Deep Machine Learning for Medical Diagnosis, Application to Lung Cancer Detection: A Review
Abstract
1. Introduction
- A Unique Grouping of Articles: Unlike traditional reviews that often categorize research based on methods or results, this review adopts a distinctive approach by grouping articles based on the types of databases utilized, offering a fresh perspective on the research landscape. This approach allows for a more comprehensive and nuanced understanding of the research landscape. By examining the types of databases that researchers are using, it’s therefore possible to gain insights into the availability, quality, and representativeness of data and the limitations of the current research. This information can be used to identify gaps in the literature, suggest new avenues for research, and develop more robust, and reliable research methods.
- Detailed Presentation of Widely-Used Databases: This review provides an extensive examination of the most commonly used databases in lung cancer detection, shedding light on their features, applications, and significance.
- Integration of Recent Research: In a rapidly evolving field, this review incorporates recent articles on the subject, ensuring a contemporary understanding of the latest developments and trends.
2. Methodology
3. Performance Metrics
4. Datasets
5. Deep Learning Approach for Lung Cancer Diagnosis
5.1. Deep Learning Techniques Using Public Databases
5.1.1. Deep Learning Techniques for Lung Cancer Using LIDC Dataset
5.1.2. Deep Learning Techniques for Lung Cancer Using LUNA16 Dataset
5.1.3. Deep Learning Techniques for Lung Cancer Using NLST Dataset
5.1.4. Deep Learning Techniques for Lung Cancer Using TCGA Dataset
5.1.5. Deep Learning Techniques for Lung Cancer Using JSRT Dataset
5.1.6. Deep Learning Techniques for Lung Cancer Using Kaggle DSB Dataset
5.1.7. Deep Learning Techniques for Lung Cancer Using Decathlon Dataset
5.1.8. Deep Learning Techniques for Lung Cancer Using Tianchi Dataset
5.1.9. Deep Learning Techniques for Lung Cancer Using Peking University Cancer Hospital Dataset
5.1.10. Deep Learning Techniques for Lung Cancer Using MSKCC Dataset
5.1.11. Deep Learning Techniques for Lung Cancer Using SEER Dataset
5.1.12. Deep Learning Techniques for Lung Cancer Using TCIA Dataset
5.2. Deep Learning Technics Using Proprietary Datasets
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ANN | Artificial Neural Network |
| AUC | Area Under the (ROC) Curve |
| BERT | Bidirectional Encoder Representation from Transformers |
| CT | Computed Tomography |
| DCNN | Deep Convolutional Neural Network |
| DL | Deep Learning |
| DNN | Deep Neural Network |
| GAN | Generative Adversarial Networks |
| IoU | Intersection Over Union |
| ML | Machine Learning |
| PET | Positron Emission Tomography |
| RNN | Recurrent Neural Network |
| SEA | Sparse AutoEncoder |
| ViT | Vision Transformer |
Appendix A. Details informations about Metrics and Results of Works Using Private/Public Datasets
| Metric | Definition | Note | Task |
|---|---|---|---|
| Accuracy | Quantifies the model’s effectiveness in correctly classifying both positive and negative instances | Classification | |
| Sensitivity (Recall) | Measures the proportion of actual positive cases that are correctly identified | Classification | |
| Specificity | Measures the proportion of actual negative cases that are correctly identified | Classification | |
| Precision | Measures the proportion of positive identifications that are actually correct | Classification | |
| F1-score | Harmonic mean of Precision and Recall, provides a balanced measure | Classification, Segmentation | |
| ROC | Receiver Operating Characteristic Curve | Graphical representation of Sensitivity vs (1-Specificity) | Classification |
| Dice (DSC) | Measures the similarity of two sets, commonly used for image segmentation | Segmentation | |
| IOU | Intersection over Union, measures the overlap between two sets | Segmentation |
| Ref. | Methodology | Dataset | Results(%) | Tasks |
|---|---|---|---|---|
| [33] | Application of CNN-ResNet50 combined with RBF SVM to 11 datasets generated by different deep extractors. | LIDC | Accuracy = 88.41, AUC = 93.19 | Classification |
| [34] | CNN has been trained with a learning rate of 0.01 and a batch size of 32. There are two convolution operations, each with 32 filters and a kernel size of 5. An aggregate layer with a kernel size of 2 is used to prevent excessive motion. | LIDC | Accuracy = 84.15, Sensitivity = 83.96 | Classification |
| [35] | DenseNet121 uses identity connections between layers, giving each layer access to the characteristics of all previous layers. This increases the use of information from all layers without increasing the complexity of the model. | LIDC | Accuracy = 87.67, Specificity = 93.38, Precision = 87.88, AUC = 93.79 | Classification |
| [37] | ConRad is designed to extract various features from cancer images using both biomarkers predicted by CBM and radiomic features. CBM predicts biomarkers such as subtlety, calcification, sphericity, margin, lobulation, spiculation, texture and diameter. | LIDC | AUC = 96.10 | Classification |
| [39] | DBN models the joint distribution of lung nodule images and hidden neural network layers. It is built iteratively using RBMs stacked on top of each other. Each MBR consists of a visible and a hidden layer. RBMs are trained using stochastic gradient descent and the contrastive divergence algorithm. | LIDC | Sensitivity = 73.40, Specificty = 82.20 | Classification |
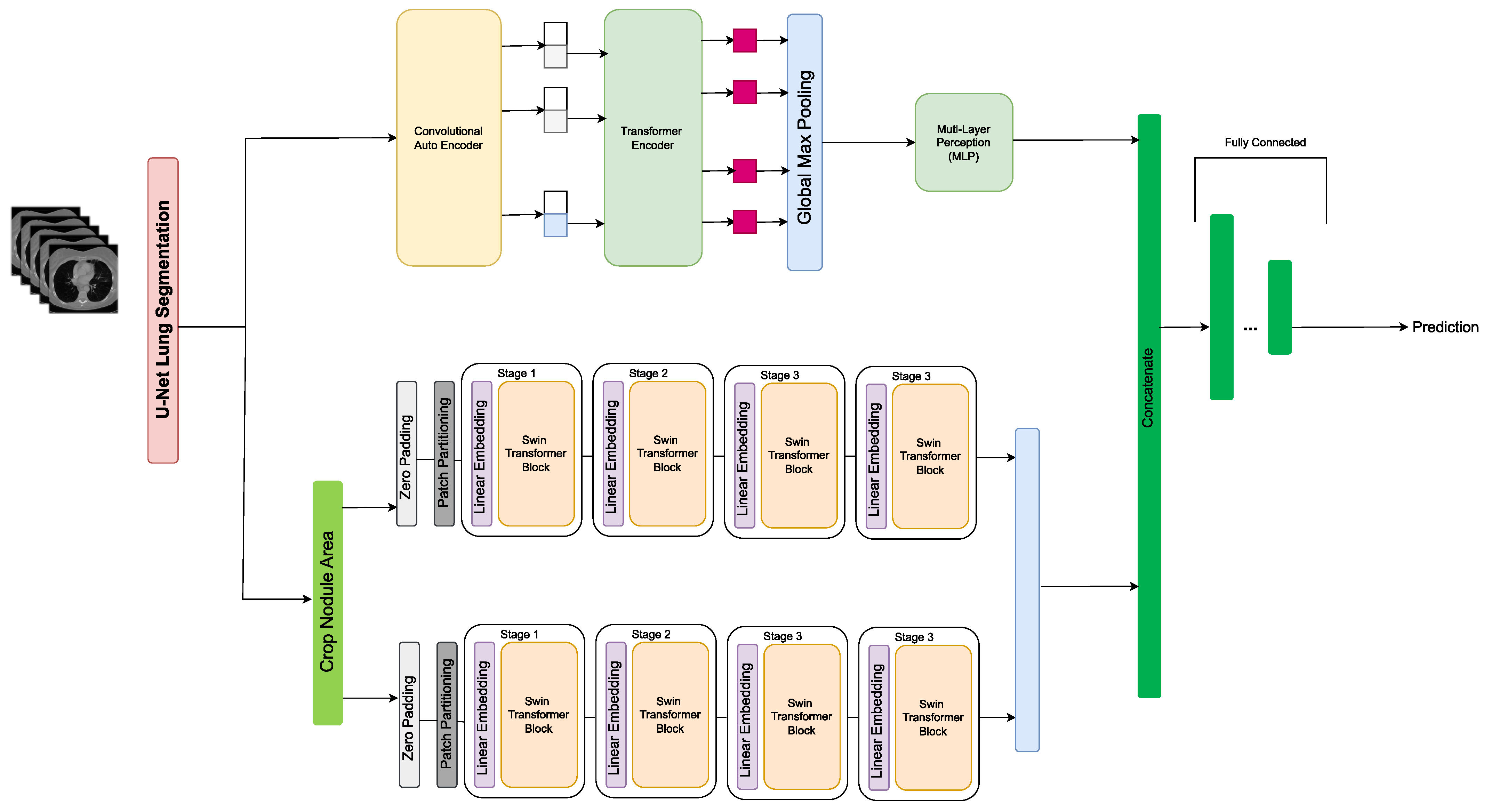
| [40] | CAET-SWin (Transformer) combines spatial and temporal features extracted using two parallel self-attention mechanisms to perform malignancy prediction based on CT images. It takes advantage of the 3D structure of unthin CT scans by simultaneously extracting inter-slice and intra-slice features. The extracted features are then merged to form the final output. | LIDC | Accuracy = 82.65, Sensitivity = 83.66, Specificity = 81.66 | Classification |
| [41] | LungNet extracts distinctive features from CT scans. It includes three 3D convolution layers with dimensions 16 × 3 × 3 and a 3D maximum aggregate layer with kernel size = 2, stride = 2. Three fully connected layers with decreasing feature vectors (128, 64, and 64) were combined to reduce the dimensionality of the learning convergence characteristics of the model. | LIDC | AUC = 85.00 | Classification |
| [43] | ResNet50 was used as a feature extractor. The extracted features were converted into feature vectors. Each nodule image was transformed into a digital vector representing the features extracted by the model. These feature vectors served as input data for the RBF SVM classifier. | LIDC | AUC = 93.10 | Detection |
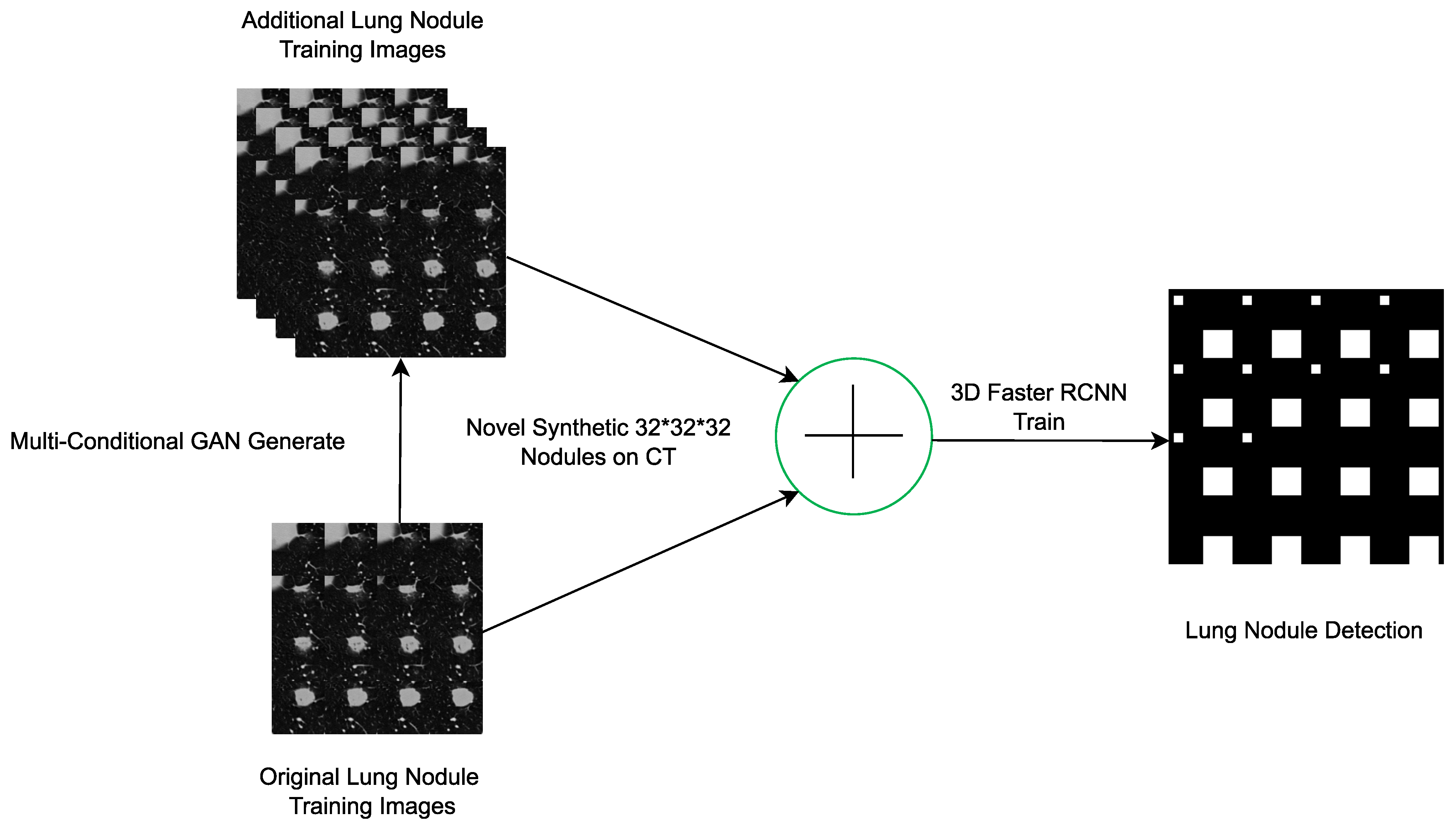
| [45] | 3D-CGAN generates realistic images of lung nodules with various conditions such as size, ground attenuation and presence of surrounding tissue. CGAN consists of a generator and two discriminators (context and node). The generator takes as input noise regions (noise bins) of fixed size and generates realistic lung nodes adhering to specified size and damping conditions. It also uses context information, such as surrounding tissue. | LIDC | CPM = 55.00 | Detection |
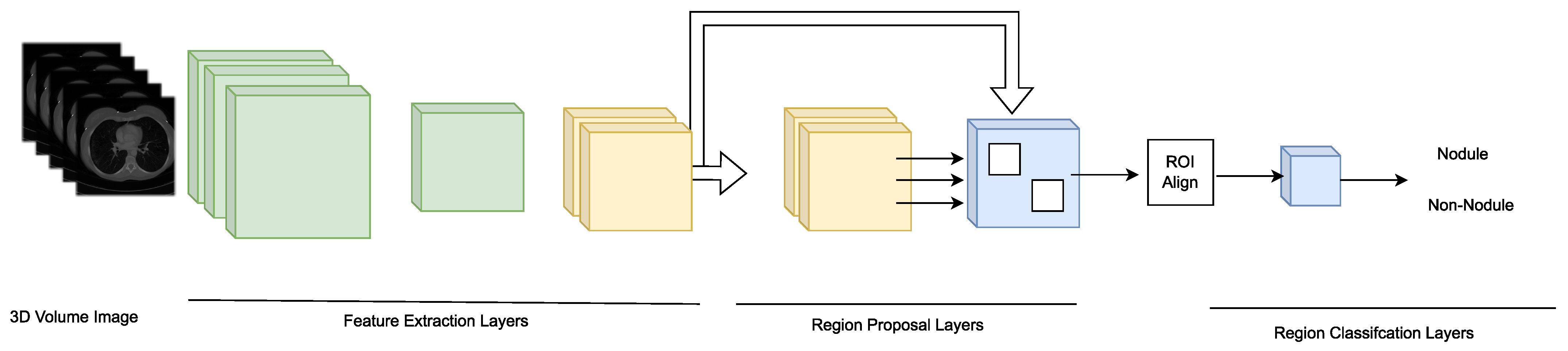
| [46] | A recognition system based on the Faster R-CNN model with a regional recommendation network consisting of 27 convolutional layers is proposed. The model uses 3D convolutional layers to extract three-dimensional information from chest CT images. | LIDC | Sensitivity = 96.00 | Detection |
| [47] | CNN classifies INCs into lung nodules through convolutional layers. Each layer applies filters to extract important information from the input image. These filters detect certain patterns in an image, such as edges, shapes or textures. | LIDC | Sensitivity = 94.01, AUC = 82.00 | Detection |
| [36] | ShCNN is trained using a WSLnO-based deformable model for lung region segmentation. The results show that the ShCNN model based on the proposed method outperforms several other existing methods, including CNN, IPCT+NN, dictionary-based segmentation+ShCNN and the deformable model based on WCBA+ShCNN. | LIDC | Precision = 93.03 | Segmentation |
| [48] | A CNN based on the VGG16Net architecture is trained to classify thoracic CT slices. It uses a full convolutional layer structure and a convolutional layer structure + a global average clustering layer (Conv + GAP), resulting in a FC layer. A nodule activation map (NAM) generated by a weighted average of the activation maps with weights learned in the FC layer. | LIDC | Accuracy = 86.60 | Segmentation |
| [49] | iW-Net works in 2 ways. Automatic segmentation: iW-Net receives as input a cube of fixed dimensions centered on the nodule, identified either manually by the user or automatically by the system. The network proposes an initial segmentation of the nodule. Interactive segmentation: If the user is not satisfied with the proposed segmentation, he can adjust it by manually inserting the ends of a line representing the nodule’s diameter. iW-Net then integrates this information to refine the segmentation. | LIDC | IoU = 55.00 | Segmentation |
| [51] | U-Net’s symmetrical architecture, consisting of encoder and decoder blocks with jump connections, enables detailed low-level information to be retained and combined with high-level information. This improves the accuracy of nodular pixel localization in the image. | LIDC | DSC = 83.00 | Segmentation |
| [53] | NoduleNet combines node detection, false positive reduction and node segmentation in a unified framework trained for multiple tasks. This unified approach improves model performance by solving several aspects of the node detection problem. | LIDC | CPM = 87.27, DSC = 83.10 | Segmentation |
| [52] | MV-CNN through Multivue structure incorporates three branches processing axial, coronal and sagittal views of CT images separately. This multiview approach enables the model to capture 3D information without requiring the input of a full 3D volume, thus reducing data redundancy and increasing efficiency. | LIDC | DSC = 77.67, ASD = 24.00 | Segmentation |
| [62] | 3D-MSViT processes information at different scales, capturing both fine details and global characteristics of nodules through the Patch Embedding Block. It processes the patch features at each scale dimension of CT images individually using the Local Transformer Block. Feature maps of different scales are scaled to uniform resolution and merged into a unified representation using the Global Transformer Block. | LUNA16, LIDC | Sensitivity = 97.81 | Classification |
| [60] | 3D ECA-ResNet introduces residual connections (skip connections), effectively alleviates the problem of gradient disappearance, enabling feature reuse and faster information transmission. It emphasizes channel information by explicitly modeling the correlation between them. It adaptively adjusts the feature channel, thereby strengthening the feature extraction capability of the network. | LUNA16, LIDC-IDRI | Accuracy = 94.89 | Classification |
| [57] | Swin Transformer transforms CT images into non-overlapping blocks through patching operations, including embedding, merging, and masking patches. This method allows efficient processing of images that are not naturally sequential, as is the case with CT images. Introducing connections between non-overlapping windows in consecutive Swin Transformer blocks improves network modeling capabilities. | LUNA16 | Accuracy = 82.26 | Classification |
| [59] | Dilated SegNet helps enlarge the receptive field of filters with its dilated convolution layers. This helps capture broader features of CT images, which is effectively useful for detecting smaller nodules. | LUNA16 | DSC = 89.00 | Segmentation |
| [56] | The framework uses an adapted version of Faster R-CNN which includes 2 RPNs and a deconvolution layer, to detect nodule candidates. Multiple CNNs are trained sequentially each handling more difficult cases than the previous model. This boosting approach helps increase sensitivity for detection of such small lung nodules. | LUNA16 | Sensitivity = 86.42 | Segmentation |
| [63] | A 3D CNN model is used to analyze full CT volumes end-to-end. This in-depth analysis can detect cancer candidate regions in CT volumes. This allows potentially cancerous areas to be precisely located, facilitating a more targeted assessment. An additional CNN model is developed to predict cancer risk based on the outputs of ROI detection models and full volume analysis. This model can also incorporate regions from previous CT scans of the patient, allowing for longitudinal comparison and more accurate risk assessment. | NLST | AUC = 94.40 | Classification |
| [66] | Time-distance ViT is proposed to interpret temporal distances in longitudinal and irregularly sampled medical images. The method uses continuous time vector embeddings to integrate temporal information into image analysis. Time encoding is performed with sinusoids at different frequencies, allowing a linear representation of temporal distances. TEM is used to modulate self-attention weights based on the time elapsed between image acquisitions. | NLST | AUC = 78.60 | Classification |
| [68] | CXR-LC based on a CNN, it combines radiographic images with basic information (age, gender, smoking status). It also leverages transfer learning from the Inception v4 network to predict all-cause mortality in the PLCO trial. | NLST | AUC = 75.50 | Classification |
| [64] | The proposed model is based on the VGG16 2D CNN architecture using transfer learning which made it possible to take advantage of a rich and varied knowledge base for a specific task, thus improving the accuracy and speed of learning of the model. The adaptation of this architecture made it possible to benefit from its 13 convolutional layers and its 4 pooling layers for efficient extraction of features from lung images. | NLST | Accuracy = 90.40, F1-score = 90.10 | Classification |
| [70] | FDTrans implements a preprocessing process to convert histopathological images to YCbCr color space and then to spatial spectrum via DCT. This step captures relevant frequency information, essential for distinguishing the subtle details of cancerous tissue. CSAM reallocates weights between low- and high-frequency information channels. CDTB processes features of Y, Cb and Cr channels, capturing long-term dependencies and global contextual connections between different components of images. | TCGA | AUC = 93.16 | Classification |
| [69] | Gene Transformer combines multi-head attention mechanics with 1D layers. Multi-head attention allows the model to simultaneously process complex genomic information from thousands of genes from different patient samples. the attention mechanism sequentially selects subsets of genes and reveals a set of scores defining the importance of each gene for subtype classification, focusing only on genes relevant to a task. | TCGA | Accuracy = 100, Precision = 100, Recall = 100, F1-Score = 100 | Classification |
| [74] | The study explores the use of DenseNet121 combined with transfer learning. The structure of DenseNet121 solves the problem of the disappearing gradient. The model also provides CAMs to identify the location of lung nodules. This allows visualization of the most salient regions on the images used to identify the output class. | JSRT | Specificity = 74.96 | Classification |
| [75] | U-Net is used to accurately segment the left and right lung fields in standard CXRs. This segmentation allows the lungs to be isolated from the heart and other parts in the images, thus leading to a more focused analysis of suspicious lesions and lung nodules. | JSRT | Accuracy = 96.00 | Segmentation |
| [76] | The study compares the approaches and performances of award-winning algorithms developed during the Kaggle Data Science Bowl. U-Net has been commonly used for lung nodule segmentation. The study found substantial performance variations in the public and final test sets. Transfer learning has been used in most classification algorithms, highlighting U-Net’s ability to adapt and learn from pre-existing data, which is essential when working with limited datasets or specific. | DSB | Logloss = 39.97 | Segmentation |
| [77] | The VGG-like 3D multipath network takes advantage of multiple paths to process 3D volumetric data which allows a better understanding of the spatial structure of lung nodules. The network is able to distinguish not only the presence of lung nodules but also classify their level of malignancy, an essential step in the early diagnosis and treatment planning of lung cancer. | LIDC, LUNA16, DSB | Accuracy = 95.60, Logloss = 38.77, DSC = 90.00 | Segmentation |
| [80] | TSFMUNet integrates a transformer module to process anisotropic data in CT images. This integration allows the model to more effectively adapt to variations in slice spacing, a common challenge in medical images. The model encodes information along the Z axis by representing the feature maps of a cut as a weighted sum of these maps and the feature maps of neighboring cuts. | MSD | Dice = 87.17 | Segmentation |
| [78] | The system consists of two main components: a segmentation part based on UNETR and a classification part based on a self-supervised network. UNETR uses transformers as an encoder to efficiently capture global multiscale information, thereby learning sequential representations of the input volume. For the classification of segmented nodules, the system uses a self-supervised architecture. This architecture focuses on predicting the same class for two different perspectives of the same sample, allowing labels and representations to be learned simultaneously in a single end-to-end process. | MSD | Accuracy = 98.77, Accuracy = 97.83 | Classification, Segmentation |
| [81] | Proposes an integrated framework for the detection of pulmonary nodules from low-dose CT scans using a model based on a 3D CNN and a 3D RPN network. This approach combines the steps of nodule screening and false positive reduction into a single jointly trained model. 3D RPN is adapted from the Faster-RCNN model for generating nodule candidates. | Tianchi | CPM = 86.60 | Detection |
| [82] | Amalgamated-CNN before segmenting CT images uses a non-sharpening mask to enhance the signal from nodules. It includes three separate CNN networks (CNN-1, CNN-2, CNN-3) with different input sizes and number of layers. This multi-layer design allows the model to process and analyze images at different scales. It uses AdaBoost classifier to merge the results. | Tianchi, LUNA16 | Sensitivity = 85.10 | Segmentation |
| [83] | DCNN consists of two main steps: detection of nodule candidates and reduction of false positives. The model uses a 3D version of the Faster R-CNN network, inspired by U-Net. A 3D DCNN classifier is then used to finely discriminate between true nodules and false positives. | Tianchi | CPM = 81.50 | Segmentation |
| [87] | BERT-BTN is used for clinical entity extraction from Chinese CT reports. BERT is used to learn deep semantic representations of characters which is essential for understanding the complex context of CT reports. A BiLSTM layer is used after BERT to capture nested structures and latent dependencies of each character in reports. | PUCH | Macro-F1 = 85.96 | Named Entity Recognition |
| [89] | Clinical Transformer is an adaptation of the Transformer architecture for precision medicine aimed at modeling the relationships between molecular and clinical measurements and survival of cancer patients. It aims to model how a biomarker in the context of other clinical or molecular features can influence patient survival particularly in immunotherapy treatment. | MSKCC | C-Index = 61.00 | Predicting survival in cancer patients |
| [90] | ANN demonstrated superior ability to model complex, nonlinear decision boundaries although it had difficulty clearly distinguishing between survival classes of less than 6 months and 0.5 to 2 years. | SEER | Accuracy = 71.18 | Classification |
| [91] | DETR is based on Transformers and aims to fully automate the anatomical localization of lung cancer in PET/CT images. It integrates global attention to the entire image allowing better localization and classification of tumors. | TCIA | IoU = 80.00, Accuracy = 97.00 | Segmentation Classification |
| Ref. | Method | Dataset Size | Performance Metrics | Task |
|---|---|---|---|---|
| [92] | Retina U-Net is suitable for the detection of primary lung tumors and associated metastases at all stages on FDG-PET/CT images. it has been adapted and trained to specifically detect T, N, and M lesions on these images. It contains additional branches in the lower levels of the decoder for end-to-end object classification and bounding box regression. | 364 FDG-PET/CTs | Sensitivity = 86.20 | Classification |
| [93] | DCNN coupled with transfer learning with VGG16 and finetuning were used by converting 3D images into 2D. The optimization of the hyperparameters of the DCNN was carried out by random search which made it possible to identify the most effective parameters for this specific classification task. | 1236 patients | Accuracy = 68.00 | Classification |
| [94] | ODNN combined with LDA was used to extract deep features from lung CT images and reduce their dimensionality. It was optimized using MGSA to improve lung cancer classification. This optimization made it possible to refine the structure of the network and improve its classification performance. | 50 lung cancer CT images | Sensitivity = 96.20 | Classification |
| [95] | ResNet is used to analyze exosomes in blood plasma via SERS. The model was trained to distinguish between exosomes derived from normal cells and those from lung cancer cell lines. | 2150 cell-derived exosome data | Accuracy = 95.00 AUC = 91.20 | Classification |
| [96] | A system including a deep learning model is developed to improve lung cancer screening in rural China, using mobile CT scanners. The model was designed to identify suspicious nodule candidates from LDCT images. After nodule detection, the model evaluates the probability of malignancy of each nodule. | 12,360 participants | Recall = 95.07 | Classification |
| [98] | RCNN combined with transformers is applied to syndrome differentiation in traditional Chinese medicine (TCM) for lung cancer diagnosis. The model was designed to process unstructured medical records. The use of data augmentation helped improve the performance of the RCNN model. The integration of transformers made it possible to efficiently process long and complex text sequences. | 1206 clinical records | F1-score = 86.50 | Classification |
| [99] | 3D U-Net with dual inputs is used for both PET and CT. It features two parallel convolution paths for independent feature extraction from PET and CT images at multiple resolution levels, thereby optimizing the analysis of features specific to each type of imaging. The features extracted from the convolution arms were concatenated and fed the deconvolution path via skip connections. | 290 pairs of PET and CT | Mean DSC = 92.00 | Segmentation |
| [100] | Dual-input U-Net implements a two-step approach. In the first step a global U-Net processes the 3D PET/CT volume to extract the preliminary tumor area while in the second step a regional U-Net refines the segmentation on slices selected by the global U-Net. | 887 patients | N/A | Segmentation |
| [103] | 3D MAU-Net is an adaptation of U-Net. At the bottleneck of the U-Net, a dual attention module was integrated to model semantic interdependencies in the spatial and channel dimensions respectively. The model proposes a multiple attention module to adaptively recalibrate and fuse the multi-scale features from the dual attention module of the previous feature maps of the decoder and the corresponding features of the encoder. | 322 CT images | DSC = 86.67 | Segmentation |
| [105] | 3D U-Net is intended for automatic measurement of bone mineral density (BMD) using LDCT scanners for opportunistic osteoporosis screening. Using 3DU-Net combined with dense connections achieved a balance between performance and computation. | 200 annotated LDCT scans, 374 independent LDCT scans | AUC = 92.70 | Segmentation |
| [106] | Unlike detection methods that provide a bounding box or classification methods that determine malignancy from a single image, this CNN model is based on an encoder-decoder architecture that reduces the resolution of the feature map and improves the robustness of the model to noise and overfitting. | 629 radiographs with 652 nodules/masses | DSC = 52.00 | Segmentation |
| [107] | Mask-RCNN is an evolution of the Faster-RCNN model which replaces the ROI pooling layer of Faster-RCNN with a more efficient ROI Alignment layer. This modification allows a more precise correspondence between the output pixels and the input pixels thus effectively preserving the spatial data contained in the image. | 90 patients | Sensitivity = 82.91 | Segmentation |
| [108] | ResNet-50 is used to analyze maximum intensity projection (MIP) images from PET in lung cancer patients by extracting features from MIP images of anterior and lateral views. These MIP images are generated from 3D PET volumes, ResNet-50 being a 2D model avoids the difficulty of directly processing 3D volumetric data. | N/A | chi-square = 23.6 | Segmentation |
| [109] | HFFM combines the advantages of 2D and 3D neural networks. 3D CNN is capable of learning three-dimensional information from CT images while 2D CNN focuses on obtaining detailed semantic information. This combination allows the model to capture both the complex spatial features and fine details of CT images. | 60 patients | Dice = 87.60 | Segmentation |
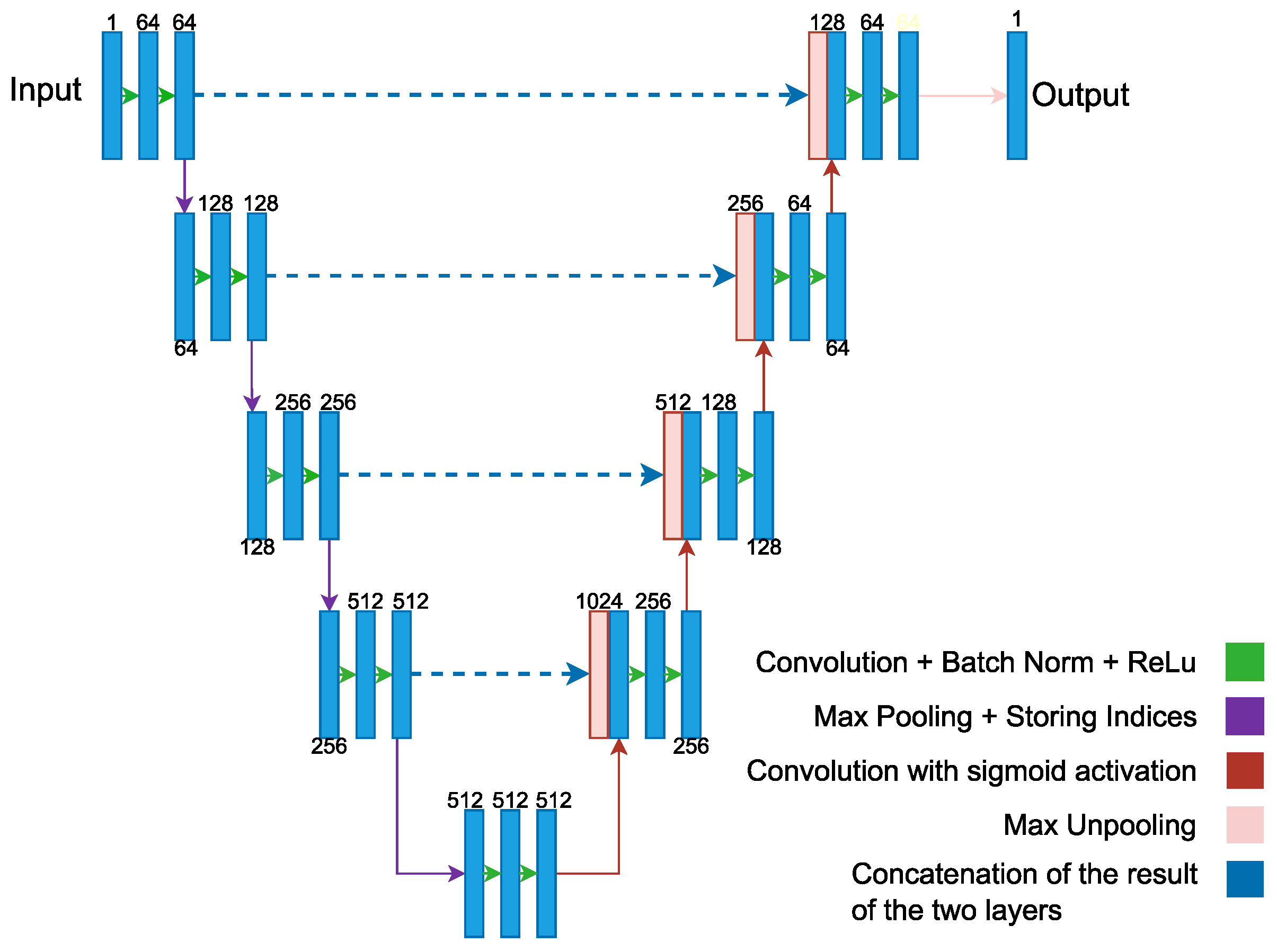
| [110] | SegNet uses a decoding process that is based on the pooling indices calculated during the maximum pooling stage of the corresponding encoder. This approach allows for efficient nonlinear sampling and eliminates the need to learn upsamples. | 240 participants | Sensitivity = 98.33 | Segmentation |
| [113] | The CNN used provided a significant improvement in the reproducibility of radiomic characteristics between the different reconstruction kernels. While the concordance of the radiomic features between the B30f and B50f cores was initially low, the application of the CNN improved them thus making the radiomic features much more reliable and comparable between the different cores. | 104 patients | CCC = 92.00 | Radiomic |
| [114] | BERT and RoBERTa were developed to extract information on social and behavioral determinants of health (SBDoH) from unstructured clinical text in electronic health records (EHR). | 161,933 clinical notes | Strict-F1-score = 87.91 Lenient-F1-score = 89.99 | Extract SBDoH |
| [115] | The deep learning-based CAD system significantly increased the diagnostic yield for detecting new visible lung metastases. The actual positive detection rate was higher in the CAD-assisted interpretation group than in the conventional interpretation group. | 2916 chest radiographs from 1521 patients | 80.00 | Evaluate deep learning-based CAD system |
References
- World Health Organization. Cancer. 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 18 September 2023).
- Freitas, C.; Sousa, C.; Machado, F.; Serino, M.; Santos, V.; Cruz-Martins, N.; Teixeira, A.; Cunha, A.; Pereira, T.; Oliveira, H.P.; et al. The role of liquid biopsy in early diagnosis of lung cancer. Front. Oncol. 2021, 11, 634316. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Li, J.; Pei, Y.; Khurram, R.; Rehman, K.u.; Rasool, A.B. State-of-the-Art challenges and perspectives in multi-organ cancer diagnosis via deep learning-based methods. Cancers 2021, 13, 5546. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.Y.; Chao, H.S.; Chen, Y.M. Application of artificial intelligence in lung cancer. Cancers 2022, 14, 1370. [Google Scholar]
- Dodia, S.; Annappa, B.; Mahesh, P.A. Recent advancements in deep learning based lung cancer detection: A systematic review. Eng. Appl. Artif. Intell. 2022, 116, 105490. [Google Scholar]
- Riquelme, D.; Akhloufi, M.A. Deep learning for lung cancer nodules detection and classification in CT scans. Ai 2020, 1, 28–67. [Google Scholar]
- Zareian, F.; Rezaei, N. Application of Artificial Intelligence in Lung Cancer Detection: The Integration of Computational Power and Clinical Decision-Making; Springer International Publishing: Edinburgh, UK, 2022; pp. 1–14. [Google Scholar]
- Huang, S.; Yang, J.; Shen, N.; Xu, Q.; Zhao, Q. Artificial intelligence in lung cancer diagnosis and prognosis: Current application and future perspective. Semin. Cancer Biol. 2023, 89, 30–37. [Google Scholar] [CrossRef]
- Qureshi, R.; Zou, B.; Alam, T.; Wu, J.; Lee, V.; Yan, H. Computational methods for the analysis and prediction of EGFR-mutated lung cancer drug resistance: Recent advances in drug design, challenges and future prospects. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 20, 238–255. [Google Scholar]
- Al-Tashi, Q.; Saad, M.B.; Muneer, A.; Qureshi, R.; Mirjalili, S.; Sheshadri, A.; Le, X.; Vokes, N.I.; Zhang, J.; Wu, J. Machine Learning Models for the Identification of Prognostic and Predictive Cancer Biomarkers: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 7781. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021, 88, 105906. [Google Scholar] [CrossRef]
- Cheng, P.M.; Montagnon, E.; Yamashita, R.; Pan, I.; Cadrin-Chenevert, A.; Perdigón Romero, F.; Chartrand, G.; Kadoury, S.; Tang, A. Deep learning: An update for radiologists. Radiographics 2021, 41, 1427–1445. [Google Scholar] [PubMed]
- Swift, A.; Heale, R.; Twycross, A. What are sensitivity and specificity? Evid.-Based Nurs. 2020, 2–4. [Google Scholar] [CrossRef]
- Mandrekar, J.N. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.; Goadrich, M. The relationship between Precision-Recall and ROC curves. In Proceedings of the 23rd International Conference on Machine learning; 2006; pp. 233–240. [Google Scholar]
- Goutte, C.; Gaussier, E. A probabilistic interpretation of precision, recall and F-score, with implication for evaluation. In Proceedings of the European Conference on Information Retrieval, Santiago de Compostela, Spain, 21–23 March 2005; pp. 345–359. [Google Scholar]
- Janssens, A.C.J.; Martens, F.K. Reflection on modern methods: Revisiting the area under the ROC Curve. Int. J. Epidemiol. 2020, 49, 1397–1403. [Google Scholar] [CrossRef] [PubMed]
- Zou, K.H.; Warfield, S.K.; Bharatha, A.; Tempany, C.M.; Kaus, M.R.; Haker, S.J.; Wells III, W.M.; Jolesz, F.A.; Kikinis, R. Statistical validation of image segmentation quality based on a spatial overlap index1: Scientific reports. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef]
- Sun, C.; Shrivastava, A.; Singh, S.; Gupta, A. Revisiting unreasonable effectiveness of data in deep learning era. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 843–852. [Google Scholar]
- Prior, F.; Smith, K.; Sharma, A.; Kirby, J.; Tarbox, L.; Clark, K.; Bennett, W.; Nolan, T.; Freymann, J. The public cancer radiology imaging collections of The Cancer Imaging Archive. Sci. Data 2017, 4, 1–7. [Google Scholar] [CrossRef]
- Setio, A.; Traverso, A.; De Bel, T.; Berens, M.S.; Bogaard, C.v.d.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Lung Nodule Analysis 2016 (LUNA16) Dataset. 2016. Available online: https://luna16.grand-challenge.org/ (accessed on 27 March 2023).
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Yanagita, S.; Imahana, M.; Suwa, K.; Sugimura, H.; Nishiki, M. Image Format Conversion to DICOM and Lookup Table Conversion to Presentation Value of the Japanese Society of Radiological Technology (JSRT) Standard Digital Image Database. Nihon Hoshasen Gijutsu Gakkai Zasshi 2016, 72, 1015–1023. [Google Scholar] [CrossRef]
- Team, N.L.S.T.R. National Lung Screening Trial (NLST) dataset. N. Engl. J. Med. 2011, 395–409. [Google Scholar]
- Samstein, R.M.; Lee, C.H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. Review The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. Onkol. 2015, 2015, 68–77. [Google Scholar] [CrossRef]
- National Cancer Institute. Surveillance, Epidemiology, and End Results Program (SEER) Database. 2023. Available online: https://seer.cancer.gov/data/ (accessed on 28 March 2023).
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; Van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar]
- Cancer Imaging Archive. A Large-Scale CT and PET/CT Dataset for Lung Cancer Diagnosis (Lung-PET-CT-Dx). 2013. Available online: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=70224216 (accessed on 28 March 2023). [CrossRef]
- Tianchi, A. Tianchi Medical AI Competition [Season 1]: Intelligent Diagnosis of Pulmonary Nodules. 2017. Available online: https://tianchi.aliyun.com/competition/entrance/231601/information (accessed on 6 April 2023).
- Booz Allen, K. Kaggle Data Science Bowl 2017. 2017. Available online: https://www.kaggle.com/c/data-science-bowl-2017 (accessed on 6 April 2023).
- Da Nóbrega, R.V.M.; Peixoto, S.A.; da Silva, S.P.P.; Rebouças Filho, P.P. Lung nodule classification via deep transfer learning in CT lung images. In Proceedings of the 2018 IEEE 31st International Symposium on Computer-Based Medical Systems (CBMS), Karlstad, Sweden, 18–21 June 2018; pp. 244–249. [Google Scholar]
- Song, Q.; Zhao, L.; Luo, X.; Dou, X. Using deep learning for classification of lung nodules on computed tomography images. J. Healthc. Eng. 2017, 2017, 8314740. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, H.; Yoon, S.W.; Won, D.; Srihari, K. Lung nodule diagnosis on 3D computed tomography images using deep convolutional neural networks. Procedia Manuf. 2019, 39, 363–370. [Google Scholar] [CrossRef]
- Shetty, M.V.; Tunga, S. Optimized Deformable Model-based Segmentation and Deep Learning for Lung Cancer Classification. J. Med. Investig. 2022, 69, 244–255. [Google Scholar]
- Brocki, L.; Chung, N.C. Integration of Radiomics and Tumor Biomarkers in Interpretable Machine Learning Models. Cancers 2023, 15, 2459. [Google Scholar] [CrossRef] [PubMed]
- Brocki, L.; Chung, N.C. ConRad. Available online: https://github.com/lenbrocki/ConRad (accessed on 2 May 2023).
- Hua, K.L.; Hsu, C.H.; Hidayati, S.C.; Cheng, W.H.; Chen, Y.J. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. Oncotargets Ther. 2015, 8, 2015–2022. [Google Scholar]
- Khademi, S.; Heidarian, S.; Afshar, P.; Naderkhani, F.; Oikonomou, A.; Plataniotis, K.; Mohammadi, A. Spatio-Temporal Hybrid Fusion of CAE and SWIn Transformers for Lung Cancer Malignancy Prediction. arXiv 2022, arXiv:2210.15297. [Google Scholar]
- Mukherjee, P.; Zhou, M.; Lee, E.; Schicht, A.; Balagurunathan, Y.; Napel, S.; Gillies, R.; Wong, S.; Thieme, A.; Leung, A.; et al. A shallow convolutional neural network predicts prognosis of lung cancer patients in multi-institutional computed tomography image datasets. Nat. Mach. Intell. 2020, 2, 274–282. [Google Scholar] [CrossRef]
- Mukherjee, P.; Zhou, M.; Lee, E.; Gevaert, O. LungNet: A Shallow Convolutional Neural Network Predicts Prognosis of Lung Cancer Patients in Multi-Institutional CT-Image Data. 2020. Available online: https://codeocean.com/capsule/5978670/tree/v1 (accessed on 18 March 2023).
- da Nobrega, R.V.M.; Reboucas Filho, P.P.; Rodrigues, M.B.; da Silva, S.P.; Dourado Junior, C.M.; de Albuquerque, V.H.C. Lung nodule malignancy classification in chest computed tomography images using transfer learning and convolutional neural networks. Neural Comput. Appl. 2020, 32, 11065–11082. [Google Scholar]
- Ridnik, T.; Ben-Baruch, E.; Noy, A.; Zelnik-Manor, L. Imagenet-21k pretraining for the masses. arXiv 2021, arXiv:2104.10972. [Google Scholar]
- Han, C.; Kitamura, Y.; Kudo, A.; Ichinose, A.; Rundo, L.; Furukawa, Y.; Umemoto, K.; Li, Y.; Nakayama, H. Synthesizing diverse lung nodules wherever massively: 3D multi-conditional GAN-based CT image augmentation for object detection. In Proceedings of the 2019 International Conference on 3D Vision (3DV), Quebec, QC, Canada, 16–19 September 2019; pp. 729–737. [Google Scholar]
- Katase, S.; Ichinose, A.; Hayashi, M.; Watanabe, M.; Chin, K.; Takeshita, Y.; Shiga, H.; Tateishi, H.; Onozawa, S.; Shirakawa, Y.; et al. Development and performance evaluation of a deep learning lung nodule detection system. BMC Med. Imaging 2022, 22, 203. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Huo, Y.; Liang, Z.; Li, L. Expert knowledge-infused deep learning for automatic lung nodule detection. J.-Ray Sci. Technol. 2019, 27, 17–35. [Google Scholar]
- Feng, X.; Yang, J.; Laine, A.F.; Angelini, E.D. Discriminative localization in CNNs for weakly-supervised segmentation of pulmonary nodules. In Proceedings of the Medical Image Computing and Computer Assisted Intervention- MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, 11–13 September 2017; Proceedings, Part III 20. pp. 568–576. [Google Scholar]
- Aresta, G.; Jacobs, C.; Araújo, T.; Cunha, A.; Ramos, I.; van Ginneken, B.; Campilho, A. iW-Net: An automatic and minimalistic interactive lung nodule segmentation deep network. Sci. Rep. 2019, 9, 11591. [Google Scholar] [CrossRef] [PubMed]
- Aresta, G.; Jacobs, C.; Araújo, T.; Cunha, A.; Ramos, I.; van Ginneken, B.; Campilho, A. iW-Net: Source Code. 2019. Available online: https://github.com/gmaresta/iW-Net (accessed on 25 March 2023).
- Rocha, J.; Cunha, A.; Mendonça, A.M. Conventional filtering versus u-net based models for pulmonary nodule segmentation in ct images. J. Med. Syst. 2020, 44, 81. [Google Scholar]
- Wang, S.; Zhou, M.; Gevaert, O.; Tang, Z.; Dong, D.; Liu, Z.; Jie, T. A multi-view deep convolutional neural networks for lung nodule segmentation. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Republic of Korea, 11–15 July 2017; pp. 1752–1755. [Google Scholar]
- Tang, H.; Zhang, C.; Xie, X. Nodulenet: Decoupled false positive reduction for pulmonary nodule detection and segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part VI 22. pp. 266–274. [Google Scholar]
- Tang, H.; Zhang, C. LungNet Code. Github. 2019. Available online: https://github.com/uci-cbcl/NoduleNet (accessed on 20 March 2023).
- Xiao, N.; Qiang, Y.; Bilal Zia, M.; Wang, S.; Lian, J. Ensemble classification for predicting the malignancy level of pulmonary nodules on chest computed tomography images. Oncol. Lett. 2020, 20, 401–408. [Google Scholar] [CrossRef]
- Xie, H.; Yang, D.; Sun, N.; Chen, Z.; Zhang, Y. Automated pulmonary nodule detection in CT images using deep convolutional neural networks. Pattern Recognit. 2019, 85, 109–119. [Google Scholar] [CrossRef]
- Sun, R.; Pang, Y.; Li, W. Efficient Lung Cancer Image Classification and Segmentation Algorithm Based on an Improved Swin Transformer. Electronics 2023, 12, 1024. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 10347–10357. [Google Scholar]
- Agnes, S.A.; Anitha, J. Appraisal of deep-learning techniques on computer-aided lung cancer diagnosis with computed tomography screening. J. Med. Phys. 2020, 45, 98. [Google Scholar]
- Yuan, H.; Wu, Y.; Dai, M. Multi-Modal Feature Fusion-Based Multi-Branch Classification Network for Pulmonary Nodule Malignancy Suspiciousness Diagnosis. J. Digit. Imaging 2022, 36, 617–626. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 11534–11542. [Google Scholar]
- Mkindu, H.; Wu, L.; Zhao, Y. 3D multi-scale vision transformer for lung nodule detection in chest CT images. Signal Image Video Process. 2023, 17, 2473–2480. [Google Scholar] [CrossRef]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Bates, J.H.; Matthew Kinsey, C. Discriminating TB lung nodules from early lung cancers using deep learning. Bmc Med. Inform. Decis. Mak. 2022, 22, 1–7. [Google Scholar]
- National Institute of Allergy and Infectious Diseases. National Institute of Allergy and Infectious Disease (NIAID) TB Portal. 2021. Available online: https://tbportals.niaid.nih.gov/ (accessed on 27 March 2023).
- Li, T.Z.; Xu, K.; Gao, R.; Tang, Y.; Lasko, T.A.; Maldonado, F.; Sandler, K.; Landman, B.A. Time-distance vision transformers in lung cancer diagnosis from longitudinal computed tomography. arXiv 2022, arXiv:2209.01676. [Google Scholar]
- Li, T. Time Distance Transformer Code. 2023. Available online: https://github.com/tom1193/time-distance-transformer (accessed on 18 September 2023).
- Lu, M.T.; Raghu, V.K.; Mayrhofer, T.; Aerts, H.J.; Hoffmann, U. Deep learning using chest radiographs to identify high-risk smokers for lung cancer screening computed tomography: Development and validation of a prediction model. Ann. Intern. Med. 2020, 173, 704–713. [Google Scholar]
- Khan, A.; Lee, B. Gene transformer: Transformers for the gene expression-based classification of lung cancer subtypes. arXiv 2021, arXiv:2108.11833. [Google Scholar]
- Cai, M.; Zhao, L.; Hou, G.; Zhang, Y.; Wu, W.; Jia, L.; Zhao, J.; Wang, L.; Qiang, Y. FDTrans: Frequency Domain Transformer Model for predicting subtypes of lung cancer using multimodal data. Comput. Biol. Med. 2023, 158, 106812. [Google Scholar] [CrossRef]
- Primakov, S.P.; Ibrahim, A.; van Timmeren, J.E.; Wu, G.; Keek, S.A.; Beuque, M.; Granzier, R.W.; Lavrova, E.; Scrivener, M.; Sanduleanu, S.; et al. Automated detection and segmentation of non-small cell lung cancer computed tomography images. Nat. Commun. 2022, 13, 3423. [Google Scholar] [CrossRef]
- Padhani, A.; Ollivier, L. The RECIST criteria: Implications for diagnostic radiologists. Br. J. Radiol. 2001, 74, 983–986. [Google Scholar]
- Primakov. DuneAI-Automated-Detection-and-Segmentation-of-non-Small-Cell-Lung-Cancer-Computed-Tomography-Images. 2022. Available online: https://github.com/primakov/DuneAI-Automated-detection-and-segmentation-of-non-small-cell-lung-cancer-computed-tomography-images (accessed on 15 March 2023).
- Ausawalaithong, W.; Thirach, A.; Marukatat, S.; Wilaiprasitporn, T. Automatic lung cancer prediction from chest X-ray images using the deep learning approach. In Proceedings of the 2018 11th Biomedical Engineering International Conference (BMEiCON), Chiang Mai, Thailand, 21–24 November 2018; pp. 1–5. [Google Scholar]
- Gordienko, Y.; Gang, P.; Hui, J.; Zeng, W.; Kochura, Y.; Alienin, O.; Rokovyi, O.; Stirenko, S. Deep learning with lung segmentation and bone shadow exclusion techniques for chest X-ray analysis of lung cancer. In Advances in Computer Science for Engineering and Education 13; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; pp. 638–647. [Google Scholar]
- Yu, K.H.; Lee, T.L.M.; Yen, M.H.; Kou, S.; Rosen, B.; Chiang, J.H.; Kohane, I.S. Reproducible machine learning methods for lung cancer detection using computed tomography images: Algorithm development and validation. J. Med. Internet Res. 2020, 22, e16709. [Google Scholar] [CrossRef]
- Tekade, R.; Rajeswari, K. Lung cancer detection and classification using deep learning. In Proceedings of the 2018 Fourth International Conference on Computing Communication Control and Automation (ICCUBEA), Pune, India, 6–18 August 2018; pp. 1–5. [Google Scholar]
- Said, Y.; Alsheikhy, A.; Shawly, T.; Lahza, H. Medical Images Segmentation for Lung Cancer Diagnosis Based on Deep Learning Architectures. Diagnostics 2023, 13, 546. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Guo, D.; Terzopoulos, D. A transformer-based network for anisotropic 3D medical image segmentation. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 8857–8861. [Google Scholar]
- Tang, H.; Liu, X.; Xie, X. An end-to-end framework for integrated pulmonary nodule detection and false positive reduction. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 859–862. [Google Scholar]
- Huang, W.; Xue, Y.; Wu, Y. A CAD system for pulmonary nodule prediction based on deep three-dimensional convolutional neural networks and ensemble learning. PLoS ONE 2019, 14, e0219369. [Google Scholar] [CrossRef]
- Tang, H.; Kim, D.R.; Xie, X. Automated pulmonary nodule detection using 3D deep convolutional neural networks. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 523–526. [Google Scholar]
- Fischer, A.M.; Yacoub, B.; Savage, R.H.; Martinez, J.D.; Wichmann, J.L.; Sahbaee, P.; Grbic, S.; Varga-Szemes, A.; Schoepf, U.J. Machine learning/deep neuronal network: Routine application in chest computed tomography and workflow considerations. J. Thorac. Imaging 2020, 35, S21–S27. [Google Scholar] [PubMed]
- Mohit, B. Named entity recognition. In Natural Language Processing of Semitic Languages; Springer: Berlin/Heidelberg, Germany, 2014; pp. 221–245. [Google Scholar]
- Zeng, D.; Liu, K.; Lai, S.; Zhou, G.; Zhao, J. Relation classification via convolutional deep neural network. In Proceedings of the COLING 2014, The 25th International Conference on Computational Linguistics: Technical Papers, Dublin, Ireland, 23–29 August 2014; pp. 2335–2344. [Google Scholar]
- Zhang, H.; Hu, D.; Duan, H.; Li, S.; Wu, N.; Lu, X. A novel deep learning approach to extract Chinese clinical entities for lung cancer screening and staging. BMC Med. Inform. Decis. Mak. 2021, 21, 214. [Google Scholar]
- Tenney, I.; Das, D.; Pavlick, E. BERT rediscovers the classical NLP pipeline. arXiv 2019, arXiv:1905.05950. [Google Scholar]
- Kipkogei, E.; Arango Argoty, G.A.; Kagiampakis, I.; Patra, A.; Jacob, E. Explainable Transformer-Based Neural Network for the Prediction of Survival Outcomes in Non-Small Cell Lung Cancer (NSCLC). medRxiv 2021. [Google Scholar] [CrossRef]
- Doppalapudi, S.; Qiu, R.G.; Badr, Y. Lung cancer survival period prediction and understanding: Deep learning approaches. Int. J. Med. Inform. 2021, 148, 104371. [Google Scholar]
- Barbouchi, K.; El Hamdi, D.; Elouedi, I.; Aïcha, T.B.; Echi, A.K.; Slim, I. A transformer-based deep neural network for detection and classification of lung cancer via PET/CT images. Int. J. Imaging Syst. Technol. 2023. [Google Scholar] [CrossRef]
- Weikert, T.; Jaeger, P.; Yang, S.; Baumgartner, M.; Breit, H.; Winkel, D.; Sommer, G.; Stieltjes, B.; Thaiss, W.; Bremerich, J.; et al. Automated lung cancer assessment on 18F-PET/CT using Retina U-Net and anatomical region segmentation. Eur. Radiol. 2023, 33, 4270–4279. [Google Scholar]
- Nishio, M.; Sugiyama, O.; Yakami, M.; Ueno, S.; Kubo, T.; Kuroda, T.; Togashi, K. Computer-aided diagnosis of lung nodule classification between benign nodule, primary lung cancer, and metastatic lung cancer at different image size using deep convolutional neural network with transfer learning. PLoS ONE 2018, 13, e0200721. [Google Scholar] [CrossRef]
- Lakshmanaprabu, S.; Mohanty, S.N.; Shankar, K.; Arunkumar, N.; Ramirez, G. Optimal deep learning model for classification of lung cancer on CT images. Future Gener. Comput. Syst. 2019, 92, 374–382. [Google Scholar]
- Shin, H.; Oh, S.; Hong, S.; Kang, M.; Kang, D.; Ji, Y.g.; Choi, B.H.; Kang, K.W.; Jeong, H.; Park, Y.; et al. Early-stage lung cancer diagnosis by deep learning-based spectroscopic analysis of circulating exosomes. ACS Nano 2020, 14, 5435–5444. [Google Scholar] [PubMed]
- Shao, J.; Wang, G.; Yi, L.; Wang, C.; Lan, T.; Xu, X.; Guo, J.; Deng, T.; Liu, D.; Chen, B.; et al. Deep learning empowers lung cancer screening based on mobile low-dose computed tomography in resource-constrained sites. Front.-Biosci.-Landmark 2022, 27, 212. [Google Scholar] [CrossRef]
- Su, X.L.; Wang, J.W.; Che, H.; Wang, C.F.; Jiang, H.; Lei, X.; Zhao, W.; Kuang, H.X.; Wang, Q.H. Clinical application and mechanism of traditional Chinese medicine in treatment of lung cancer. Chin. Med. J. 2020, 133, 2987–2997. [Google Scholar]
- Liu, Z.; He, H.; Yan, S.; Wang, Y.; Yang, T.; Li, G.Z. End-to-end models to imitate traditional Chinese medicine syndrome differentiation in lung cancer diagnosis: Model development and validation. JMIR Med. Inform. 2020, 8, e17821. [Google Scholar]
- Wang, S.; Mahon, R.; Weiss, E.; Jan, N.; Taylor, R.; McDonagh, P.; Quinn, B.; Yuan, L. Automated Lung Cancer Segmentation Using a Dual-Modality Deep Learning Network with PET and CT Images. Int. J. Radiat. Oncol. Biol. Phys. 2022, 114, e557–e558. [Google Scholar] [CrossRef]
- Park, J.; Kang, S.K.; Hwang, D.; Choi, H.; Ha, S.; Seo, J.M.; Eo, J.S.; Lee, J.S. Automatic Lung Cancer Segmentation in [18F] FDG PET/CT Using a Two-Stage Deep Learning Approach. Nucl. Med. Mol. Imaging 2022, 57, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, J.; Tan, T.; Teng, X.; Sun, X.; Zhao, H.; Liu, L.; Xiao, Y.; Lee, B.; Li, Y.; et al. Deep learning methods for lung cancer segmentation in whole-slide histopathology images—the acdc @ lunghp challenge 2019. IEEE J. Biomed. Health Inform. 2020, 25, 429–440. [Google Scholar] [CrossRef]
- Li, Z. Automatic Cancer Detection and Classification in Whole-Slide Lung Histopathology Challenge. 2023. Available online: https://acdc-lunghp.grand-challenge.org/ (accessed on 18 September 2023).
- Chen, W.; Yang, F.; Zhang, X.; Xu, X.; Qiao, X. MAU-Net: Multiple attention 3D U-Net for lung cancer segmentation on CT images. Procedia Comput. Sci. 2021, 192, 543–552. [Google Scholar] [CrossRef]
- Fu, J.; Liu, J.; Tian, H.; Li, Y.; Bao, Y.; Fang, Z.; Lu, H. Dual attention network for scene segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 3146–3154. [Google Scholar]
- Pan, Y.; Shi, D.; Wang, H.; Chen, T.; Cui, D.; Cheng, X.; Lu, Y. Automatic opportunistic osteoporosis screening using low-dose chest computed tomography scans obtained for lung cancer screening. Eur. Radiol. 2020, 30, 4107–4116. [Google Scholar] [CrossRef]
- Shimazaki, A.; Ueda, D.; Choppin, A.; Yamamoto, A.; Honjo, T.; Shimahara, Y.; Miki, Y. Deep learning-based algorithm for lung cancer detection on chest radiographs using the segmentation method. Sci. Rep. 2022, 12, 727. [Google Scholar] [CrossRef]
- Feng, J.; Jiang, J. Deep learning-based chest CT image features in diagnosis of lung cancer. Comput. Math. Methods Med. 2022, 2022, 4153211. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; Choi, H.; Paeng, J.C.; Cheon, G.J.; Kang, K.W. Deep Learning-Based Feature Extraction from Whole-Body PET/CT Employing Maximum Intensity Projection Images: Preliminary Results of Lung Cancer Data. Nucl. Med. Mol. Imaging 2023, 57, 216–222. [Google Scholar] [PubMed]
- Yan, S.; Huang, Q.; Yu, S.; Liu, Z. Computed tomography images under deep learning algorithm in the diagnosis of perioperative rehabilitation nursing for patients with lung cancer. Sci. Program. 2022, 2022, 8685604. [Google Scholar] [CrossRef]
- Chen, X.; Duan, Q.; Wu, R.; Yang, Z. Segmentation of lung computed tomography images based on SegNet in the diagnosis of lung cancer. J. Radiat. Res. Appl. Sci. 2021, 14, 396–403. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Choe, J.; Lee, S.M.; Do, K.H.; Lee, G.; Lee, J.G.; Lee, S.M.; Seo, J.B. Deep learning–based image conversion of CT reconstruction kernels improves radiomics reproducibility for pulmonary nodules or masses. Radiology 2019, 292, 365–373. [Google Scholar] [CrossRef]
- Yu, Z.; Yang, X.; Dang, C.; Wu, S.; Adekkanattu, P.; Pathak, J.; George, T.J.; Hogan, W.R.; Guo, Y.; Bian, J.; et al. A study of social and behavioral determinants of health in lung cancer patients using transformers-based natural language processing models. In Proceedings of the AMIA Annual Symposium Proceedings, San Diego, CA, USA, 30 October–3 November 2021; p. 1225. [Google Scholar]
- Hwang, E.J.; Lee, J.S.; Lee, J.H.; Lim, W.H.; Kim, J.H.; Choi, K.S.; Choi, T.W.; Kim, T.H.; Goo, J.M.; Park, C.M. Deep learning for detection of pulmonary metastasis on chest radiographs. Radiology 2021, 301, 455–463. [Google Scholar] [CrossRef]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef]







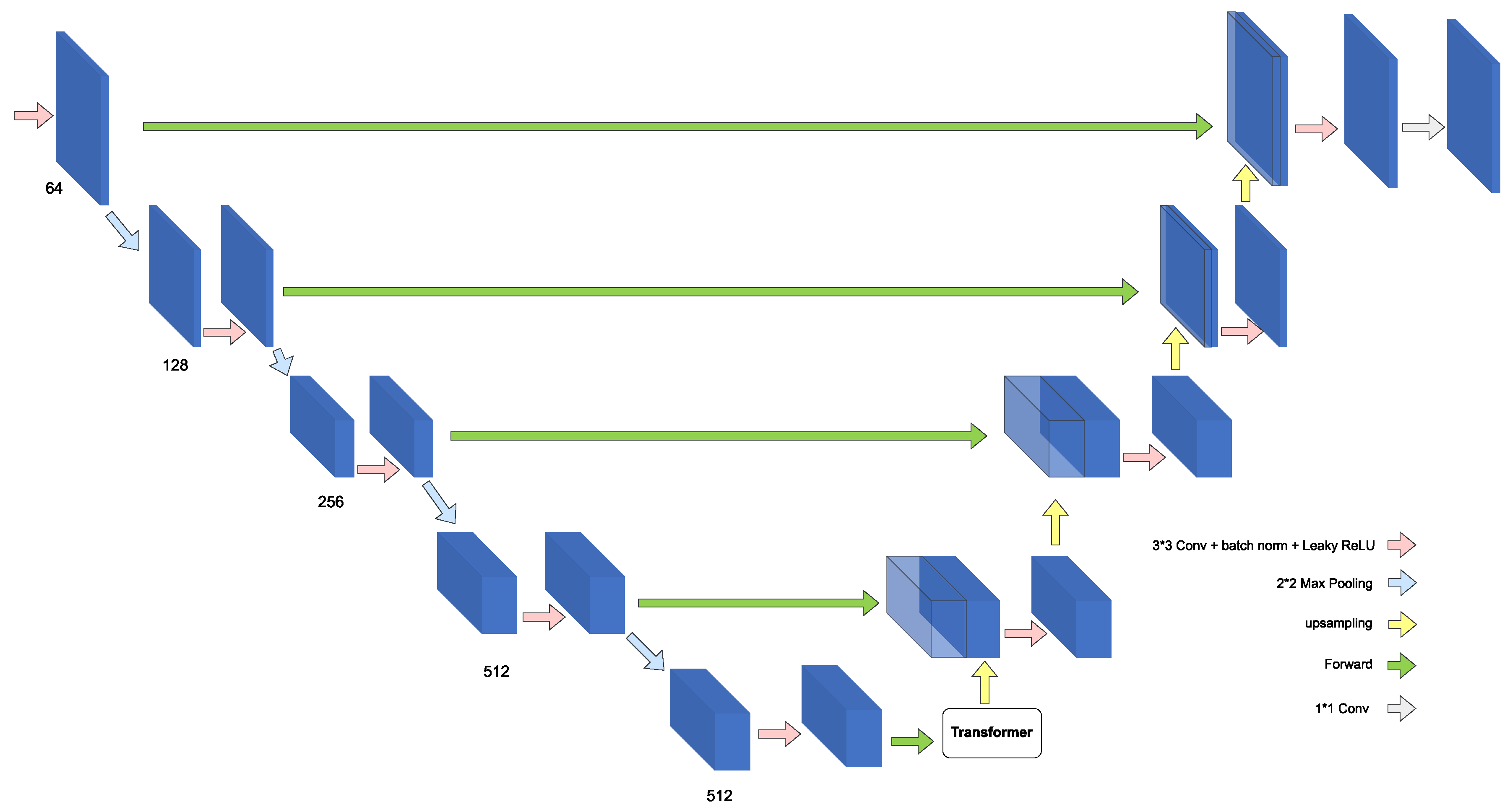
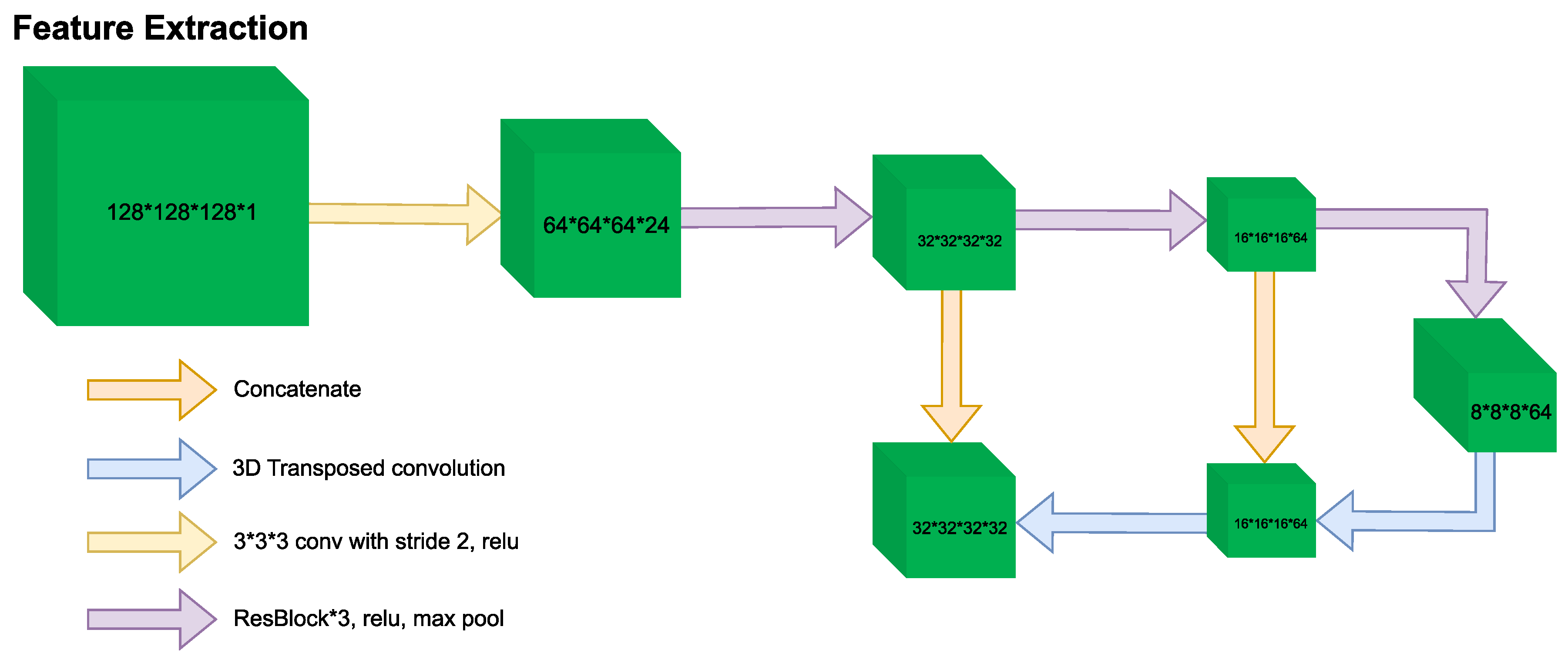

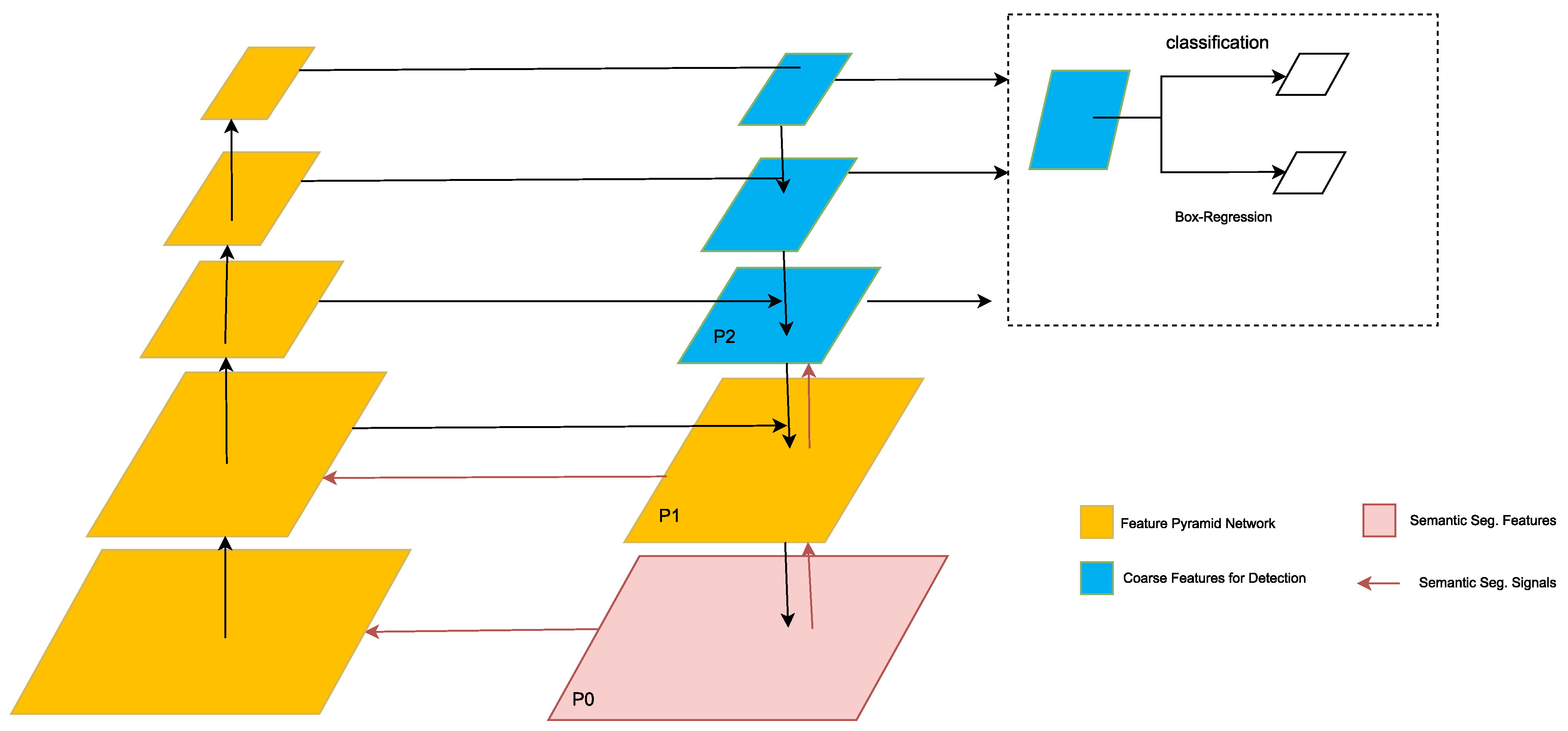

| Ref. | Dataset Description | Number of Images | Data Type | File Format | Label Type |
|---|---|---|---|---|---|
| [21] | LIDC-IDRI is a comprehensive collection of lung CT scans and annotations designed to support the development of computer-aided diagnostic systems for lung cancer. | 1010 patients | CT | Nifti, DICOM | Detection |
| [22] | LUNA16 derived from the LIDC database, specializing in lung nodule detection. | 888 patients, 1186 nodules | CT | DICOM | Detection Segmentation |
| [23] | ChestX-Ray8 aims to enable the detection and localization of diseases, by providing a large-scale database annotated with NLP and specialists for clinical challenges. | 108,948 front view X-ray images of 32,717 patients | Chest X-ray | DICOM | Classification Localization |
| [24] | JSRT is composed of digital radiographs of Japanese patients with a resolution of 2048 × 2048 pixels, provides detailed annotations of lung nodules. | 247 chest radiographs | CT | Big-endian raw | Segmentation Classification |
| [25] | NLST contains data from a large clinical trial conducted by the National Cancer Institute to assess low-dose CT screening for lung cancer. | Information on 53,000 individuals | CT, Chest X-ray | DICOM | Classification |
| [26] | MSKCC contains digitized histopathology slide images with resolution of 512 × 512 pixels and an 8-bit color depth from patients with different cancer types that have been manually annotated by pathologists to identify regions of cancerous tissue. | 25,000 digitized slides, age, sex, cancer type | CT | TSV, SEG | Prediction |
| [27] | TCGA includes subtypes such as lung adenocarcinoma and lung squamous cell carcinoma, and is designed to support cancer research and diagnostic and therapeutic development. | Data for over 11,429 patient sample | CT | DCM | Classification |
| [28] | Managed by the National Cancer Institute, the SEER dataset includes more than 40 years of cancer incidence and survival data from registries across the U.S., analyzing cancer trends, risk factors and treatment outcomes to inform cancer research and guide health policy decisions. | 9.5 million cancer cases | CT | N/A | Segmentation Classification |
| [29] | MSD contains 3D medical images covering 10 imaging modalities and 10 anatomical structures to support research and evaluation of segmentation algorithms in medical image analysis. It was used to develop the MSD Challenge, which tests the ability of machine learning algorithms to generalize to various semantic segmentation tasks. | 2633 scans | CT, MRI, US | Nifti | Segmentation |
| [30] | TCIA (Lung) has been created to promote research in the field of medical image analysis, in particular in the development and evaluation of computer-assisted diagnostic systems for cancer offering a wide variety of images. | 48,723 scans | CT | Nifti, DICOM | Segmentation Classification |
| [31] | Tianchi encourages early detection of cancer, by providing various medical images with radiologist labels indicating the presence, location, size and malignancy of the nodule. The dataset contains various imaging parameters and patient demographics to extract lung nodule characteristics. | 800 scans | CT | MHD | Detection |
| [32] | DSB contained more 256 × 256 pixel grayscale microscope images of cells, labeled with annotations indicating the presence of cancer, to challenge participants to develop image recognition algorithms for cancer detection. The various images of cell cultures, tissues and blood samples presented complexities such as variation in cell size, shape and texture for the classification task. | 800 scans | CT | DICOM | Classification |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gayap, H.T.; Akhloufi, M.A. Deep Machine Learning for Medical Diagnosis, Application to Lung Cancer Detection: A Review. BioMedInformatics 2024, 4, 236-284. https://doi.org/10.3390/biomedinformatics4010015
Gayap HT, Akhloufi MA. Deep Machine Learning for Medical Diagnosis, Application to Lung Cancer Detection: A Review. BioMedInformatics. 2024; 4(1):236-284. https://doi.org/10.3390/biomedinformatics4010015
Chicago/Turabian StyleGayap, Hadrien T., and Moulay A. Akhloufi. 2024. "Deep Machine Learning for Medical Diagnosis, Application to Lung Cancer Detection: A Review" BioMedInformatics 4, no. 1: 236-284. https://doi.org/10.3390/biomedinformatics4010015
APA StyleGayap, H. T., & Akhloufi, M. A. (2024). Deep Machine Learning for Medical Diagnosis, Application to Lung Cancer Detection: A Review. BioMedInformatics, 4(1), 236-284. https://doi.org/10.3390/biomedinformatics4010015







