Abstract
Chest X-ray (CXR) is one of the most common radiological examinations for both nonemergent and emergent clinical indications, but human error or lack of prioritization of patients can hinder timely interpretation. Deep learning (DL) algorithms have proven to be useful in the assessment of various abnormalities including tuberculosis, lung parenchymal lesions, or pneumothorax. The deep learning–based automatic detection algorithm (DLAD) was developed to detect visual patterns on CXR for 12 preselected findings. To evaluate the proposed system, we designed a single-site retrospective study comparing the DL algorithm with the performance of five differently experienced radiologists. On the assessed dataset (n = 127) collected from the municipal hospital in the Czech Republic, DLAD achieved a sensitivity (Se) of 0.925 and specificity (Sp) of 0.644, compared to bootstrapped radiologists’ Se of 0.661 and Sp of 0.803, respectively, with statistically significant difference. The negative likelihood ratio (NLR) of the proposed software (0.12 (0.04–0.32)) was significantly lower than radiologists’ assessment (0.42 (0.4–0.43), p < 0.0001). No critical findings were missed by the software.
1. Introduction
CXR is one of the key tools for the early detection and evaluation of respiratory disease and other acute pulmonary findings [1,2,3]. The current model, which could be described as first in, first out (FIFO), suggests that patients with more significant or urgent needs are not prioritized for radiographic description [4]. Despite the widespread use of patient prioritization tools (PPTs) in healthcare services [5,6,7], the existing literature has focused primarily on the emergency department setting [8,9,10].
Proposed DLAD is a software tool that identifies and prioritizes scans with acute findings for evaluating radiologists. The aim of this study is to verify the effectiveness of the software before deploying to clinical workplace, to validate the initial results of the prioritization model for patient triage based on features extracted by using DL and to monitor the possible changes in the user workflow.
2. Background
The use of DL in radiology has been a subject of active interest and research in recent years. Approximately 2,500,000 CXRs were performed in the Czech Republic in 2020, making it the most commonly used radiological examination after dental X-rays [11].
2.1. Literature Review Methodology
The aim of the initial literature review was to establish the basic premises of computer-aided detection/diagnosis, to analyze the methodology of software evaluation, and to define the area of interest for comparison of the DLAD software with similar medical devices. The primary sources of initial review include the website Grand Challenge: AI for Radiology (https://grand-challenge.org/aiforradiology/ (accessed on 22 November 2022)). As a secondary source, we used the free full-text archive PubMed (https://pubmed.ncbi.nlm.nih.gov/ (accessed on 22 November 2022)) with the keywords “artificial intelligence”, “deep learning”, “computer-aided diagnosis”, “chest X-ray”, “chest radiograph” and “detection”. Given the reported inaccuracy of publicly available datasets [12] and their unsuitability for designing clinically robust models [13], we decided to exclude studies that leveraged these data as a training or test set. The complete analysis of relevant studies for comparison with the DLAD system is available in the Table A1.
2.2. Related Works
Previous research suggests that DL algorithms can perform as well as, or in some cases better than, a radiologist in recognizing certain findings on a CXR [14,15,16]. Currently, the most promising results of AI versus radiologist are in detecting isolated findings such as tuberculosis [17,18] or lung parenchymal lesions [19,20,21,22,23]. DL models can achieve solid results compared to a radiologist in recognizing individual pathologies, but to date only few studies [24,25,26,27] have compared DL-based solutions to a radiologist in a setting where a central reader determining ground truth has access to clinical information about the patient as it would in real life.
A practical approach is to ask how AI can help the radiologist. Recent studies have shown that complex DL models have significantly improved chest X-ray interpretation by radiologists and have been well received by clinicians [25,28]. Another promising real-world use case for DL in the context of CXR is triage (prioritization) of patients to reduce report turnaround time for critical findings [29].
3. Software
DLAD (Carebot AI CXR v1.22) is a software that assists radiologists in the interpretation of CXRs in posterior–anterior (PA) or anterior–posterior (AP) projections. By utilizing DL algorithms, the solution automatically detects abnormality based on visual patterns for the following findings: atelectasis, consolidation, cardiomegaly, mediastinal widening, pneumoperitoneum, pneumothorax, pulmonary edema, pulmonary lesion, bone fracture, hilar enlargement, subcutaneous emphysema, and pleural effusion. (Standardized descriptions of individual findings were determined by discussion with individual annotators involved in the development of the DLAD software. The exact specifications are included in the user manual provided to all collaborating radiologists.)
3.1. Model Architecture
For image recognition and classification tasks, various convolutional neural network (CNN) architectures (shown in Figure 1) have proven to be successful. The standard architecture includes several convolutional layers that segment the image into small pieces that can be easily processed [30]. Each image is first segmented into light or dark (or specifically coloured) areas, edges in different orientations, patterns, etc., then fused into simple shapes, and finally merged into recognizable complex features in subsequent layers [31].

Figure 1.
The CNN architecture consists of convolutional and pooling layers and fully connected output layer at the end to provide the final prediction.
When training conventional CNNs, data scientists initially train the model with different hyperparameters to see which combinations perform the best. The optimal model is frequently determined either through a straightforward validation on test data or by more rigorous cross-validation [32,33]. Model soups, a recent discovery by [34], are formed by averaging the weights of several fine-tuned models rather than combining each of their separate outputs. The outcome is a single model that represents the average of various models with diverse hyperparameter configurations [34]. Additionally, model soups boost resilience in the same way that ensemble approaches do [35]. The proposed DLAD (Carebot AI CXR v1.22) leverages the novel model soup approach. The in-depth architecture of the deep neural networks used is not provided due to the commercial nature of the software.
3.2. Datasets
Anonymized CXRs from sites in Europe, Asia, and North America were used in the development of DLAD. The use of CXR scans from multiple workplaces is intended to reflect the variability in the quality of screening between hospitals and local population characteristics [36,37]. This approach allows the DL model to adapt to new conditions [38,39]. Patients under 18 years of age were excluded from the dataset, as were images of poor quality or incorrect projection. Collected DICOM images were annotated by a team of 22 radiologists with experience ranging from one to more than 10 years. The consensus on the normal/abnormal label, with regard to abovementioned 12 preselected findings, from the three annotating radiologists was required to establish ground truth. In case of disagreement between annotating radiologists for the label normal/abnormal, the image was not included in the training set.
3.3. Internal Test
For the initial internal test, we used 397 CXRs that were not observed during the model training. Selected images were retrospectively assessed by three independent radiologists. The ground truth was determined by 100% agreement. The results of the internal test are shown in Table 1. Figure 2 shows an image involving a rib fracture that DLAD incorrectly classified as a scan without any abnormality.

Table 1.
Performance of DLAD during internal test.

Figure 2.
Confusion matrix and a false negative image that DLAD incorrectly classified as a true negative during the internal test. The CXR scan in high quality can be found in the Appendix A (Figure A1).
Of the selected images, 213 (53.65%) were reported as normal and 184 (46.35%) as abnormal. DLAD software correctly interpreted 338 images (85.1%), with only one image resolving in a false negative outcome (FNR = 0.0054). A higher false positive rate (FPR = 0.2582) is an expected occurrence. Considering that DLAD software is intended to serve as a decision support system, this outcome is considered rather desirable.
4. Methodology
The purpose of our study is to provide evidence that the investigated pre-certification medical device (shown in Figure 3) meets the requirements in accordance with its intended use. Given this, a retrospective study was performed to evaluate the clinical effectiveness on prospectively collected CXRs. The evaluation of DLAD performance compared to that of radiologists in standard clinical practice was set as the primary endpoint of the study. (The methodology and statistical evaluation was designed in collaboration with the Institute of Biostatistics and Analysis, Ltd.)

Figure 3.
User interface of DLAD software implemented in PACS (CloudPACS by OR-CZ).
4.1. Data Source
To collect the CXR data for the retrospective study, we addressed a municipal hospital in the Czech Republic that provides healthcare services to up to 130,000 residents of a medium-sized city (approximately 70,000 inhabitants) and the surrounding area. Pediatric CXR images (under 18 years of age), scans with technical problems (poor image quality, rotation), and images in lateral projection were excluded from the analysis. In total, 127 anonymized CXR images (Table 2) were prospectively collected between August 15 and 17, 2022, and subsequently submitted to five independent radiologists of varying experience for annotation. The selected radiologists were asked to assess whether the CXR image shows any of the 12 abnormalities mentioned in the Software section. The assessed annotators had no knowledge of the patient’s history or previous or follow-up examinations. This enabled an objective comparison of the results with the DLAD software’s assessment. The radiologists were differently experienced: #5f0 and #442 were junior radiologists with less than five years of experience, #c8a was a radiologist with more than five years of experience, and #630 and #24a were heads of the radiology department with more than 10 and 20 years of experience, respectively.

Table 2.
Patient demographic data and findings prevalence.
4.2. Ground Truth
A head of the radiology department with more than 20 years of experience and knowledge of the local specifics (awareness of the utilized X-ray machines and their quality) was appointed as a central reader to determine the ground truth. Central reader used the patient’s previous and follow-up examinations of various modalities (computed tomography, ultrasound, magnetic resonance imaging) and, where appropriate, the collateral patient records from other examining physicians (history, spirometry, etc.). The availability of supporting information has an impact on reducing uncertainty [37,40,41].
4.3. Objectives
The primary objective was to evaluate the performance of the DLAD compared to radiologists’ assessment in routine clinical practice. Our secondary objective was the comparison of performance of the DLAD and individual radiologists with different experience.
4.4. Statistical Analysis
DLAD performance was quantified by means of sensitivity (Se) and specificity (Sp), positive (PLR) and negative likelihood ratio (NLR), and positive (PPV) and negative predictive value (NPV). Se and Sp are related to the rate of true positive and false positive cases, respectively. Their mutual relations are expressed by PLR = Se/(1 − Sp) and NLR = (1 − Se)/Sp. The likelihood ratios (LRs) depend only on Se and Sp and are equivalent to the relative risk. Higher PLR and lower NLR are desirable. Predictive values (PVs) indicate the clinical accuracy of the diagnostic test. PV depends on Se and Sp and also on the prevalence of the disease in the population. A paired design was applied to the data, i.e., all images were evaluated by both DLAD and radiologist, and compared with the status given by ground truth. Furthermore, balanced accuracy (BA = (Se + Sp)/2 and F1 score were calculated and compared.
The primary objective was to compare DLAD performance against clinical practice. As all images were evaluated by all assessed radiologists, the clinical practice was simulated by generating 10,000 randomly selected subsets of size n = 127 by using bootstrap, with each image in each dataset evaluated by a randomly selected radiologist. Radiologists were randomly assigned, all with equal probability. The statistics were calculated in these datasets and the average performance in clinical practice was expressed and compared with DLAD results. The average Se, Sp, and PVs were compared by using a one-sample binomial test against the DLAD values; LRs were compared by using a one-sample t-test.
The analysis of the secondary objective (comparison of (1) DLAD against the (2) individual radiologists) consisted of the estimation of the parameters above and their statistical comparison by using confidence intervals (CI) and p-values. The procedure to compare the statistics consisted of (i) solving the global hypothesis test to an error calculating the Wald test statistic (e.g., and vs. and/or ) and if significant (ii) solving the two individual hypothesis tests (e.g., and ) along with a multiple comparison method (e.g., McNemar with continuity correction for Se and Sp, Holm method for LRs, and weighted generalized score statistics for PVs, respectively) to an error. Differences among radiologists and DLAD were visualized by using forest plots. Analysis was done in R software using the compbdt package [42]. All tests were performed as two-tailed at the 5% significance level.
5. Results
A total of 127 images with established ground truth were evaluated: 40 (31.5%) with a finding and 87 (68.5%) without any finding. The DLAD correctly identified 37 images as abnormal and 56 images as normal (73.2% in total). A total of 31 (24.4%) normal images were incorrectly classified as abnormal. The higher false positive rate was expected since DLAD was trained to assign even suspect findings as abnormal. Another three (2.4%, Figure A2, Figure A3, Figure A4) images were incorrectly classified as without any finding, even though they were with findings (false negative rate) (Table 3).

Table 3.
DLAD vs. Ground truth.
Applying the DLAD software, sensitivity reached a value of 0.925 (95% CI 0.804–0.977) and specificity 0.644 (0.539–0.737). In bootstrap-simulated clinical practice (where the images were evaluated by randomly selected radiologists with different experience), the sensitivity was 0.661 (0.572–0.743), and the specificity was 0.803 (0.723–0.868). DLAD showed a statistically higher sensitivity (p < 0.0001) and a statistically lower specificity (p = 0.0001), i.e., DLAD assessed more images without any finding an abnormality. The DLAD PLR (2.6 (1.9–3.5)) was statistically significantly lower (i.e., worse) than radiologists’ assessment (3.59 (3.44–3.73), p < 0.0001) and the NLR (0.12 (0.04–0.32)) was significantly lower (i.e., better) than radiologists’ assessment (0.42 (0.4–0.43), p < 0.0001). The PPV, i.e., the probability of a positive finding if an image is actually abnormal, was 0.544 (0.427–0.657) for DLAD and 0.614 (0.524–0.699) for radiologist assessment. There was no statistically significant difference in this parameter (p = 0.13). The NPV (the probability that a patient is without any finding when the image was classified as normal) was 0.949 (0.863–0.985) for DLAD and 0.843 (0.767–0.901) for radiologist assessment, and this difference was statistically significant (p < 0.0001) (Table 4).

Table 4.
Performance of DLAD compared to radiologists’ assessment.
Significant differences were observed among the radiologists (Table 5). Both sensitivity and specificity were significantly different in all radiologists with varying experience compared to DLAD (global p-value < 0.0001). For three radiologists, the DLAD had a significantly higher sensitivity and would therefore probably help to identify patients with a finding who the radiologists determined to be without any finding (Table 5, Figure 4). On the contrary, the specificity was significantly worse with DLAD, with the exception of one radiologist (#442), who assessed many normal images as abnormal. However, because DLAD software is intended to be assistive, this result was expected and the final decision is the doctor’s responsibility. Although the PPV on average did not differ significantly from the DLAD classification, worse values for individual radiologists (except that of radiologist #442 with a very good assessment of abnormal images but with low specificity) were found (Table 6, Figure 4).

Table 5.
Labeling of images by individual radiologists with respect to ground truth.

Figure 4.
Forest plots for performance of DLAD and individual radiologists (A) sensitivity and specificity, (B) likelihood ratios, (C) predictive values.

Table 6.
Performance of individual radiologists compared to DLAD.
All radiologists could benefit from the proposed DLAD. All of them assessed some images as normal although they were abnormal and DLAD classified them as abnormal. For individual radiologists, it was 3, 4, 12, 15, and even 26 images, which would be prioritized and the doctor would check the image again, carefully and/or consult with a more experienced colleague (Table 5).
6. Discussion
To be able to evaluate the potential benefits of DLAD in prioritizing and assessing patient scans, we compared the CAD with five differently experienced radiologists, and additionally simulated standard clinical practice by generating 10,000 randomly selected subsets of size n = 127 by using a bootstrap, with each image in the dataset being assessed by a randomly selected radiologist. In our test, the proposed DLAD system was able to achieve promising sensitivity and reasonable specificity compared to the analyzed commercial and academic applications. The performance achieved (particularly the high sensitivity compared to the evaluating radiologists as well as to similar solutions mentioned below) indicates that the DL algorithm learned to detect individual visual patterns on CXR, including on a distribution of data that was not observed beforehand.
Multiple pathologies with similar sample size (n = 370) were addressed in [43], which applied commercial software (Arterys Chest & MSK AI) to detect fractures (n = 4), nodules (n = 24), opacities (n = 105), pleural effusion (n = 89), and pneumothorax (n = 22). The ground truth was defined by a three-fourths (3/4) consensus of the readers. On this sample, the software achieved overall sensitivity/specificity 0.988/0.438, for fractures 0.667/0.9499, nodules 0.64/0.842, opacities 0.9615/0.4804, pleural effusion 0.921/0.872, and pneumothorax 1.0/0.758. Another commercially available solution (Lunit INSIGHT CXR) was investigated in [44], wherein three major thoracic abnormalities (nodule/mass (n = 80), consolidation (n = 31), and pneumothorax (n = 35)) were investigated on a similarly sized test set (n = 244). In addition, as in our case, the study compared the assessment of CAD systems and annotators: radiologists, nonradiologists, and clinicians. The software areas under the ROC curve (AUCs) for nodule/mass, consolidation, and pneumothorax were 0.988, 1.000, and 0.999, respectively. For the image classification, the overall area under the ROC curve (AUC) of the pooled physicians was 0.868 without CAD and 0.911 with CAD. Ref. [45] focused on comparison of the DL algorithm (Qure.ai qXR) with radiological assessment, wherein the performance was analyzed in four pathologies (pulmonary opacities (n = 336), pleural effusion (n = 136), hilar prominence (n = 134), and enlarged cardiac silhouette (n = 122)) on a smaller dataset (n = 874). The AUC for the DL algorithm and test radiologists ranged between 0.837 and 0.929 and between 0.693 and 0.923, respectively. The DL algorithm had the lowest AUC (0.758) for assessing changes in pulmonary opacities over follow-up CXR. The evaluation of the same solution (Qure.ai qXR) for the detection of multiple pathologies (blunted costophrenic angle, cardiomegaly, cavity, consolidation, fibrosis, hilar enlargement, nodule, opacity, and pleural effusion) was addressed in [46], which evaluated the proposed DL algorithm on two different datasets: a large-scale dataset (n = 100,000) whose ground truth was based on existing radiological reports, and a smaller one (n = 2000) that was evaluated by three radiologist majority vote. On the smaller dataset, the proposed system demonstrated an AUC of 0.92 (CI 0.91–0.94) for detection of abnormal scans, and AUC of 0.96 (0.94–0.98), 0.96 (0.94–0.98), 0.95 (0.87–1), 0.95 (0.92–0.98), 0.93 (0.90–0.96), 0.89 (0.83–0.94), 0.91 (0.87–0.96), 0.94 (0.93–0.96), 0.98 (0.97–1) for the detection of blunted costophrenic angle, cardiomegaly, cavity, consolidation, fibrosis, hilar enlargement, nodule, opacity, and pleural effusion. The AUCs were similar on the larger dataset except for detecting normal results where the AUC was 0.86 (0.85–0.86). Classification between normal and abnormal CXRs to reduce the time and cost associated with reporting normal studies was addressed in [47] by using Qure.ai qXR commercial solution. For the retrospectively collected dataset (n = 430, 285 abnormal, 145 normal), a radiologist with eight years of experience, with access to existing reports, established the ground truth. The DL algorithm achieved sensitivity 0.9719 (0.945–0.9878) and specificity 0.683 (0.6–0–758) with 46 FPs and 8 FNs. Outside of commercial applications, it is interesting to mention [29], which applied the DL algorithm to a large-scale test set (n = 15,887) with the goal of triaging normal CXRs. The ground truth was determined by NLP extraction from the original radiology reports. Normal CXRs were detected by the proposed system with a sensitivity of 0.71, specificity of 0.95, PPV of 0.73, and NPV of 0.94.
7. Limitations
Unlike multiple commercially available solutions that have been trained to recognize individual findings, the proposed DLAD (Carebot AI CXR v1.22) was trained to identify abnormal scans. We took this step to facilitate the clinical use of the algorithm in different sites, regardless of the local prevalence of the disease. One of the limitations of this study (and the studies mentioned above) is the lack of a more comprehensive way of assessing the correctness of the pathologies’ localization. Although the measurement of sensitivity and specificity deviations allows us to investigate primarily the presence of pathology, the emphasis in clinical practice is on the correct localization of findings and follow-up clinical workflow. We plan to address this issue in a future study.
The purpose of the present research was the preclinical evaluation of a DLAD currently undergoing regulatory procedure according to the EU MDR 2017/745. This study does not aim to present new DL approaches or architectures but investigates the first deployment of DLAD in the healthcare system of the Czech Republic. As stated in the Literature Review Methodology, the proposed DLAD did not leverage or was not compared with models that utilize publicly available data. DLADs based on publicly available datasets, such as ChestXray14, CheXpert, or COVIDx CXR-2, may serve to present the novel methodology, but they are by no means safe or robust for use in actual clinical practice [12,13]. These datasets contain inaccurate ground truth, poor-quality images, inappropriate file formats, and even initial problem descriptions. This problem is widely known and is one of the reasons why independent preclinical and clinical evaluation of medical software exists in the first place.
8. Conclusions
CAD systems can improve radiologists’ workflow by prioritizing the worklist according to anticipated imaging findings and increasing overall diagnostic sensitivity with no acute findings being missed. The proposed DLAD software showed statistically significantly better sensitivity, negative likelihood ratio, and negative predictive value, and on the contrary, lower specificity and positive likelihood ratio. When we focused on individual radiologists with different specializations and lengths of practice, varying results were found. However, no trend was found according to the radiologist’s experience, and it is therefore not possible to clearly decide which radiologists benefit the most from the proposed software. In general, the benefit of DLAD was indirectly demonstrated. Although it assesses more false positive images due to its threshold settings, it would alert to a finding in a CXR that a doctor evaluated as normal. This proves the contribution of the model in real practice as a support tool for identifying abnormal findings. However, a primary evaluation by a doctor is absolutely essential.
Author Contributions
Conceptualization, D.K. and A.C.; methodology, D.K. and P.O.; software, D.K., M.B. and R.H.; data curation, D.K., A.C. and R.H.; writing—D.K., A.C., M.B., K.K. and P.O.; validation, A.C., M.P. and P.O.; project administration, D.K.; funding acquisition, D.K. All authors have read and agreed to the published version of the manuscript.
Funding
The research was funded by Carebot, Ltd.
Institutional Review Board Statement
This study was made possible by the “Agreement for Cooperation on the Development of the Software Carebot”, which was signed on 19 September 2022 by Carebot, OR-CZ and Havířov Hospital.
Informed Consent Statement
Patient consent was waived due to Regulation 2016/679 of the European Parliament and of the Council of 27 April 2016 on the protection of natural persons with regard to the processing of personal data and on the free movement of such data, and repealing Directive 95/46/EC (General Data Protection Regulation), i.e. the CXR images were stripped of all direct or indirect identifiers without the possibility of retrospective patient identification.
Data Availability Statement
Data from this study can be provided by Carebot, Ltd. to independent researchers. Please contact the author for more information, if required.
Conflicts of Interest
In relation to this study, We declare the following conflicts of interest: The study was funded by Carebot, Ltd. The authors are employees of Carebot, Ltd.
Abbreviations
The following abbreviations are used in this manuscript:
| AI | Artificial Intelligence |
| CADe/CADx | Computer-Aided Detection/Diagnosis |
| CI | Confidence Interval |
| CNN | Convolutional Neural Network |
| CXR | Chest X-ray |
| DL | Deep Learning |
| DLAD | Deep Learning–based Automatic Detection Algorithm |
| MD | Medical Device |
| Se | Sensitivity |
| Sp | Specificity |
| FP | False Positive |
| FN | False Negative |
| LR | Likelihood Ratios |
| PLR | Positive Likelihood Ratio |
| NLR | Negative Likelihood Ratio |
| PV | Predictive Values |
| PPV | Positive Predictive Value |
| NPV | Negative Predictive Value |
| BA | Balanced Accuracy |
Appendix A

Figure A1.
False Negative image incorrectly classified by DLAD software during internal software validation. The software failed to detect a rib fracture.
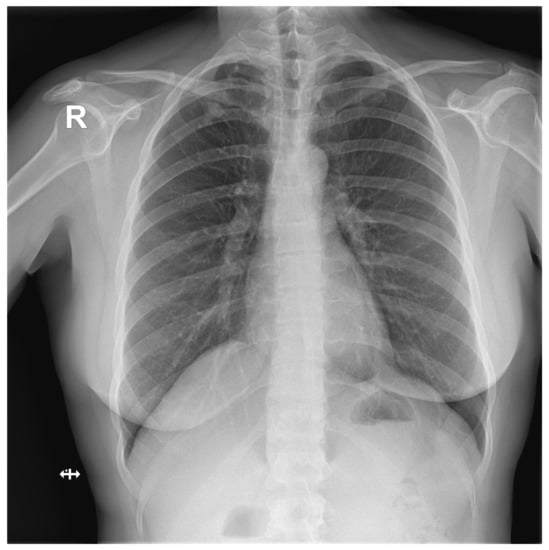
Figure A2.
False Negative image #1 incorrectly classified by DLAD software during the retrospective study. The software failed to detect a lesion in lung parenchyma. Assessed radiologists #5f0 and #24a also incorrectly classified this image as Normal.

Figure A3.
False Negative image #2 incorrectly classified by DLAD software during the retrospective study. The software failed to detect a lesion in lung parenchyma. Assessed radiologists #5f0, #442 and #c8a also incorrectly classified this image as Normal.

Figure A4.
False Negative image #3 incorrectly classified by DLAD software during the retrospective study. The software failed to detect a rib fracture. Assessed radiologists #5f0 and #630 also incorrectly classified this image as Normal. Assessed radiologist #442 classified the image correctly as Abnormal but failed to locate a rib fracture.

Table A1.
Analysis of existing solutions and publications on CXR computer-aided detection.
Table A1.
Analysis of existing solutions and publications on CXR computer-aided detection.
| Study | Target Population | Number of Patients, Scans | Used Software | Ground Truth | Statistical Results |
|---|---|---|---|---|---|
| [17] | suspected TB | 317 patients | qXR (Qure.ai) | microbiological confirmation, 1 radiologist | qXR Se 0.71, Sp 0.80, radiologist Se 0.56 and Sp 0.80, AUC for confirmed TB 0.81, detection of pleural effusion and “cavity” type. For TB in qXR AUC 0.94 and 0.84, others 0.75–0.94, radiologist Se 0.56, Sp 0.80, low AUC 0.75 for hilar lymphadenopathy and 0.76 consolidation, largest for cardiomegaly 0.94 |
| [15] | suspected TB | 1032 patients | 12 different solutions including qXR and Lunit | 2 doctors focused on TB | Expert Se 0.955, Sp 0.422. With setting this Se, qXR and Lunit had Sp 0.487 and 0.387, DeepTek SP 0.463, Delft imaging 0.453, JF Healthcare 0.41, Oxipit 0.408, InferVision 0.265, Artelus 0.231, Dr CADx 0.121, SemanticMD 0.101, EPCON 0.093, COTO 0.063. AUROC for qXR and Delft Imaging 0.82, PR AUC 0.41 and 0.39, DeepTek Genki AUROC 0.78, Lunit 0.82, JF Healthcare similar. |
| [48] | drug-resistant TB | 311 patients, 346 images | qXR (Qure.ai) | with initial identification by a radiologist, with possible comparison in CT | Correlation of radiologist and qXR in hilar lymphadenopathy, pleural effusion cavity, and atelectasis, but not in nodules. Se/Sp hilar lymphadenopathy 0.621/0.741, cavity 0.75/0.821, atelectasis 0.194/0.727, pleural effusion 0.6/0.949, nodule 0.597/0.742. |
| [18] | undergoing medical screening during military service (asymptomatic) | 19,686 patients, 20,135 images | Lunit INSIGHT CXR | microbiological confirmation, identification by a radiologist | AUC Lunit for pulmonary TB 0.999, for other abnormalities 0.967, Se for high Se for nodule 1.0, for high Sp 1.0, radiologist 0.8, and other abnormalities Se 0.921, 0.679, 0.821, Sp nodules 0.959, 0.997 and 0.997, others 0.960, 0.997, 0.998. |
| [20] | suspected lung cancer | 1512 images | red dot (behold.ai) | biopsy | Of the urgent ones given by the red dot radiologist, he evaluated 15% as non-urgent and 85% as urgent, the non-urgent red dot ones were determined as non-urgent, he just evaluated more of them as urgent |
| [23] | suspected lung cancer | 400 images | red dot (behold.ai) | 3 radiologists | Average radiologist Se 0.78, Sp 0.96, behold.ai Se 0.80, Sp 0.93, overall improvement of 3.67–13.33% confidence percentage, radiologist agreement improved to 94%, and missed tumors reduced by 60%. |
| [21] | high risk (smokers) with lung screening | 5485 patients | Lunit INSIGHT CXR | certified radiologist, GT if cancer for confirmed within one year | Lunit AUC 0.93 for chest radiographs, 0.99 for digital and 0.86 for CT, Se 0.862 and Sp 0.85, cancer detection Se 0.76, radiological 0.80. |
| [22] | various databases | 378 patients, 434 images | Lunit INSIGHT CXR | 2 radiology residents and 2 chest radiologists | Se 0.883, Sp 0.8618, Lunit AUC abnormalities 0.872, nodules Se 0.891. |
| [49] | various databases | 3790 patients, 3887 images | red dot (behold.ai) | 2 radiologists, 3rd arbitrator | Normal with an accuracy of 0.977, 84.6% of them were identified by radiologists as borderline. 13.5% missed abnormality by radiologists. |
| [45] | multiple radiological abnormalities (14), from the database of Wang et al. 2017 | 724 patients, 874 images | qXR (Qure.ai) | 4 radiologists + 2 as GT | AUC qXR 0.837–0.929, radiologist 0.693 and 0.923 |
| [44] | multiple radiographic abnormalities (3) | 244 images | Lunit INSIGHT CXR | clinicians (3 groups-GP, radio, non-radio) | AUC 0.993, Se 97.26, Sp 92.86, reliability 0.9549, AUC for nodules, consolidation, pneumothorax 0.988, 1 and 0.999. AUC of radiologists, non-radiologists and clinicians without Lunit 0.931, 0.915 and 0.769. With the help of Lunit AUC increased to 0.959, 0.944 and 0.894. |
| [46] | multiple radiographic abnormalities (9) | 100,000 images from 89,354 patients + 2000 images | qXR (Qure.ai) | comparison with the agreement of 3 radiologists on the 2000 and reports of different on 100,000 scans | AUC for smaller dataset 0.92, different for individual abnormalities, similar for large, individual abnormalities AUC 0.98–0.89 |
| [50] | various databases + TB | 1444 patients | ResNet-based DLAD | training set 2 radiologists, GT PCR, culture | in the second session, with the use of DLAD, all values increased, but significantly for non-radiologists: AUROC doctor only non-radiologists 0.746, with p-value 0.023, AUROC 0.664 with p-value 0.0088, Se 0.723, Sp 0.67, TDR 0.582. Certified radiologists AUROC 0.946, p = 0.0082, AUROC 0.9, p = 0.0003, Se 0.906, Sp 0.948, TDR 0.797, chest radiologists AUROC 0.971 p = 0.0218, AUROC 0.925, p = 0.0001, Se 0.952, Sp 0.930, TD 0.870. With DLAD non-radiologists 0.850, AUROC 0.781 with p-value 0.0236, |Se 0.848, SP 0.800, TDR 0.724, certified radiologists AUROC 0.961, p = 0.0606, AUROC 0.924, p = 0.0353, Se 0.930, Sp 0.954, TDR 0.849. Chest radiologists AUROC 0.977 p = 0.1623, AUROC 0.942, p = 0.0036, Se 0.964, Sp 0.936, TD 0.897. |
| [19] | various hospital datasets for nodule detection | 600 images for internal and 693 for external validation | ResNet-based DLAD | 5 radiologists | Internal validation: AUROC 0.96, External validation: AUROCs 0.92, 0.99, 0.94, and 0.96, and JAFROC FOMs were 0.870, 0.924, 0.831, and 0.880 for Seoul National University Hospital, Boramae Hospital, National Cancer Center, and University of California San Francisco Medical Center. Nodule-detection false-positive rate of DLAD 0.02–0.34 in external datasets, for radiograph classification performance were Se 0.79, 0.911, 0.712 and 0.88, Sp 0.95, 0.98, 1.0 and 0.93, and for nodules Se 0.699, 0.82, 0.696 and 0.75. |
| [51] | population from Oulu Hospital, Finland, detection of multiple findings | 9579 images | ChestLink | 2 certified radiologists + original radiologists report | As a result, 9 false negative cases evaluated by ChestLink. Oxipit Se 0.998%, Sp 0.364. |
| [43] | 5 pathologies | 370 images | Arterys Chest AI | 4 radiologists | Overall Se/Sp 0.988/0.4384. Se/Sp for fractures 0.667/0.9499, nodules 0.64/0.8417, opacities 0.9615/0.4804, pleural effusion 0.9213/0.8716, pneumothorax 1.0/0.7576. |
| [52] | retrospective diagnosis of COVID | 279 images | Lunit INSIGHT CXR | CT or 3 radiologists. | Lunit AUROC/Se/Sp 0.921, 0.956, 0.887. Radiologist AUROC/Se/Sp0.941, 0.912, 0.969. |
| [47] | identification of normal/abnormal | 430 images | DLAD | experienced radiologist + reference of an existing report | Se 0.9719, Sp 0.6828, 46 FP, 8 FN (3 clinically insignificant, 5 clinically significant). |
| [14] | 72 findings | 1998 images | VGG16, ResNet-50 | triple consensus dataset | AI AUC 0.772 on test, 0.807 on train, Se 0.716, PPV 0.730, Sp 0.980. Radiologist Se 0.720, PPV 0.682, Sp 0.973. |
| [29] | multiple abnormalities | 15,887 images | DLAD | 2 radiologists with 3 years of experience, in case of disagreement another radiologist with 10 years of experience | DLAD normal radiograph Se 0.71, Sp 0.95, for critical radiograph Se 0.65, Sp 0.94. |
References
- Moncada, D.C.; Rueda, Z.V.; Macías, A.; Suárez, T.; Ortega, H.; Vélez, L.A. Reading and interpretation of chest X-ray in adults with community-acquired pneumonia. Braz. J. Infect. Dis. 2011, 15, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, W. Chest X-ray interpretation: Not just black and white. Nursing2020 2014, 44, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, A.; Chung, M.; Bernheim, A.; Eber, C. Portable chest X-ray in coronavirus disease-19 (COVID-19): A pictorial review. Clin. Imaging 2020, 64, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Baltruschat, I.; Steinmeister, L.; Nickisch, H.; Saalbach, A.; Grass, M.; Adam, G.; Knopp, T.; Ittrich, H. Smart chest X-ray worklist prioritization using artificial intelligence: A clinical workflow simulation. Eur. Radiol. 2021, 31, 3837–3845. [Google Scholar] [CrossRef] [PubMed]
- Mills, A.F.; Argon, N.T.; Ziya, S. Resource-based patient prioritization in mass-casualty incidents. Manuf. Serv. Oper. Manag. 2013, 15, 361–377. [Google Scholar] [CrossRef]
- Sung, I.; Lee, T. Optimal allocation of emergency medical resources in a mass casualty incident: Patient prioritization by column generation. Eur. J. Oper. Res. 2016, 252, 623–634. [Google Scholar] [CrossRef]
- Déry, J.; Ruiz, A.; Routhier, F.; Bélanger, V.; Côté, A.; Ait-Kadi, D.; Gagnon, M.P.; Deslauriers, S.; Lopes Pecora, A.T.; Redondo, E.; et al. A systematic review of patient prioritization tools in non-emergency healthcare services. Syst. Rev. 2020, 9, 1–14. [Google Scholar] [CrossRef]
- Schull, M.J.; Guttmann, A.; Leaver, C.A.; Vermeulen, M.; Hatcher, C.M.; Rowe, B.H.; Zwarenstein, M.; Anderson, G.M. Prioritizing performance measurement for emergency department care: Consensus on evidencebased quality of care indicators. Can. J. Emerg. Med. 2011, 13, 300–309. [Google Scholar] [CrossRef]
- Ashour, O.M.; Okudan Kremer, G.E. Dynamic patient grouping and prioritization: A new approach to emergency department flow improvement. Health Care Manag. Sci. 2016, 19, 192–205. [Google Scholar] [CrossRef]
- Ding, Y.; Park, E.; Nagarajan, M.; Grafstein, E. Patient prioritization in emergency department triage systems: An empirical study of the Canadian triage and acuity scale (CTAS). Manuf. Serv. Oper. Manag. 2019, 21, 723–741. [Google Scholar] [CrossRef]
- Ústav zdravotnických informací a statistiky České republiky (ÚZIS). Medical Equipment of Health Establishments of Czech Republic in Year 2020. 2021. Available online: https://www.uzis.cz/res/f/008364/ai-2021-02-t1-pristrojove-vybaveni-zz-2020.pdf (accessed on 22 November 2022).
- Oakden-Rayner, L. Exploring large-scale public medical image datasets. Acad. Radiol. 2020, 27, 106–112. [Google Scholar] [CrossRef]
- Hryniewska, W.; Bombiński, P.; Szatkowski, P.; Tomaszewska, P.; Przelaskowski, A.; Biecek, P. Checklist for responsible deep learning modeling of medical images based on COVID-19 detection studies. Pattern Recognit. 2021, 118, 108035. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.T.; Wong, K.C.; Gur, Y.; Ansari, N.; Karargyris, A.; Sharma, A.; Morris, M.; Saboury, B.; Ahmad, H.; Boyko, O.; et al. Comparison of chest radiograph interpretations by artificial intelligence algorithm vs radiology residents. JAMA Netw. Open 2020, 3, e2022779. [Google Scholar] [CrossRef] [PubMed]
- Codlin, A.J.; Dao, T.P.; Vo, L.N.Q.; Forse, R.J.; Van Truong, V.; Dang, H.M.; Nguyen, L.H.; Nguyen, H.B.; Nguyen, N.V.; Sidney-Annerstedt, K.; et al. Independent evaluation of 12 artificial intelligence solutions for the detection of tuberculosis. Sci. Rep. 2021, 11, 23895. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, K.G.; de Rooij, M.; Schalekamp, S.; van Ginneken, B.; Rutten, M.J. How does artificial intelligence in radiology improve efficiency and health outcomes? Pediatr. Radiol. 2021, 52, 2087–2093. [Google Scholar] [CrossRef] [PubMed]
- Nash, M.; Kadavigere, R.; Andrade, J.; Sukumar, C.A.; Chawla, K.; Shenoy, V.P.; Pande, T.; Huddart, S.; Pai, M.; Saravu, K. Deep learning, computer-aided radiography reading for tuberculosis: A diagnostic accuracy study from a tertiary hospital in India. Sci. Rep. 2020, 10, 210. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, S.; Hwang, E.J.; Goo, J.M.; Lee, W.Y.; Lee, S.; Kim, H.; Andrews, J.R.; Park, C.M. Deep learning–based automated detection algorithm for active pulmonary tuberculosis on chest radiographs: Diagnostic performance in systematic screening of asymptomatic individuals. Eur. Radiol. 2021, 31, 1069–1080. [Google Scholar] [CrossRef]
- Nam, J.G.; Park, S.; Hwang, E.J.; Lee, J.H.; Jin, K.N.; Lim, K.Y.; Vu, T.H.; Sohn, J.H.; Hwang, S.; Goo, J.M.; et al. Development and validation of deep learning–based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Radiology 2019, 290, 218–228. [Google Scholar] [CrossRef]
- Hussein, M.; Brozik, J.; Hopewell, H.; Patel, H.; Rasalingham, S.; Dillard, L.; Morgan, T.N.; Tappouni, R.; Malik, Q.; Lucas, E.; et al. Artificial intelligence: A potential prioritisation tool for chest radiographs with suspected thoracic malignancy. Lung Cancer 2020, 139, S25. [Google Scholar] [CrossRef]
- Yoo, H.; Kim, K.H.; Singh, R.; Digumarthy, S.R.; Kalra, M.K. Validation of a deep learning algorithm for the detection of malignant pulmonary nodules in chest radiographs. JAMA Netw. Open 2020, 3, e2017135. [Google Scholar] [CrossRef]
- Koo, Y.H.; Shin, K.E.; Park, J.S.; Lee, J.W.; Byun, S.; Lee, H. Extravalidation and reproducibility results of a commercial deep learning-based automatic detection algorithm for pulmonary nodules on chest radiographs at tertiary hospital. J. Med Imaging Radiat. Oncol. 2021, 65, 15–22. [Google Scholar] [CrossRef]
- Tam, M.; Dyer, T.; Dissez, G.; Morgan, T.N.; Hughes, M.; Illes, J.; Rasalingham, R.; Rasalingham, S. Augmenting lung cancer diagnosis on chest radiographs: Positioning artificial intelligence to improve radiologist performance. Clin. Radiol. 2021, 76, 607–614. [Google Scholar] [CrossRef]
- Liu, X.; Faes, L.; Kale, A.U.; Wagner, S.K.; Fu, D.J.; Bruynseels, A.; Mahendiran, T.; Moraes, G.; Shamdas, M.; Kern, C.; et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit. Health 2019, 1, e271–e297. [Google Scholar] [CrossRef]
- Qin, Z.Z.; Sander, M.S.; Rai, B.; Titahong, C.N.; Sudrungrot, S.; Laah, S.N.; Adhikari, L.M.; Carter, E.J.; Puri, L.; Codlin, A.J.; et al. Using artificial intelligence to read chest radiographs for tuberculosis detection: A multi-site evaluation of the diagnostic accuracy of three deep learning systems. Sci. Rep. 2019, 9, 15000. [Google Scholar] [CrossRef]
- Castiglioni, I.; Ippolito, D.; Interlenghi, M.; Monti, C.B.; Salvatore, C.; Schiaffino, S.; Polidori, A.; Gandola, D.; Messa, C.; Sardanelli, F. Artificial intelligence applied on chest X-ray can aid in the diagnosis of COVID-19 infection: A first experience from Lombardy, Italy. Eur. Radiol. Exp. 2021, 5, 7. [Google Scholar] [CrossRef]
- Sun, J.; Peng, L.; Li, T.; Adila, D.; Zaiman, Z.; Melton, G.B.; Ingraham, N.; Murray, E.; Boley, D.; Switzer, S.; et al. A prospective observational study to investigate performance of a chest X-ray artificial intelligence diagnostic support tool across 12 US hospitals. arXiv 2021, arXiv:2106.02118v2. [Google Scholar]
- Lee, S.; Shin, H.J.; Kim, S.; Kim, E.K. Successful Implementation of an Artificial Intelligence-Based Computer-Aided Detection System for Chest Radiography in Daily Clinical Practice. Korean J. Radiol. 2022, 23, 847–852. [Google Scholar] [CrossRef]
- Annarumma, M.; Withey, S.J.; Bakewell, R.J.; Pesce, E.; Goh, V.; Montana, G. Automated triaging of adult chest radiographs with deep artificial neural networks. Radiology 2019, 291, 196. [Google Scholar] [CrossRef]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a convolutional neural network. In Proceedings of the 2017 International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Advances in Neural Information Processing Systems; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2012; Volume 25. [Google Scholar]
- Refaeilzadeh, P.; Tang, L.; Liu, H. Cross-validation. Encycl. Database Syst. 2009, 5, 532–538. [Google Scholar]
- Ramezan, C.A.; Warner, T.A.; Maxwell, A.E. Evaluation of sampling and cross-validation tuning strategies for regional-scale machine learning classification. Remote Sens. 2019, 11, 185. [Google Scholar] [CrossRef]
- Wortsman, M.; Ilharco, G.; Gadre, S.Y.; Roelofs, R.; Gontijo-Lopes, R.; Morcos, A.S.; Namkoong, H.; Farhadi, A.; Carmon, Y.; Kornblith, S.; et al. Model soups: Averaging weights of multiple fine-tuned models improves accuracy without increasing inference time. In Proceedings of the International Conference on Machine Learning, Baltimore, MA, USA, 17–23 July 2022; pp. 23965–23998. [Google Scholar]
- Suzuki, K.; Matsuzawa, T. Model Soups for Various Training and Validation Data. AI 2022, 3, 796–808. [Google Scholar] [CrossRef]
- Balabanova, Y.; Coker, R.; Fedorin, I.; Zakharova, S.; Plavinskij, S.; Krukov, N.; Atun, R.; Drobniewski, F. Variability in interpretation of chest radiographs among Russian clinicians and implications for screening programmes: Observational study. BMJ 2005, 331, 379–382. [Google Scholar] [CrossRef]
- Whaley, J.S.; Pressman, B.D.; Wilson, J.R.; Bravo, L.; Sehnert, W.J.; Foos, D.H. Investigation of the variability in the assessment of digital chest X-ray image quality. J. Digit. Imaging 2013, 26, 217–226. [Google Scholar] [CrossRef]
- Ravnik, D.; Jerman, T.; Pernuš, F.; Likar, B.; Špiclin, Ž. Dataset variability leverages white-matter lesion segmentation performance with convolutional neural network. In Proceedings of the Medical Imaging 2018: Image Processing, Houston, TX, USA, 10–15 February 2018; Volume 10574, pp. 388–396. [Google Scholar]
- Alvarez-Estevez, D.; Fernández-Varela, I. Addressing database variability in learning from medical data: An ensemble-based approach using convolutional neural networks and a case of study applied to automatic sleep scoring. Comput. Biol. Med. 2020, 119, 103697. [Google Scholar] [CrossRef]
- Abboud, S.; Weiss, F.; Siegel, E.; Jeudy, J. TB or Not TB: Interreader and intrareader variability in screening diagnosis on an iPad versus a traditional display. J. Am. Coll. Radiol. 2013, 10, 42–44. [Google Scholar] [CrossRef]
- Ekpo, E.; Egbe, N.; Akpan, B. Radiographers’ performance in chest X-ray interpretation: The Nigerian experience. Br. J. Radiol. 2015, 88, 20150023. [Google Scholar] [CrossRef]
- Roldán-Nofuentes, J.A. Compbdt: An R program to compare two binary diagnostic tests subject to a paired design. BMC Med. Res. Methodol. 2020, 20, 143. [Google Scholar] [CrossRef]
- Arterys. Retrospective Study X-ray Chest AI Whitepaper. 2020. Available online: https://www.arterys.com/retrospective-study-x-ray-chest-ai-wp (accessed on 10 January 2023).
- Choi, S.Y.; Park, S.; Kim, M.; Park, J.; Choi, Y.R.; Jin, K.N. Evaluation of a deep learning-based computer-aided detection algorithm on chest radiographs: Case–control study. Medicine 2021, 100, e25663. [Google Scholar] [CrossRef]
- Singh, R.; Kalra, M.K.; Nitiwarangkul, C.; Patti, J.A.; Homayounieh, F.; Padole, A.; Rao, P.; Putha, P.; Muse, V.V.; Sharma, A.; et al. Deep learning in chest radiography: Detection of findings and presence of change. PLoS ONE 2018, 13, e0204155. [Google Scholar] [CrossRef]
- Putha, P.; Tadepalli, M.; Reddy, B.; Raj, T.; Chiramal, J.A.; Govil, S.; Sinha, N.; KS, M.; Reddivari, S.; Jagirdar, A.; et al. Can artificial intelligence reliably report chest x-rays?: Radiologist validation of an algorithm trained on 2.3 million x-rays. arXiv 2018, arXiv:1807.07455. [Google Scholar]
- Caring-Research. Automated classification of chest X-rays as normal/abnormal using a high sensitivity deep learning algorithm. In Proceedings of the European Congress of Radiology 2019, Vienna, Austria, 27 February–3 March 2019. [Google Scholar]
- Engle, E.; Gabrielian, A.; Long, A.; Hurt, D.E.; Rosenthal, A. Performance of Qure. ai automatic classifiers against a large annotated database of patients with diverse forms of tuberculosis. PLoS ONE 2020, 15, e0224445. [Google Scholar] [CrossRef] [PubMed]
- Dyer, T.; Dillard, L.; Harrison, M.; Morgan, T.N.; Tappouni, R.; Malik, Q.; Rasalingham, S. Diagnosis of normal chest radiographs using an autonomous deep-learning algorithm. Clin. Radiol. 2021, 76, 473.e9–473.e15. [Google Scholar] [CrossRef]
- Hwang, E.J.; Park, S.; Jin, K.N.; Im Kim, J.; Choi, S.Y.; Lee, J.H.; Goo, J.M.; Aum, J.; Yim, J.J.; Cohen, J.G.; et al. Development and validation of a deep learning–based automated detection algorithm for major thoracic diseases on chest radiographs. JAMA Netw. Open 2019, 2, e191095. [Google Scholar] [CrossRef]
- Keski-Filppula, T.; Nikki, M.; Haapea, M.; Ramanauskas, N.; Tervonen, O. Using artificial intelligence to detect chest X-rays with no significant findings in a primary health care setting in Oulu, Finland. arXiv 2022, arXiv:2205.08123. [Google Scholar]
- Jang, S.B.; Lee, S.H.; Lee, D.E.; Park, S.Y.; Kim, J.K.; Cho, J.W.; Cho, J.; Kim, K.B.; Park, B.; Park, J.; et al. Deep-learning algorithms for the interpretation of chest radiographs to aid in the triage of COVID-19 patients: A multicenter retrospective study. PLoS ONE 2020, 15, e0242759. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).