Predicting and Visualizing STK11 Mutation in Lung Adenocarcinoma Histopathology Slides Using Deep Learning
Abstract
1. Introduction
2. Materials and Methods
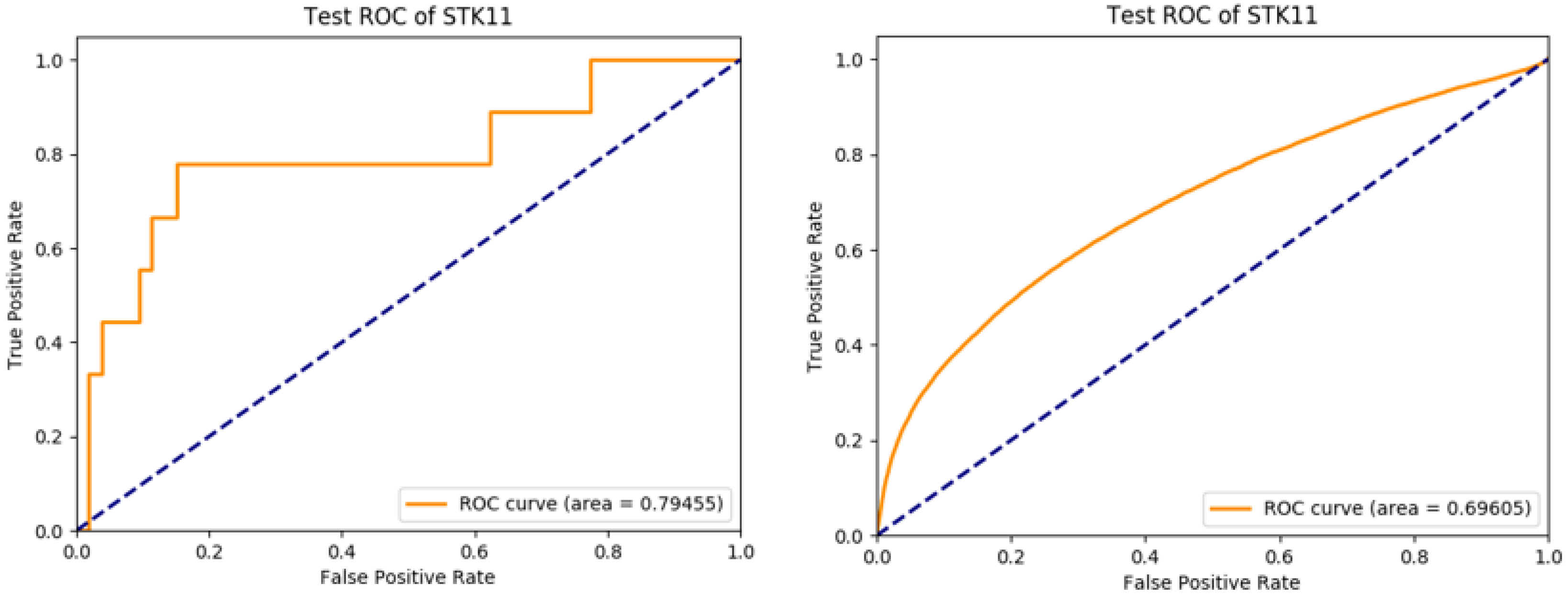
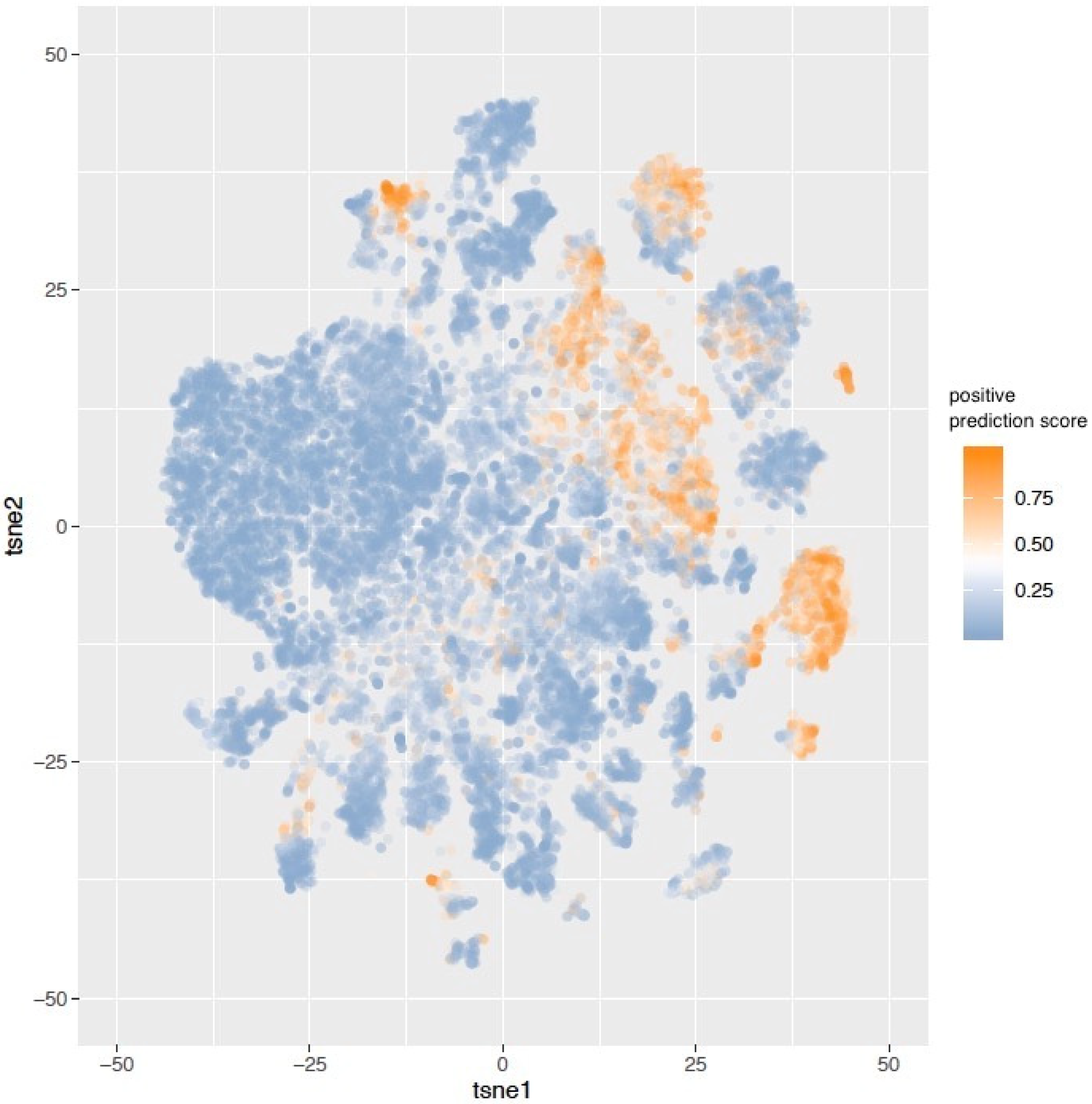
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mansuet-Lupo, A.; Alifano, M.; Pécuchet, N.; Biton, J.; Becht, E.; Goc, J.; Germain, C.; Ouakrim, H.; Régnard, J.-F.; Cremer, I.; et al. Intratumoral Immune Cell Densities Are Associated with Lung Adenocarcinoma Gene Alterations. Am. J. Respir. Crit. Care Med. 2016, 194, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, V.; Vogel, T.; Leube, B.; Driemel, C.; Goecke, T.; Möslein, G.; Royer-Pokora, B. STK11 genotyping and cancer risk in Peutz-Jeghers syndrome. J. Med. Genet. 2005, 42, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.-Y.; Zhang, C.; Li, Y.-F.; Su, J.; Xie, Z.; Liu, S.-Y.; Yan, L.-X.; Chen, Z.-H.; Yang, X.-N.; Lin, J.-T.; et al. Genetic and Immune Profiles of Solid Predominant Lung Adenocarcinoma Reveal Potential Immunotherapeutic Strategies. J. Thorac. Oncol. 2018, 13, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Skoulidis, F.; Albacker, L.; Hellmann, M.; Awad, M.; Gainor, J.; Goldberg, M.; Schrock, A.; Gay, L.; Elvin, J.; Ross, J.; et al. MA 05.02 STK11/LKB1 Loss of Function Genomic Alterations Predict Primary Resistance to PD-1/PD-L1 Axis Blockade in KRAS-Mutant NSCLC. J. Thorac. Oncol. 2017, 12, S1815. [Google Scholar] [CrossRef][Green Version]
- Gillette, M.A.; Satpathy, S.; Cao, S.; Dhanasekaran, S.M.; Vasaikar, S.V.; Krug, K.; Petralia, F.; Li, Y.; Liang, W.-W.; Reva, B.; et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell 2020, 182, 200–225.e35. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.; Liu, W.; DeLair, D.; Razavian, N.; Fenyö, D. Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models. Cell Rep. Med. 2021, 2, 100400. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.H.; Nomikou, S.; Coudray, N.; Jour, G.; Dawood, Z.; Hong, R.; Esteva, E.; Sakellaropoulos, T.; Donnelly, D.; Moran, U.; et al. Deep Learning and Pathomics Analyses Reveal Cell Nuclei as Important Features for Mutation Prediction of BRAF-Mutated Melanomas. J. Investig. Dermatol. 2021, in press. [CrossRef]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Wang, L.-B.; Karpova, A.; Gritsenko, M.A.; Kyle, J.E.; Cao, S.; Li, Y.; Rykunov, D.; Colaprico, A.; Rothstein, J.H.; Hong, R.; et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell 2021, 39, 509–528.e20. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, R.; Liu, W.; Fenyö, D. Predicting and Visualizing STK11 Mutation in Lung Adenocarcinoma Histopathology Slides Using Deep Learning. BioMedInformatics 2022, 2, 101-105. https://doi.org/10.3390/biomedinformatics2010006
Hong R, Liu W, Fenyö D. Predicting and Visualizing STK11 Mutation in Lung Adenocarcinoma Histopathology Slides Using Deep Learning. BioMedInformatics. 2022; 2(1):101-105. https://doi.org/10.3390/biomedinformatics2010006
Chicago/Turabian StyleHong, Runyu, Wenke Liu, and David Fenyö. 2022. "Predicting and Visualizing STK11 Mutation in Lung Adenocarcinoma Histopathology Slides Using Deep Learning" BioMedInformatics 2, no. 1: 101-105. https://doi.org/10.3390/biomedinformatics2010006
APA StyleHong, R., Liu, W., & Fenyö, D. (2022). Predicting and Visualizing STK11 Mutation in Lung Adenocarcinoma Histopathology Slides Using Deep Learning. BioMedInformatics, 2(1), 101-105. https://doi.org/10.3390/biomedinformatics2010006






