libraries with deep docking. Nat. Protoc. 2022, 17, 672–697. [Google Scholar] [CrossRef]
- Egwom, O.J.; Hassan, M.; Tanimu, J.J.; Hamada, M.; Ogar, O.M. An LDA–SVM Machine Learning Model for Breast Cancer Classification. BioMedInformatics 2022, 2, 345–358. [Google Scholar] [CrossRef]
- Carreras, J.; Kikuti, Y.Y.; Miyaoka, M.; Hiraiwa, S.; Tomita, S.; Ikoma, H.; Kondo, Y.; Ito, A.; Hamoudi, R.; Nakamura, N. The Use of the Random Number Generator and Artificial Intelligence Analysis for Dimensionality Reduction of Follicular Lymphoma Transcriptomic Data. BioMedInformatics 2022, 2, 268–280. [Google Scholar] [CrossRef]
- Vermeulen, C.; Pagès-Gallego, M.; Kester, L.; Kranendonk, M.E.G.; Wesseling, P.; Verburg, N.; Hamer, P.d.W.; Kooi, E.J.; Dankmeijer, L.; van der Lugt, J.; et al. Ultra-fast deep-learned CNS tumour classification during surgery. Nature 2023, 622, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Carreras, J.; Kikuti, Y.Y.; Roncador, G.; Miyaoka, M.; Hiraiwa, S.; Tomita, S.; Ikoma, H.; Kondo, Y.; Ito, A.; Shiraiwa, S.; et al. High Expression of Caspase-8 Associated with Improved Survival in Diffuse Large B-Cell Lymphoma: Machine Learning and Artificial Neural Networks Analyses. BioMedInformatics 2021, 1, 18–46. [Google Scholar] [CrossRef]
- Pandey, M.; Gromiha, M.M. Predicting potential residues associated with lung cancer using deep neural network. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2020, 822, 111737. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.; Gromiha, M.M. MutBLESS: A tool to identify disease-prone sites in cancer using deep learning. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2023, 1869, 166721. [Google Scholar] [CrossRef]
- Pandey, M.; Anoosha, P.; Yesudhas, D.; Gromiha, M.M. Identification of potential driver mutations in glioblastoma using machine learning. Brief. Bioinform. 2022, 23, bbac451. [Google Scholar] [CrossRef] [PubMed]
- Shihab, H.A.; Gough, J.; Cooper, D.N.; Stenson, P.D.A.; Barker, G.L.; Edwards, K.J.; Day, I.N.M.; Gaunt, T.R. Predicting the Functional, Molecular, and Phenotypic Consequences of Amino Acid Substitutions using Hidden Markov Models. Hum. Mutat. 2012, 34, 57–65. [Google Scholar] [CrossRef]
- Reva, B.; Antipin, Y.; Sander, C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011, 39, e118. [Google Scholar] [CrossRef]
- Sim, N.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Liu, X.; Shen, J.; Li, Z.; Sang, Y.; Wu, X.; Zha, Y.; Liang, W.; Wang, C.; Wang, K.; et al. Clinically Applicable AI System for Accurate Diagnosis, Quantitative Measurements, and Prognosis of COVID-19 Pneumonia Using Computed Tomography. Cell 2020, 181, 1423–1433.e11. [Google Scholar] [CrossRef] [PubMed]
- Ibrokhimov, B.; Kang, J. Deep Learning Model for COVID-19-Infected Pneumonia Diagnosis Using Chest Radiography Images. BioMedInformatics 2022, 2, 654–670. [Google Scholar] [CrossRef]
- Montazeri, M.; ZahediNasab, R.; Farahani, A.; Mohseni, H.; Ghasemian, F. Machine Learning Models for Image-Based Diagnosis and Prognosis of COVID-19: Systematic Review. JMIR Med. Inform. 2021, 9, e25181. [Google Scholar] [CrossRef]
- Rawat, P.; Sharma, D.; Pandey, M.; Prabakaran, R.; Gromiha, M.M. Understanding the mutational frequency in SARS-CoV-2 proteome using structural features. Comput. Biol. Med. 2022, 147, 105708. [Google Scholar] [CrossRef]
- Sharma, D.; Rawat, P.; Greiff, V.; Janakiraman, V.; Gromiha, M.M. Predicting the immune escape of SARS-CoV-2 neutralizing antibodies upon mutation. Biochim. Biophys. Acta Mol. Basis Dis. 2024, 1870, 166959. [Google Scholar] [CrossRef]
- Prado, J.J.; Rojas, I. Machine Learning for Diagnosis of Alzheimer’s Disease and Early Stages. BioMedInformatics 2021, 1, 182–200. [Google Scholar] [CrossRef]
- Rangaswamy, U.; Dharshini, S.P.; Yesudhas, D.; Gromiha, M. VEPAD—Predicting the effect of variants associated with Alzheimer’s disease using machine learning. Comput. Biol. Med. 2020, 124, 103933. [Google Scholar] [CrossRef]
- Kulandaisamy, A.; Parvathy Dharshini, S.A.; Gromiha, M.M. Alz-Disc: A Tool to Discriminate Disease-causing and Neutral Mutations in Alzheimer’s Disease. Comb. Chem. High Throughput Screen 2023, 26, 769–777. [Google Scholar] [PubMed]
- Keles, A.; Keles, A.; Keles, M.B.; Okatan, A. PARNet: Deep neural network for the diagnosis of Parkinson’s disease. Multimed. Tools Appl. 2023, 1–13. [Google Scholar] [CrossRef]
- Kakati, T.; Bhattacharyya, D.K.; Kalita, J.K.; Norden-Krichmar, T.M. DEGnext: Classification of differentially expressed genes from RNA-seq data using a convolutional neural network with transfer learning. BMC Bioinform. 2022, 23, 17. [Google Scholar] [CrossRef] [PubMed]
- Bostanci, E.; Kocak, E.; Unal, M.; Guzel, M.S.; Acici, K.; Asuroglu, T. Machine Learning Analysis of RNA-seq Data for Diagnostic and Prognostic Prediction of Colon Cancer. Sensors 2022, 23, 3080. [Google Scholar] [CrossRef] [PubMed]
- Filho, U.L.; Pais, T.A.; Pais, R.J. Facilitating “Omics” for Phenotype Classification Using a User-Friendly AI-Driven Platform: Application in Cancer Prognostics. BioMedInformatics 2023, 3, 1071–1082. [Google Scholar] [CrossRef]
- Li, J.; Li, X.; Ma, J.; Wang, F.; Cui, S.; Ye, Z. Computed tomography–based radiomics machine learning classifiers to differentiate type I and type II epithelial ovarian cancers. Eur. Radiol. 2023, 33, 5193–5204. [Google Scholar] [CrossRef] [PubMed]
- Krause, T.; Wassan, J.T.; Mc Kevitt, P.; Wang, H.; Zheng, H.; Hemmje, M. Analyzing Large Microbiome Datasets Using Machine Learning and Big Data. BioMedInformatics 2021, 1, 138–165. [Google Scholar] [CrossRef]
- Shen, W.X.; Liang, S.R.; Jiang, Y.Y.; Chen, Y.Z. Enhanced metagenomic deep learning for disease prediction and consistent signature recognition by restructured microbiome 2D representations. Patterns 2023, 4, 100658. [Google Scholar] [CrossRef]
- Matsuzaka, Y.; Yashiro, R. Applications of Deep Learning for Drug Discovery Systems with BigData. BioMedInformatics 2022, 2, 603–624. [Google Scholar] [CrossRef]
- Eder, M.; Moser, E.; Holzinger, A.; Jeanquartier, F. Interpretable Machine Learning with Brain Image and Survival Data. BioMedInformatics 2022, 2, 492–510. [Google Scholar] [CrossRef]
- Anand, A.; Kadian, T.; Shetty, M.K.; Gupta, A. Explainable AI decision model for ECG data of cardiac disorders. Biomed. Signal Process. Control. 2022, 75, 103584. [Google Scholar] [CrossRef]
- Lalithadevi, B.; Krishnaveni, S.; Gnanadurai, J.S.C. A Feasibility Study of Diabetic Retinopathy Detection in Type II Diabetic Patients Based on Explainable Artificial Intelligence. J. Med. Syst. 2023, 47, 85. [Google Scholar] [CrossRef]
- Moon, I.; LoPiccolo, J.; Baca, S.C.; Sholl, L.M.; Kehl, K.L.; Hassett, M.J.; Liu, D.; Schrag, D.; Gusev, A. Machine learning for genetics-based classification and treatment response prediction in cancer of unknown primary. Nat. Med. 2023, 29, 2057–2067. [Google Scholar] [CrossRef]
- Ultsch, A.; Hoffmann, J.; Röhnert, M.A.; Von Bonin, M.; Oelschlägel, U.; Brendel, C.; Thrun, M.C. An Explainable AI System for the Diagnosis of High-Dimensional Biomedical Data. BioMedInformatics 2024, 4, 197–218. [Google Scholar] [CrossRef]
- Ramírez-Mena, A.; Andrés-León, E.; Alvarez-Cubero, M.J.; Anguita-Ruiz, A.; Martinez-Gonzalez, L.J.; Alcala-Fdez, J. Explainable artificial intelligence to predict and identify prostate cancer tissue by gene expression. Comput. Methods Programs Biomed. 2023, 240, 107719. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Das, A. Peripheral blood mononuclear cell derived biomarker detection using eXplainable Artificial Intelligence (XAI) provides better diagnosis of breast cancer. Comput. Biol. Chem. 2023, 104, 107867. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Chauhan, A.; Shetty, M.K.; Gupta, M.D.; Gupta, A. ECG-iCOVIDNet: Interpretable AI model to identify changes in the ECG signals of post-COVID subjects. Comput. Biol. Med. 2022, 146, 105540. [Google Scholar] [CrossRef] [PubMed]
- Kırboğa, K.K.; Küçüksille, E.U. Identifying Cardiovascular Disease Risk Factors in Adults with Explainable Artificial Intelligence. Anatol. J. Cardiol. 2023, 27, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Kamal, M.S.; Northcote, A.; Chowdhury, L.; Dey, N.; Crespo, R.G.; Herrera-Viedma, E. Alzheimer’s patient analysis using image and gene expression data and explainable-AI to present associated genes. IEEE Trans. Instrum. Meas. 2021, 70, 1–7. [Google Scholar] [CrossRef]
- Lötsch, J.; Kringel, D.; Ultsch, A. Explainable Artificial Intelligence (XAI) in Biomedicine: Making AI Decisions Trustworthy for Physicians and Patients. BioMedInformatics 2022, 2, 1–17. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S. A Unified Approach to Interpreting Model Predictions. Neural Inf. Process. Syst. 2017, 30, 1–10. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should I trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gromiha, M.M.; Preethi, P.; Pandey, M. From Code to Cure: The Impact of Artificial Intelligence in Biomedical Applications. BioMedInformatics 2024, 4, 542-548. https://doi.org/10.3390/biomedinformatics4010030
Gromiha MM, Preethi P, Pandey M. From Code to Cure: The Impact of Artificial Intelligence in Biomedical Applications. BioMedInformatics. 2024; 4(1):542-548. https://doi.org/10.3390/biomedinformatics4010030
Chicago/Turabian StyleGromiha, M. Michael, Palanisamy Preethi, and Medha Pandey. 2024. "From Code to Cure: The Impact of Artificial Intelligence in Biomedical Applications" BioMedInformatics 4, no. 1: 542-548. https://doi.org/10.3390/biomedinformatics4010030
APA StyleGromiha, M. M., Preethi, P., & Pandey, M. (2024). From Code to Cure: The Impact of Artificial Intelligence in Biomedical Applications. BioMedInformatics, 4(1), 542-548. https://doi.org/10.3390/biomedinformatics4010030





