Artificial Intelligence: The Milestone in Modern Biomedical Research
Abstract
1. Introduction
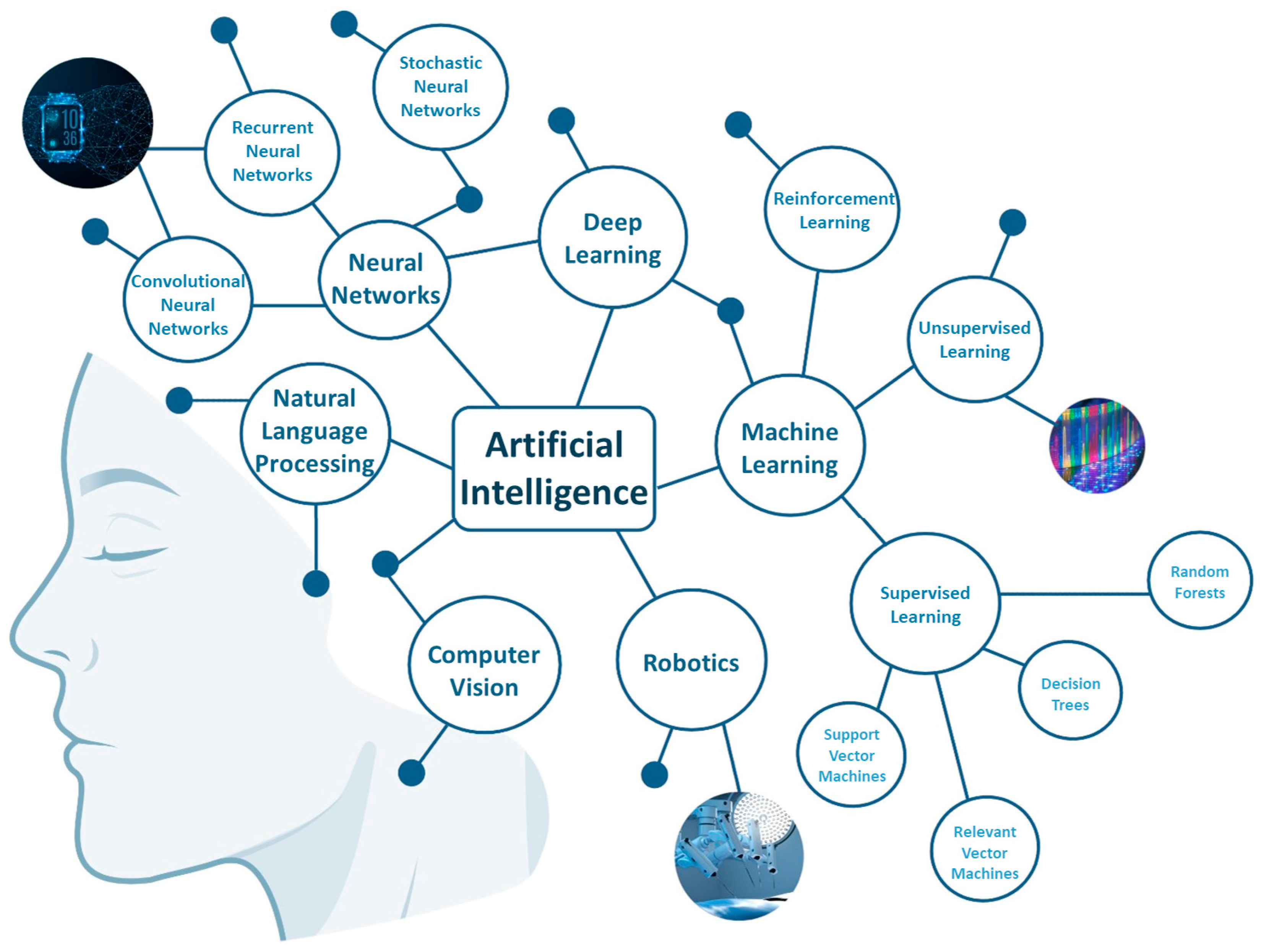
2. Deciphering the Main Fields of AI
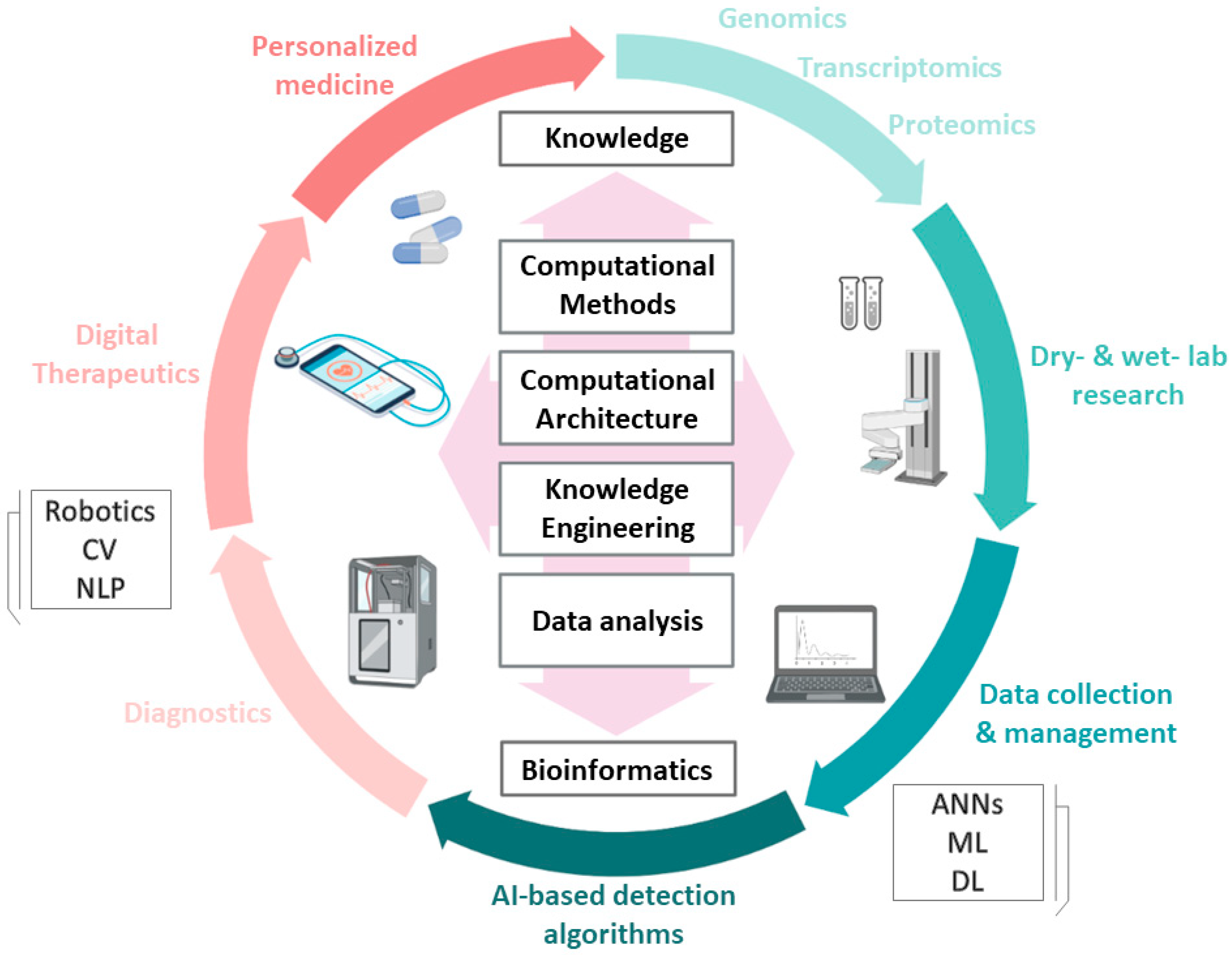
3. AI in Life Sciences
3.1. Applications of AI in Biology
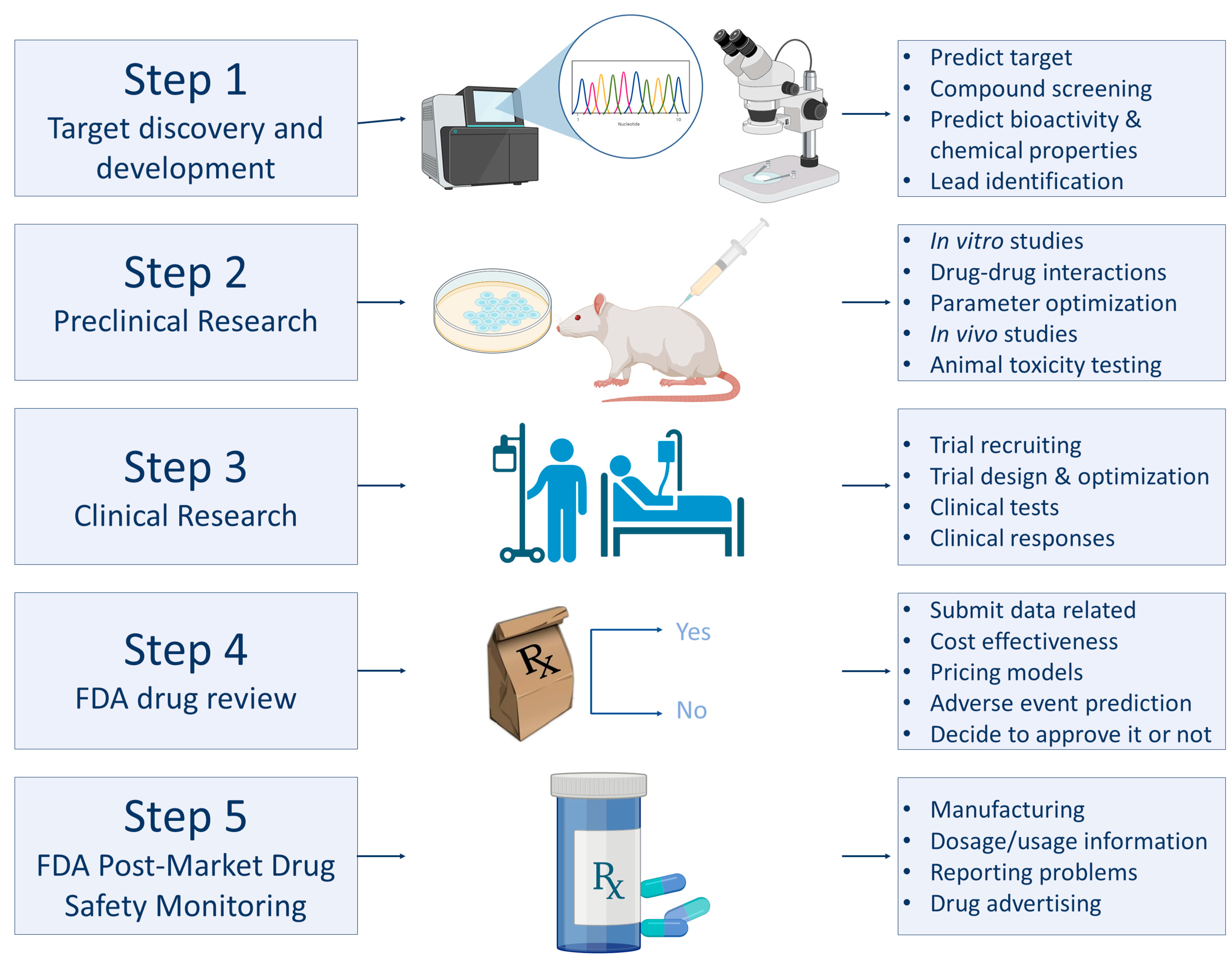
3.2. Applications of AI in Medicine
| AI Algorithm | Description | Applications | Reference | |
|---|---|---|---|---|
| Medical image analysis | Arterys Cardio DL | Automated analysis of cardiac MRI scans | Radiology Cardiology | [106] |
| Arterys Oncology DL | Detection of liver lesions and lung nodules on MRI and CT scans | Radiology Oncology | [107] | |
| EchoMD Automated Ejection Fraction Software | Echocardiogram analysis for evaluation of the left ventricular ejection fraction | Radiology Cardiology | [108] | |
| HealthPNX | Assessment of chest X-rays for signs indicative of pneumothorax | Radiology | [109] | |
| icobrain | Analysis of brain CT and MRI scans of patients with brain injuries and neurological disorders | Radiology Neurology | [110,111] | |
| IDx-DR | Analysis of retinal images for diabetic retinopathy detection | Ophthalmology | [112] | |
| OsteoDetect | Analysis of wrist X-rays for fracture diagnosis | Radiology | [113] | |
| ProFound™ AI Software V2.1 | Analysis of breast density and detection of malignant lesions using mammograms | Radiology Oncology | [107,114] | |
| QuantX™ | Diagnosis of breast lesions through MRI scans analysis | Radiology Oncology | [115] | |
| Viz LVO | Detection of signs of stroke using CT angiography scans | Radiology Neurology | [116] | |
| Surgical robotics | Da Vinci® Surgical System | Telemanipulated minimally invasive robotic system with robotic arms that translate user’s hand movements, providing precision and filtering of tremors | Prostatectomies, gynecological, urological, gastrointestinal, cardiothoracic surgeries | [94] |
| Flex® robotic System | Flexible robotic endoscope that allows surgeons to access and visualize anatomical areas which normally are inaccessible by minimally invasive approaches | Minimally invasive surgeries of larynx, oropharynx and hypopharynx | [117] | |
| FreeHand® v1.2 | Robotic camera controller that provides steadier images, better control of camera movements and reduced surgical time | Minimally invasive and laparoscopic surgeries (gynecological, urological, thoracic, general) | [118] | |
| NAVIO™ Surgical System | Handheld robotic system that offers real-time bone mapping, planning of implant positioning and robotic-assisted bone preparation | Knee arthroplasty | [119] | |
| NeoGuide™ Endoscopy System | Computer-assisted colonoscope that enables visualization of the lower gastrointestinal (GI) tract, adjusting to colon’s shape and thus decreasing looping | Colonoscopy | [120] | |
| Senhance® Surgical System | Remotely controlled robotic system with three robotic arms, providing improved visualization and haptic feedback | Laparoscopic surgeries (abdominal, gynecological, urological) | [121] | |
| Sensei® X Robotic Catheter System | Remotely manipulated cardiac catheter that translates surgeon’s hand motions, providing stability during catheter positioning | Catheter positioning | [122] | |
| Remote patient monitoring | ADAMM Intelligent Asthma Monitoring | Wearable device attached to the torso that monitors asthma symptoms and alerts in cases of significant deviations | Pulmonology | [123] |
| CardioMEMS™ HF System | Wireless monitoring system that records pulmonary artery (PA) pressure in patients with heart failure through an implantable PA sensor. The daily data are sent and assessed by the healthcare provider | Cardiology | [124] | |
| Confirm Rx™ Insertable Cardiac Monitor | Insertable device that monitors patients’ heart rhythms, detects signs of arrythmia and transmits data to clinicians | Cardiology | [125] | |
| ReDS™ System | Device that rapidly and non-invasively measures the absolute fluid content in the lungs of heart failure patients. | Cardiology | [126] | |
| Triggerfish® | Contact lens, embedded with a microsensor, that monitors (for 24 h) ocular dimensional alterations and thus intraocular pressure variations in patients with glaucoma | Ophthalmology | [127] | |
| Personal biosensing devices | Eversense® E3 Continuous Glucose Monitoring (CGM) System | A system utilizing an implantable glucose sensor that regularly monitors glucose levels, a transmitter worn externally and a mobile application for real-time data display | Clinical Chemistry | [128] |
| Fitbit | Smartwatch that monitors physical activities, sleep and can detect signs of atrial fibrillation | Cardiology | [129] | |
| KardiaMobile® 6L | Portable heart monitor that detects heart arryhthmias | Cardiology | [130] | |
| Study Watch with Irregular Pulse Monitor | Wearable device that records biometric information, such as heart’s electrical activity, and detects irregular heart rates | Cardiology | [131] |
4. Challenges and Future Directions
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Green, E.D.; Watson, J.D.; Collins, F.S. Human Genome Project: Twenty-five years of big biology. Nature 2015, 526, 29–31. [Google Scholar] [CrossRef]
- Collins, F.S.; Morgan, M.; Patrinos, A. The Human Genome Project: Lessons from large-scale biology. Science 2003, 300, 286–290. [Google Scholar] [CrossRef]
- Biswas, N.; Chakrabarti, S. Artificial Intelligence (AI)-Based Systems Biology Approaches in Multi-Omics Data Analysis of Cancer. Front. Oncol. 2020, 10, 588221. [Google Scholar] [CrossRef]
- Branco, I.; Choupina, A. Bioinformatics: New tools and applications in life science and personalized medicine. Appl. Microbiol. Biotechnol. 2021, 105, 937–951. [Google Scholar] [CrossRef]
- Leite, M.L.; de Loiola Costa, L.S.; Cunha, V.A.; Kreniski, V.; de Oliveira Braga Filho, M.; da Cunha, N.B.; Costa, F.F. Artificial intelligence and the future of life sciences. Drug Discov. Today 2021, 26, 2515–2526. [Google Scholar] [CrossRef]
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69S, S36–S40. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Kishore, S.; Pandey, D.K. Artificial Intelligence in Biological Sciences. Life 2022, 12, 1430. [Google Scholar] [CrossRef]
- Savage, N. Breaking into the black box of artificial intelligence. Nature 2022. [Google Scholar] [CrossRef]
- Chen, C.; Wu, T.; Guo, Z.; Cheng, J. Combination of deep neural network with attention mechanism enhances the explainability of protein contact prediction. Proteins 2021, 89, 697–707. [Google Scholar] [CrossRef]
- Canzoneri, R.; Lacunza, E.; Abba, M.C. Genomics and bioinformatics as pillars of precision medicine in oncology. Medicina 2019, 79, 587–592. [Google Scholar]
- Tran, K.A.; Kondrashova, O.; Bradley, A.; Williams, E.D.; Pearson, J.V.; Waddell, N. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med. 2021, 13, 152. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, S.L.; Nir, G.; Salcudean, S.E. A new era: Artificial intelligence and machine learning in prostate cancer. Nat. Rev. Urol. 2019, 16, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Sirsat, M.S.; Ferme, E.; Camara, J. Machine Learning for Brain Stroke: A Review. J. Stroke Cerebrovasc. Dis. 2020, 29, 105162. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Cai, N.; Pacheco, P.P.; Narrandes, S.; Wang, Y.; Xu, W. Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genom. Proteom. 2018, 15, 41–51. [Google Scholar] [CrossRef]
- Eraslan, G.; Avsec, Z.; Gagneur, J.; Theis, F.J. Deep learning: New computational modelling techniques for genomics. Nat. Rev. Genet. 2019, 20, 389–403. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Ghahramani, Z. Probabilistic machine learning and artificial intelligence. Nature 2015, 521, 452–459. [Google Scholar] [CrossRef]
- Renganathan, V. Overview of artificial neural network models in the biomedical domain. Bratisl. Lek. Listy 2019, 120, 536–540. [Google Scholar] [CrossRef]
- Vogels, T.P.; Rajan, K.; Abbott, L.F. Neural network dynamics. Annu. Rev. Neurosci. 2005, 28, 357–376. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, H.; Yang, Z.; Wang, J.; Sun, Y.; Xu, B.; Zhao, Z. Neural network-based approaches for biomedical relation classification: A review. J. Biomed. Inform. 2019, 99, 103294. [Google Scholar] [CrossRef] [PubMed]
- Albaradei, S.; Thafar, M.; Alsaedi, A.; Van Neste, C.; Gojobori, T.; Essack, M.; Gao, X. Machine learning and deep learning methods that use omics data for metastasis prediction. Comput. Struct. Biotechnol. J. 2021, 19, 5008–5018. [Google Scholar] [CrossRef] [PubMed]
- Zaharchuk, G.; Gong, E.; Wintermark, M.; Rubin, D.; Langlotz, C.P. Deep Learning in Neuroradiology. AJNR Am. J. Neuroradiol. 2018, 39, 1776–1784. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, A.N.; Kambhampati, C.; Monson, J.R.; Drew, P.J. Artificial intelligence in medicine. Ann. R. Coll. Surg. Engl. 2004, 86, 334–338. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, B.; Wang, C.; Wang, Z.; Liu, B.; Fang, T. Computer Vision-Based Construction Process Sensing for Cyber-Physical Systems: A Review. Sensors 2021, 21, 5468. [Google Scholar] [CrossRef]
- Kreimeyer, K.; Foster, M.; Pandey, A.; Arya, N.; Halford, G.; Jones, S.F.; Forshee, R.; Walderhaug, M.; Botsis, T. Natural language processing systems for capturing and standardizing unstructured clinical information: A systematic review. J. Biomed. Inform. 2017, 73, 14–29. [Google Scholar] [CrossRef]
- Wu, S.; Roberts, K.; Datta, S.; Du, J.; Ji, Z.; Si, Y.; Soni, S.; Wang, Q.; Wei, Q.; Xiang, Y.; et al. Deep learning in clinical natural language processing: A methodical review. J. Am. Med. Inform. Assoc. 2020, 27, 457–470. [Google Scholar] [CrossRef]
- Shimizu, H.; Nakayama, K.I. Artificial intelligence in oncology. Cancer Sci. 2020, 111, 1452–1460. [Google Scholar] [CrossRef]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-Generation Machine Learning for Biological Networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef]
- Tang, B.; Pan, Z.; Yin, K.; Khateeb, A. Recent Advances of Deep Learning in Bioinformatics and Computational Biology. Front. Genet. 2019, 10, 214. [Google Scholar] [CrossRef]
- Larranaga, P.; Calvo, B.; Santana, R.; Bielza, C.; Galdiano, J.; Inza, I.; Lozano, J.A.; Armananzas, R.; Santafe, G.; Perez, A.; et al. Machine learning in bioinformatics. Brief. Bioinform. 2006, 7, 86–112. [Google Scholar] [CrossRef] [PubMed]
- Mathe, C.; Sagot, M.F.; Schiex, T.; Rouze, P. Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res. 2002, 30, 4103–4117. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Jongeneel, C.V. Searching the expressed sequence tag (EST) databases: Panning for genes. Brief. Bioinform. 2000, 1, 76–92. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M.; International Nucleotide Sequence Database Collaboration. The sequence read archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef]
- Swan, A.L.; Mobasheri, A.; Allaway, D.; Liddell, S.; Bacardit, J. Application of machine learning to proteomics data: Classification and biomarker identification in postgenomics biology. OMICS 2013, 17, 595–610. [Google Scholar] [CrossRef]
- Lise, S.; Buchan, D.; Pontil, M.; Jones, D.T. Predictions of hot spot residues at protein-protein interfaces using support vector machines. PLoS ONE 2011, 6, e16774. [Google Scholar] [CrossRef]
- Preto, A.J.; Matos-Filipe, P.; de Almeida, J.G.; Mourao, J.; Moreira, I.S. Predicting Hot Spots Using a Deep Neural Network Approach. Methods Mol. Biol. 2021, 2190, 267–288. [Google Scholar] [CrossRef]
- Lee, E.S.; Durant, T.J.S. Supervised machine learning in the mass spectrometry laboratory: A tutorial. J. Mass Spectrom. Adv. Clin. Lab. 2022, 23, 1–6. [Google Scholar] [CrossRef]
- Yates, J.R.; Ruse, C.I.; Nakorchevsky, A. Proteomics by mass spectrometry: Approaches, advances, and applications. Annu. Rev. Biomed. Eng. 2009, 11, 49–79. [Google Scholar] [CrossRef]
- Domon, B.; Aebersold, R. Mass spectrometry and protein analysis. Science 2006, 312, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Sadygov, R.G.; Cociorva, D.; Yates, J.R., 3rd. Large-scale database searching using tandem mass spectra: Looking up the answer in the back of the book. Nat. Methods 2004, 1, 195–202. [Google Scholar] [CrossRef]
- Wei, Y.; Varanasi, R.S.; Schwarz, T.; Gomell, L.; Zhao, H.; Larson, D.J.; Sun, B.; Liu, G.; Chen, H.; Raabe, D.; et al. Machine-learning-enhanced time-of-flight mass spectrometry analysis. Patterns 2021, 2, 100192. [Google Scholar] [CrossRef]
- Moreira, I.S.; Koukos, P.I.; Melo, R.; Almeida, J.G.; Preto, A.J.; Schaarschmidt, J.; Trellet, M.; Gumus, Z.H.; Costa, J.; Bonvin, A. SpotOn: High Accuracy Identification of Protein-Protein Interface Hot-Spots. Sci. Rep. 2017, 7, 8007. [Google Scholar] [CrossRef]
- Qiao, Y.; Xiong, Y.; Gao, H.; Zhu, X.; Chen, P. Protein-protein interface hot spots prediction based on a hybrid feature selection strategy. BMC Bioinform. 2018, 19, 14. [Google Scholar] [CrossRef]
- Meyer, M.J.; Beltran, J.F.; Liang, S.; Fragoza, R.; Rumack, A.; Liang, J.; Wei, X.; Yu, H. Interactome INSIDER: A structural interactome browser for genomic studies. Nat. Methods 2018, 15, 107–114. [Google Scholar] [CrossRef]
- Gaulton, K.J.; Nammo, T.; Pasquali, L.; Simon, J.M.; Giresi, P.G.; Fogarty, M.P.; Panhuis, T.M.; Mieczkowski, P.; Secchi, A.; Bosco, D.; et al. A map of open chromatin in human pancreatic islets. Nat. Genet. 2010, 42, 255–259. [Google Scholar] [CrossRef]
- Muerdter, F.; Boryn, L.M.; Arnold, C.D. STARR-seq—Principles and applications. Genomics 2015, 106, 145–150. [Google Scholar] [CrossRef]
- Bianco, S.; Rodrigue, S.; Murphy, B.D.; Gevry, N. Global Mapping of Open Chromatin Regulatory Elements by Formaldehyde-Assisted Isolation of Regulatory Elements Followed by Sequencing (FAIRE-seq). Methods Mol. Biol. 2015, 1334, 261–272. [Google Scholar] [CrossRef]
- Kaur, H.; Singh, Y.; Singh, S.; Singh, R.B. Gut microbiome-mediated epigenetic regulation of brain disorder and application of machine learning for multi-omics data analysis. Genome 2021, 64, 355–371. [Google Scholar] [CrossRef]
- Gou, W.; Ling, C.W.; He, Y.; Jiang, Z.; Fu, Y.; Xu, F.; Miao, Z.; Sun, T.Y.; Lin, J.S.; Zhu, H.L.; et al. Interpretable Machine Learning Framework Reveals Robust Gut Microbiome Features Associated With Type 2 Diabetes. Diabetes Care 2021, 44, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Cammarota, G.; Ianiro, G.; Ahern, A.; Carbone, C.; Temko, A.; Claesson, M.J.; Gasbarrini, A.; Tortora, G. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 635–648. [Google Scholar] [CrossRef] [PubMed]
- De Vos, W.M.; de Vos, E.A. Role of the intestinal microbiome in health and disease: From correlation to causation. Nutr. Rev. 2012, 70 (Suppl. 1), S45–S56. [Google Scholar] [CrossRef]
- Marya, N.B.; Powers, P.D.; Chari, S.T.; Gleeson, F.C.; Leggett, C.L.; Abu Dayyeh, B.K.; Chandrasekhara, V.; Iyer, P.G.; Majumder, S.; Pearson, R.K.; et al. Utilisation of artificial intelligence for the development of an EUS-convolutional neural network model trained to enhance the diagnosis of autoimmune pancreatitis. Gut 2021, 70, 1335–1344. [Google Scholar] [CrossRef]
- Reiman, D.; Metwally, A.; Yang, D. Using convolutional neural networks to explore the microbiome. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2017, 2017, 4269–4272. [Google Scholar] [CrossRef]
- Reiman, D.; Farhat, A.M.; Dai, Y. Predicting Host Phenotype Based on Gut Microbiome Using a Convolutional Neural Network Approach. Methods Mol. Biol. 2021, 2190, 249–266. [Google Scholar] [CrossRef]
- Janssens, Y.; Nielandt, J.; Bronselaer, A.; Debunne, N.; Verbeke, F.; Wynendaele, E.; Van Immerseel, F.; Vandewynckel, Y.P.; De Tre, G.; De Spiegeleer, B. Disbiome database: Linking the microbiome to disease. BMC Microbiol. 2018, 18, 50. [Google Scholar] [CrossRef]
- Cheng, L.; Qi, C.; Zhuang, H.; Fu, T.; Zhang, X. gutMDisorder: A comprehensive database for dysbiosis of the gut microbiota in disorders and interventions. Nucleic Acids Res. 2020, 48, D554–D560. [Google Scholar] [CrossRef]
- Dai, D.; Zhu, J.; Sun, C.; Li, M.; Liu, J.; Wu, S.; Ning, K.; He, L.J.; Zhao, X.M.; Chen, W.H. GMrepo v2: A curated human gut microbiome database with special focus on disease markers and cross-dataset comparison. Nucleic Acids Res. 2022, 50, D777–D784. [Google Scholar] [CrossRef]
- Shi, W.; Qi, H.; Sun, Q.; Fan, G.; Liu, S.; Wang, J.; Zhu, B.; Liu, H.; Zhao, F.; Wang, X.; et al. gcMeta: A Global Catalogue of Metagenomics platform to support the archiving, standardization and analysis of microbiome data. Nucleic Acids Res. 2019, 47, D637–D648. [Google Scholar] [CrossRef]
- Reiman, D.; Metwally, A.A.; Sun, J.; Dai, Y. PopPhy-CNN: A Phylogenetic Tree Embedded Architecture for Convolutional Neural Networks to Predict Host Phenotype From Metagenomic Data. IEEE J. Biomed. Health Inform. 2020, 24, 2993–3001. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Woloszynek, S.; Agbavor, F.; Mell, J.C.; Sokhansanj, B.A.; Rosen, G.L. Learning, visualizing and exploring 16S rRNA structure using an attention-based deep neural network. PLoS Comput. Biol. 2021, 17, e1009345. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Pan, Y.; Hao, M.; Wang, Y.; Bryant, S.H. PubChem applications in drug discovery: A bibliometric analysis. Drug Discov. Today 2014, 19, 1751–1756. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Nowotka, M.M.; Gaulton, A.; Mendez, D.; Bento, A.P.; Hersey, A.; Leach, A. Using ChEMBL web services for building applications and data processing workflows relevant to drug discovery. Expert Opin. Drug Discov. 2017, 12, 757–767. [Google Scholar] [CrossRef]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schioth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef]
- Yu, W.; MacKerell, A.D., Jr. Computer-Aided Drug Design Methods. Methods Mol. Biol. 2017, 1520, 85–106. [Google Scholar] [CrossRef]
- Martinelli, D.D. Generative machine learning for de novo drug discovery: A systematic review. Comput. Biol. Med. 2022, 145, 105403. [Google Scholar] [CrossRef]
- Baptista, D.; Ferreira, P.G.; Rocha, M. Deep learning for drug response prediction in cancer. Brief. Bioinform. 2021, 22, 360–379. [Google Scholar] [CrossRef]
- Verma, J.; Khedkar, V.M.; Coutinho, E.C. 3D-QSAR in drug design—A review. Curr. Top. Med. Chem. 2010, 10, 95–115. [Google Scholar] [CrossRef]
- Wang, T.; Wu, M.B.; Lin, J.P.; Yang, L.R. Quantitative structure-activity relationship: Promising advances in drug discovery platforms. Expert Opin. Drug Discov. 2015, 10, 1283–1300. [Google Scholar] [CrossRef] [PubMed]
- Basak, S.C. Some Comments on the Three-Pronged Chemobiodescriptor Approach to QSAR—A Historical View of the Emerging Integration. Curr. Comput. Aided Drug Des. 2021, 17, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Carracedo-Reboredo, P.; Linares-Blanco, J.; Rodriguez-Fernandez, N.; Cedron, F.; Novoa, F.J.; Carballal, A.; Maojo, V.; Pazos, A.; Fernandez-Lozano, C. A review on machine learning approaches and trends in drug discovery. Comput. Struct. Biotechnol. J. 2021, 19, 4538–4558. [Google Scholar] [CrossRef] [PubMed]
- Dara, S.; Dhamercherla, S.; Jadav, S.S.; Babu, C.M.; Ahsan, M.J. Machine Learning in Drug Discovery: A Review. Artif. Intell. Rev. 2022, 55, 1947–1999. [Google Scholar] [CrossRef]
- Kaul, V.; Enslin, S.; Gross, S.A. History of artificial intelligence in medicine. Gastrointest. Endosc. 2020, 92, 807–812. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Mazurowski, M.A.; Buda, M.; Saha, A.; Bashir, M.R. Deep learning in radiology: An overview of the concepts and a survey of the state of the art with focus on MRI. J. Magn. Reson. Imaging 2019, 49, 939–954. [Google Scholar] [CrossRef]
- Al-Waisy, A.S.; Al-Fahdawi, S.; Mohammed, M.A.; Abdulkareem, K.H.; Mostafa, S.A.; Maashi, M.S.; Arif, M.; Garcia-Zapirain, B. COVID-CheXNet: Hybrid deep learning framework for identifying COVID-19 virus in chest X-rays images. Soft Comput. 2020, 1–16. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Lee, E.H.; Zheng, J.; Zhang, W.; Halabi, S.; Liu, C.; Deng, K.; Song, J.; Yeom, K.W. Decoding COVID-19 pneumonia: Comparison of deep learning and radiomics CT image signatures. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1478–1486. [Google Scholar] [CrossRef]
- Mambou, S.J.; Maresova, P.; Krejcar, O.; Selamat, A.; Kuca, K. Breast Cancer Detection Using Infrared Thermal Imaging and a Deep Learning Model. Sensors 2018, 18, 2799. [Google Scholar] [CrossRef]
- Devnath, L.; Summons, P.; Luo, S.; Wang, D.; Shaukat, K.; Hameed, I.A.; Aljuaid, H. Computer-Aided Diagnosis of Coal Workers’ Pneumoconiosis in Chest X-ray Radiographs Using Machine Learning: A Systematic Literature Review. Int. J. Environ. Res. Public Health 2022, 19, 6439. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Chi, J.; Liu, J.; Yang, L.; Zhang, B.; Yu, D.; Zhao, Y.; Lu, X. A survey of computer-aided diagnosis of lung nodules from CT scans using deep learning. Comput. Biol. Med. 2021, 137, 104806. [Google Scholar] [CrossRef]
- Meena, T.; Roy, S. Bone Fracture Detection Using Deep Supervised Learning from Radiological Images: A Paradigm Shift. Diagnostics 2022, 12, 2420. [Google Scholar] [CrossRef]
- Kundisch, A.; Honning, A.; Mutze, S.; Kreissl, L.; Spohn, F.; Lemcke, J.; Sitz, M.; Sparenberg, P.; Goelz, L. Deep learning algorithm in detecting intracranial hemorrhages on emergency computed tomographies. PLoS ONE 2021, 16, e0260560. [Google Scholar] [CrossRef]
- Yala, A.; Lehman, C.; Schuster, T.; Portnoi, T.; Barzilay, R. A Deep Learning Mammography-based Model for Improved Breast Cancer Risk Prediction. Radiology 2019, 292, 60–66. [Google Scholar] [CrossRef]
- Tufail, A.B.; Ma, Y.K.; Kaabar, M.K.A.; Martinez, F.; Junejo, A.R.; Ullah, I.; Khan, R. Deep Learning in Cancer Diagnosis and Prognosis Prediction: A Minireview on Challenges, Recent Trends, and Future Directions. Comput. Math. Methods Med. 2021, 2021, 9025470. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, M.; Wang, S.; Li, X.; Sun, Y. Emerging role of deep learning-based artificial intelligence in tumor pathology. Cancer Commun. 2020, 40, 154–166. [Google Scholar] [CrossRef]
- Siontis, K.C.; Noseworthy, P.A.; Attia, Z.I.; Friedman, P.A. Artificial intelligence-enhanced electrocardiography in cardiovascular disease management. Nat. Rev. Cardiol. 2021, 18, 465–478. [Google Scholar] [CrossRef]
- El-Khatib, H.; Popescu, D.; Ichim, L. Deep Learning-Based Methods for Automatic Diagnosis of Skin Lesions. Sensors 2020, 20, 1753. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Peng, L.; Varadarajan, A.V.; Keane, P.A.; Burlina, P.M.; Chiang, M.F.; Schmetterer, L.; Pasquale, L.R.; Bressler, N.M.; Webster, D.R.; et al. Deep learning in ophthalmology: The technical and clinical considerations. Prog. Retin. Eye Res. 2019, 72, 100759. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Peralta, L.F.; Bote-Curiel, L.; Picon, A.; Sanchez-Margallo, F.M.; Pagador, J.B. Deep learning to find colorectal polyps in colonoscopy: A systematic literature review. Artif. Intell. Med. 2020, 108, 101923. [Google Scholar] [CrossRef] [PubMed]
- Leung, T.; Vyas, D. Robotic Surgery: Applications. Am. J. Robot. Surg. 2014, 1, 1–64. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Morrell, A.L.G.; Morrell-Junior, A.C.; Morrell, A.G.; Mendes, J.M.F.; Tustumi, F.; DE-Oliveira-E-Silva, L.G.; Morrell, A. The history of robotic surgery and its evolution: When illusion becomes reality. Rev. Col. Bras. Cir. 2021, 48, e20202798. [Google Scholar] [CrossRef]
- Moustris, G.P.; Hiridis, S.C.; Deliparaschos, K.M.; Konstantinidis, K.M. Evolution of autonomous and semi-autonomous robotic surgical systems: A review of the literature. Int. J. Med. Robot. 2011, 7, 375–392. [Google Scholar] [CrossRef]
- Gumbs, A.A.; Grasso, V.; Bourdel, N.; Croner, R.; Spolverato, G.; Frigerio, I.; Illanes, A.; Abu Hilal, M.; Park, A.; Elyan, E. The Advances in Computer Vision That Are Enabling More Autonomous Actions in Surgery: A Systematic Review of the Literature. Sensors 2022, 22, 4918. [Google Scholar] [CrossRef]
- Pettit, R.W.; Fullem, R.; Cheng, C.; Amos, C.I. Artificial intelligence, machine learning, and deep learning for clinical outcome prediction. Emerg. Top. Life Sci. 2021, 5, 729–745. [Google Scholar] [CrossRef]
- Manickam, P.; Mariappan, S.A.; Murugesan, S.M.; Hansda, S.; Kaushik, A.; Shinde, R.; Thipperudraswamy, S.P. Artificial Intelligence (AI) and Internet of Medical Things (IoMT) Assisted Biomedical Systems for Intelligent Healthcare. Biosensors 2022, 12, 562. [Google Scholar] [CrossRef]
- Miotto, R.; Li, L.; Kidd, B.A.; Dudley, J.T. Deep Patient: An Unsupervised Representation to Predict the Future of Patients from the Electronic Health Records. Sci. Rep. 2016, 6, 26094. [Google Scholar] [CrossRef]
- Giannini, H.M.; Ginestra, J.C.; Chivers, C.; Draugelis, M.; Hanish, A.; Schweickert, W.D.; Fuchs, B.D.; Meadows, L.; Lynch, M.; Donnelly, P.J.; et al. A Machine Learning Algorithm to Predict Severe Sepsis and Septic Shock: Development, Implementation, and Impact on Clinical Practice. Crit. Care Med. 2019, 47, 1485–1492. [Google Scholar] [CrossRef]
- Rajkomar, A.; Oren, E.; Chen, K.; Dai, A.M.; Hajaj, N.; Hardt, M.; Liu, P.J.; Liu, X.; Marcus, J.; Sun, M.; et al. Scalable and accurate deep learning with electronic health records. NPJ Digit. Med. 2018, 1, 18. [Google Scholar] [CrossRef]
- Cai, X.; Perez-Concha, O.; Coiera, E.; Martin-Sanchez, F.; Day, R.; Roffe, D.; Gallego, B. Real-time prediction of mortality, readmission, and length of stay using electronic health record data. J. Am. Med. Inform. Assoc. 2016, 23, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.R.; Lu, L.; Zhang, J.Y.; Huo, T.T.; Liu, S.X.; Ye, Z.W. Application of Artificial Intelligence in Medicine: An Overview. Curr. Med. Sci. 2021, 41, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Thoren, A.; Rawshani, A.; Herlitz, J.; Engdahl, J.; Kahan, T.; Gustafsson, L.; Djarv, T. ECG-monitoring of in-hospital cardiac arrest and factors associated with survival. Resuscitation 2020, 150, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Churpek, M.M.; Yuen, T.C.; Huber, M.T.; Park, S.Y.; Hall, J.B.; Edelson, D.P. Predicting cardiac arrest on the wards: A nested case-control study. Chest 2012, 141, 1170–1176. [Google Scholar] [CrossRef] [PubMed]
- Masutani, E.M.; Bahrami, N.; Hsiao, A. Deep Learning Single-Frame and Multiframe Super-Resolution for Cardiac MRI. Radiology 2020, 295, 552–561. [Google Scholar] [CrossRef]
- Hamamoto, R.; Suvarna, K.; Yamada, M.; Kobayashi, K.; Shinkai, N.; Miyake, M.; Takahashi, M.; Jinnai, S.; Shimoyama, R.; Sakai, A.; et al. Application of Artificial Intelligence Technology in Oncology: Towards the Establishment of Precision Medicine. Cancers 2020, 12, 3532. [Google Scholar] [CrossRef]
- Asch, F.M.; Poilvert, N.; Abraham, T.; Jankowski, M.; Cleve, J.; Adams, M.; Romano, N.; Hong, H.; Mor-Avi, V.; Martin, R.P.; et al. Automated Echocardiographic Quantification of Left Ventricular Ejection Fraction Without Volume Measurements Using a Machine Learning Algorithm Mimicking a Human Expert. Circ. Cardiovasc. Imaging 2019, 12, e009303. [Google Scholar] [CrossRef]
- Adams, S.J.; Henderson, R.D.E.; Yi, X.; Babyn, P. Artificial Intelligence Solutions for Analysis of X-ray Images. Can. Assoc. Radiol. J. 2021, 72, 60–72. [Google Scholar] [CrossRef]
- Jain, S.; Vyvere, T.V.; Terzopoulos, V.; Sima, D.M.; Roura, E.; Maas, A.; Wilms, G.; Verheyden, J. Automatic Quantification of Computed Tomography Features in Acute Traumatic Brain Injury. J. Neurotrauma 2019, 36, 1794–1803. [Google Scholar] [CrossRef]
- Rakic, M.; Vercruyssen, S.; Van Eyndhoven, S.; de la Rosa, E.; Jain, S.; Van Huffel, S.; Maes, F.; Smeets, D.; Sima, D.M. icobrain ms 5.1: Combining unsupervised and supervised approaches for improving the detection of multiple sclerosis lesions. Neuroimage Clin. 2021, 31, 102707. [Google Scholar] [CrossRef] [PubMed]
- Savoy, M. IDx-DR for Diabetic Retinopathy Screening. Am. Fam. Physician 2020, 101, 307–308. [Google Scholar] [PubMed]
- Voelker, R. Diagnosing Fractures with AI. JAMA 2018, 320, 23. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Lin, L.; Wu, C.F.; Li, C.F.; Xu, R.H.; Sun, Y. Artificial intelligence for assisting cancer diagnosis and treatment in the era of precision medicine. Cancer Commun. 2021, 41, 1100–1115. [Google Scholar] [CrossRef]
- Jiang, Y.; Edwards, A.V.; Newstead, G.M. Artificial Intelligence Applied to Breast MRI for Improved Diagnosis. Radiology 2021, 298, 38–46. [Google Scholar] [CrossRef]
- Yahav-Dovrat, A.; Saban, M.; Merhav, G.; Lankri, I.; Abergel, E.; Eran, A.; Tanne, D.; Nogueira, R.G.; Sivan-Hoffmann, R. Evaluation of Artificial Intelligence-Powered Identification of Large-Vessel Occlusions in a Comprehensive Stroke Center. AJNR Am. J. Neuroradiol. 2021, 42, 247–254. [Google Scholar] [CrossRef]
- Mattheis, S.; Hasskamp, P.; Holtmann, L.; Schafer, C.; Geisthoff, U.; Dominas, N.; Lang, S. Flex Robotic System in transoral robotic surgery: The first 40 patients. Head Neck 2017, 39, 471–475. [Google Scholar] [CrossRef]
- Stolzenburg, J.U.; Franz, T.; Kallidonis, P.; Minh, D.; Dietel, A.; Hicks, J.; Nicolaus, M.; Al-Aown, A.; Liatsikos, E. Comparison of the FreeHand(R) robotic camera holder with human assistants during endoscopic extraperitoneal radical prostatectomy. BJU Int. 2011, 107, 970–974. [Google Scholar] [CrossRef]
- Battenberg, A.K.; Netravali, N.A.; Lonner, J.H. A novel handheld robotic-assisted system for unicompartmental knee arthroplasty: Surgical technique and early survivorship. J. Robot. Surg. 2020, 14, 55–60. [Google Scholar] [CrossRef]
- Eickhoff, A.; van Dam, J.; Jakobs, R.; Kudis, V.; Hartmann, D.; Damian, U.; Weickert, U.; Schilling, D.; Riemann, J.F. Computer-assisted colonoscopy (the NeoGuide Endoscopy System): Results of the first human clinical trial (“PACE study”). Am. J. Gastroenterol. 2007, 102, 261–266. [Google Scholar] [CrossRef]
- Kastelan, Z.; Knezevic, N.; Hudolin, T.; Kulis, T.; Penezic, L.; Goluza, E.; Gidaro, S.; Corusic, A. Extraperitoneal radical prostatectomy with the Senhance Surgical System robotic platform. Croat. Med. J. 2019, 60, 556–559. [Google Scholar] [CrossRef]
- Peters, B.S.; Armijo, P.R.; Krause, C.; Choudhury, S.A.; Oleynikov, D. Review of emerging surgical robotic technology. Surg. Endosc. 2018, 32, 1636–1655. [Google Scholar] [CrossRef] [PubMed]
- Rhee, H.; Belyea, M.J.; Sterling, M.; Bocko, M.F. Evaluating the Validity of an Automated Device for Asthma Monitoring for Adolescents: Correlational Design. J. Med. Internet Res. 2015, 17, e234. [Google Scholar] [CrossRef] [PubMed]
- Mangi, M.A.; Nesheiwat, Z.; Kahloon, R.; Moukarbel, G.V. CardioMEMS(TM) System in the Daily Management of Heart Failure: Review of Current Data and Technique of Implantation. Expert Rev. Med. Devices 2020, 17, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.G.; Fares, M.; Hoyt, W., Jr.; Snyder, C.S. Diagnostic Accuracy and Safety of Confirm Rx Insertable Cardiac Monitor in Pediatric Patients. Pediatr. Cardiol. 2021, 42, 142–147. [Google Scholar] [CrossRef]
- Sattar, Y.; Zghouzi, M.; Suleiman, A.M.; Sheikh, A.; Kupferman, J.; Sarfraz, A.; Arshad, J.; Mir, T.; Ullah, W.; Pacha, H.M.; et al. Efficacy of remote dielectric sensing (ReDS) in the prevention of heart failure rehospitalizations: A meta-analysis. J. Community Hosp. Intern. Med. Perspect. 2021, 11, 646–652. [Google Scholar] [CrossRef]
- Dunbar, G.E.; Shen, B.Y.; Aref, A.A. The Sensimed Triggerfish contact lens sensor: Efficacy, safety, and patient perspectives. Clin. Ophthalmol. 2017, 11, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Dehennis, A.; Mortellaro, M.A.; Ioacara, S. Multisite Study of an Implanted Continuous Glucose Sensor over 90 Days in Patients With Diabetes Mellitus. J. Diabetes Sci. Technol. 2015, 9, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Feehan, L.M.; Geldman, J.; Sayre, E.C.; Park, C.; Ezzat, A.M.; Yoo, J.Y.; Hamilton, C.B.; Li, L.C. Accuracy of Fitbit Devices: Systematic Review and Narrative Syntheses of Quantitative Data. JMIR Mhealth Uhealth 2018, 6, e10527. [Google Scholar] [CrossRef]
- Hall, A.; Mitchell, A.R.J.; Wood, L.; Holland, C. Effectiveness of a single lead AliveCor electrocardiogram application for the screening of atrial fibrillation: A systematic review. Medicine 2020, 99, e21388. [Google Scholar] [CrossRef]
- Isakadze, N.; Martin, S.S. How useful is the smartwatch ECG? Trends Cardiovasc. Med. 2020, 30, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Liu, C.; Xu, T.; Su, L.; Zhang, X. Artificial intelligence biosensors: Challenges and prospects. Biosens. Bioelectron. 2020, 165, 112412. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.; Meehan, J.; Ward, C.; Langdon, S.P.; Kunkler, I.H.; Murray, A.; Argyle, D. Implantable biosensors and their contribution to the future of precision medicine. Vet. J. 2018, 239, 21–29. [Google Scholar] [CrossRef]
- Majumder, S.; Mondal, T.; Deen, M.J. Wearable Sensors for Remote Health Monitoring. Sensors 2017, 17, 130. [Google Scholar] [CrossRef]
- Froisland, D.H.; Arsand, E. Integrating visual dietary documentation in mobile-phone-based self-management application for adolescents with type 1 diabetes. J. Diabetes Sci. Technol. 2015, 9, 541–548. [Google Scholar] [CrossRef]
- Ajami, S.; Teimouri, F. Features and application of wearable biosensors in medical care. J. Res. Med. Sci. 2015, 20, 1208–1215. [Google Scholar] [CrossRef]
- Sharma, A.; Badea, M.; Tiwari, S.; Marty, J.L. Wearable Biosensors: An Alternative and Practical Approach in Healthcare and Disease Monitoring. Molecules 2021, 26, 748. [Google Scholar] [CrossRef]
- Kulkarni, S.; Seneviratne, N.; Baig, M.S.; Khan, A.H.A. Artificial Intelligence in Medicine: Where Are We Now? Acad. Radiol. 2020, 27, 62–70. [Google Scholar] [CrossRef]
- Handelman, G.S.; Kok, H.K.; Chandra, R.V.; Razavi, A.H.; Lee, M.J.; Asadi, H. eDoctor: Machine learning and the future of medicine. J. Intern. Med. 2018, 284, 603–619. [Google Scholar] [CrossRef]
- Jiang, T.; Gradus, J.L.; Rosellini, A.J. Supervised Machine Learning: A Brief Primer. Behav. Ther. 2020, 51, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Choi, R.Y.; Coyner, A.S.; Kalpathy-Cramer, J.; Chiang, M.F.; Campbell, J.P. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl. Vis. Sci. Technol. 2020, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.H.; Beam, A.L.; Kohane, I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018, 2, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Tao, Q.; Lelieveldt, B.P.F.; van der Geest, R.J. Deep Learning for Quantitative Cardiac MRI. Am. J. Roentgenol. 2020, 214, 529–535. [Google Scholar] [CrossRef]
- Teng, Q.; Liu, Z.; Song, Y.; Han, K.; Lu, Y. A survey on the interpretability of deep learning in medical diagnosis. Multimed. Syst. 2022, 28, 2335–2355. [Google Scholar] [CrossRef] [PubMed]
- Huff, D.T.; Weisman, A.J.; Jeraj, R. Interpretation and visualization techniques for deep learning models in medical imaging. Phys. Med. Biol. 2021, 66, 04TR01. [Google Scholar] [CrossRef]
- Ward, T.M.; Mascagni, P.; Ban, Y.; Rosman, G.; Padoy, N.; Meireles, O.; Hashimoto, D.A. Computer vision in surgery. Surgery 2021, 169, 1253–1256. [Google Scholar] [CrossRef]
- Chadebecq, F.; Vasconcelos, F.; Mazomenos, E.; Stoyanov, D. Computer Vision in the Surgical Operating Room. Visc. Med. 2020, 36, 456–462. [Google Scholar] [CrossRef]
- Almujalhem, A.; Rha, K.H. Surgical robotic systems: What we have now? A urological perspective. BJUI Compass 2020, 1, 152–159. [Google Scholar] [CrossRef]
- Bitterman, D.S.; Miller, T.A.; Mak, R.H.; Savova, G.K. Clinical Natural Language Processing for Radiation Oncology: A Review and Practical Primer. Int. J. Radiat. Oncol. Biol. Phys. 2021, 110, 641–655. [Google Scholar] [CrossRef]
- Wong, A.; Plasek, J.M.; Montecalvo, S.P.; Zhou, L. Natural Language Processing and Its Implications for the Future of Medication Safety: A Narrative Review of Recent Advances and Challenges. Pharmacotherapy 2018, 38, 822–841. [Google Scholar] [CrossRef] [PubMed]
- Hassoun, S.; Jefferson, F.; Shi, X.; Stucky, B.; Wang, J.; Rosa, E. Artificial Intelligence for Biology. Integr. Comp. Biol. 2022, 61, 2267–2275. [Google Scholar] [CrossRef] [PubMed]
| Abbreviation | Definition |
|---|---|
| AI | Artificial Intelligence |
| ANNs | Artificial Neural Networks |
| CDI | Clostridium Difficile Infection |
| CNNs | Convolutional Neural Networks |
| CT | Computerized Tomography |
| CV | Computer Vision |
| DL | Deep Learning |
| DNNs | Deep Neural Networks |
| EHRs | Electronic Health Records |
| EST | Expressed Sequence Tag |
| FDA | Food & Drug Administration |
| HGP | Human Genome Project |
| INSIDER | INtegrated Structural Interactome & genomic Data browser |
| IoMT | Internet of Medical Things |
| ML | Machine Learning |
| MRI | Magnetic Resonance Imaging |
| MS | Mass Spectrometry |
| NLP | Natural Language Processing |
| NN | Neural Network |
| PA | Pulmonary Artery |
| QSAR | Quantitative Structure–Activity Relationship |
| RNNs | Recurrent Neural Networks |
| RVM | Relevant Vector Machine |
| SNNs | Stochastic Neural Networks |
| SRA | Sequence Read Archive |
| SVM | Support Vector Machine |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Athanasopoulou, K.; Daneva, G.N.; Adamopoulos, P.G.; Scorilas, A. Artificial Intelligence: The Milestone in Modern Biomedical Research. BioMedInformatics 2022, 2, 727-744. https://doi.org/10.3390/biomedinformatics2040049
Athanasopoulou K, Daneva GN, Adamopoulos PG, Scorilas A. Artificial Intelligence: The Milestone in Modern Biomedical Research. BioMedInformatics. 2022; 2(4):727-744. https://doi.org/10.3390/biomedinformatics2040049
Chicago/Turabian StyleAthanasopoulou, Konstantina, Glykeria N. Daneva, Panagiotis G. Adamopoulos, and Andreas Scorilas. 2022. "Artificial Intelligence: The Milestone in Modern Biomedical Research" BioMedInformatics 2, no. 4: 727-744. https://doi.org/10.3390/biomedinformatics2040049
APA StyleAthanasopoulou, K., Daneva, G. N., Adamopoulos, P. G., & Scorilas, A. (2022). Artificial Intelligence: The Milestone in Modern Biomedical Research. BioMedInformatics, 2(4), 727-744. https://doi.org/10.3390/biomedinformatics2040049







