Abstract
Typhoid fever caused by the bacteria Salmonella typhi gained resistance through multidrug-resistant S. typhi strains. One of the reasons behind β-lactam antibiotic resistance is -lactamase. L, D-Transpeptidases is responsible for typhoid fever as it is involved in toxin release that results in typhoid fever in humans. A molecular modeling study of these targeted proteins was carried out by various methods, such as homology modeling, active site prediction, prediction of disease-causing regions, and by analyzing the potential inhibitory activities of curcumin analogs by targeting these proteins to overcome the antibiotic resistance. The five potent drug candidate compounds were identified to be natural ligands that can inhibit those enzymes compared to controls in our research. The binding affinity of both the Go-Y032 and NSC-43319 were found against β-lactamase was −7.8 Kcal/mol in AutoDock, whereas, in SwissDock, the binding energy was −8.15 and −8.04 Kcal/mol, respectively. On the other hand, the Cyclovalone and NSC-43319 had an equal energy of −7.60 Kcal/mol in AutoDock, whereas −7.90 and −8.01 Kcal/mol in SwissDock against L, D-Transpeptidases. After the identification of proteins, the determination of primary and secondary structures, as well as the gene producing area and homology modeling, was accomplished. The screened drug candidates were further evaluated in ADMET, and pharmacological properties along with positive drug-likeness properties were observed for these ligand molecules. However, further in vitro and in vivo experiments are required to validate these in silico data to develop novel therapeutics against antibiotic resistance.
1. Introduction
Typhoid is a usual illness in economically handicapped countries where public health settings are very poor. A globally estimated 12–27 million people get stricken with typhoid fever each year, whereas the overall yearly estimated incidence lies between 292 and 395 cases per 100,000 people in Bangladesh. This infection-causing agent is an anaerobic Gram-negative rod, namely Salmonella enterica serotype Typhi (S. typhi), a highly conserved serovar subspecies of S. enterica, which is transmitted by the fecal-oral route and can infect the intestinal tract and blood [1,2,3,4,5]. S. typhi can provoke many health issues, such as fever, abdominal discomfort, and several gastrointestinal complications, such as nausea, vomiting, constipation, diarrhea, etc. The first approved antibiotics for the prevention of typhoid fever were chloramphenicol, ampicillin and cotrimoxazole [6], which have already started showing resistance and evolved multidrug resistance (MDR) S. Typhi strains over the last two decades. Due to the ever-increasing pattern of MDR in many parts of the world, combating typhoid is becoming more difficult, creating a major public health concern around the world [7].
In the early 1970s, the first MDR S. typhi strains displaying concurrent resistance to the first-line antibiotics, such as ampicillin, chloramphenicol and co-trimoxazole, were demonstrated, followed by the emergence of ciprofloxacin-resistant strains in the 1990s [8,9]. Currently, the latter is observed in more than 90% of clinical isolates from endemic areas [10,11,12]. A 15-year (1993–2013) genome-wide study on S. typhi conducted in Bangladesh using 536 medical isolates reported that these bacterial strains show resistance to ampicillin (amp), co-trimoxazole (sxt), chloramphenicol (chl), ciprofloxacin (cip), and ceftriaxone (cro) where 37.69% strains displayed co-occurring resistance towards amp, sxt, chl, and cip followed by only cip-R (R = resistant) strains to comprise 31.53% of the total. Some of the resistance genes detected in the isolates of that study were blaTEM-1B in 50.28% of amp-R, qnrS1 in 10.2% of cip-R, and tet (A, B) in 9.46% and 8.53% tet-R (tet = tetracycline) strains, respectively [13]. The presence of extended-spectrum β-lactamase (ESBL) resistance Salmonella prevalence in poultry sourced recently from super shops of five divisional megacities of Bangladesh implies its possible human transmission through contaminated foods of poultry origin and the potential health risk of the people [14,15]. Additionally, as recorded in various parts of the world, S. typhi is now increasingly developing resistance to ciprofloxacin and fluoroquinolone and has emerged as a new threat to the treatment of typhoid fever [16,17,18,19,20,21,22,23,24].
S. typhi acquires a ciprofloxacin-resistance (cip-R) property through the point mutations in quinolone resistance-determining regions (QRDR) with several positions corresponding to the genes, topoisomerase IV (parC and parE) and DNA gyrase (gyrA and gyrB) of S. typhi [25,26,27,28], whereas the acquisition of the blaTEM gene is responsible for the resistance property of S. typhi against β-lactam antibiotics through encoding the β-lactamase enzyme that hydrolyzes the peptide bond of the four-membered β-lactam ring and thus prevents β-lactam antibiotics from exerting their effect [29]. Moreover, derivatives of TEM, along with those of SHV- and CTX-M-type β-lactamase genes, comprise the family called extended-spectrum β-lactamases (ESBLs), which leads to the development of multidrug-resistant S. typhi, limiting the current treatment practices and thus posing an alarming situation in public health [30].
Typhoid toxin is a prominent feature of S. typhi that contributes potential virulence to the bacterial infection causing typhoid fever by exclusively targeting the immune system and central nervous system of the host. The presence of one type of sialic acid is necessary for its binding to the host, which is abundantly available in humans. Thus, S. typhi cannot cause typhoid in hosts other than humans [31]. The export of the toxin to the outer membrane of the bacteria begins with the secretion of the individual subunits to the periplasm via Sec machinery and their assembly into the holotoxin complex. This holotoxin is then translocated across the PG (peptidoglycan) layer from the cis side to the trans side by the action of a special type of muramidase, TtsA, which is located at the bacterial poles and requires the PG editing by the L, D-Transpeptidases, namely YcbB, for its activity. After being translocated to the trans side of the PG layer, it becomes compartmentalized in an S. typhi-containing vacuole, from where its eventual release takes place upon exposure to the antimicrobial peptide or bile salts, and in this way, the transmission continues from one infected cell to the other [32].
The function of L, D-Transpeptidases, YcbB, which is exclusively present in the bacterium S. typhi, is well-understood in the edition of PG. Glycan strands are the building blocks of the bacterial PG that are composed of N-acetylglucosamine (GlcNac) and N-acetylmuramic acid (MurNac) [33,34,35]. These building blocks make the PG by being connected by small peptides. Here, the enzyme L, D-Transpeptidases plays its role in introducing cross-links within L- and D-amino acids that comprise the peptides (Figure 1). This PG remodeling by L, D-Transpeptidases is necessary for TtsA to position the typhoid toxin for its proper release [32]. Here, L, D-Transpeptidases can be a major target for in silico studies as it plays a vital role in the secretion of typhoid toxin, and there is no effective drug available to inhibit it without exhibiting any side effects. For example, drug carbapenem and copper can inactivate L, D-Transpeptidases, yet they are associated with diarrhea, nausea, vomiting, skin rash, low blood pressure, anemia, heart problems, etc. [36]. Moreover, several antibiotics are working alone or coupled with β-lactamase inhibitors (Avibactam, Clavulanic acid (clavulanate), Relebactam, Sulbactum, Tozobactum, etc.), which have many adverse effects such as gastrointestinal complications, impairment of nervous system, hematological effects, and dermatological abnormalities, including Stevens-Johnson syndrome, toxic epidermal necrolysis, and drug-induced eosinophilia, etc. [37,38,39,40,41,42,43].
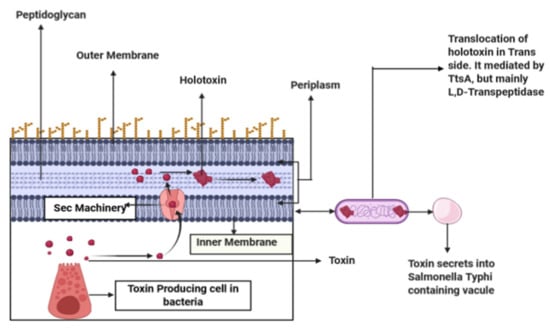
Figure 1.
A toxin produced from cell-secreted into periplasm through SEC machinery, then these toxin subunits assemble into holotoxin-translocated into trans side of bacteria mediated by TtsA. Peptidoglycan remodeling occurs through L, D-Transpeptidases, YcbB, enriched in the bacterial poles.
Curcumin, the main bioactive component of turmeric (Curcuma longa L.), has been shown to be a powerful antioxidant, anti-inflammatory, antibacterial, antifungal, and antiviral agent in many studies [44]. Curcumin has been shown to be antibacterial against Staphylococcus aureus (S. aureus). Curcumin has significantly more effective antibacterial properties when combined with other antibacterial drugs, as revealed by in vitro experiments [45]. Curcumin inhibits bacterial growth due to its structural properties and the production of anti-oxidative chemicals. Through the bacterial quorum sensing regulatory system, curcumin can decrease bacterial virulence factors, reduce bacterial biofilm formation, and restrict bacterial adherence to host receptors [46]. Curcumin’s potential antibacterial action makes it a viable option for enhancing the inhibitory impact of current antimicrobial drugs through synergism [47]. It decreased Salmonella enterica serovar Typhimurium’s motility by reducing the length of the flagellar filament (from 8 m to 5 m) and lowering its density (4 or 5 flagella/bacterium instead of 8 or 9 flagella/bacterium). Curcumin therapy reduced the proportion of flagellated bacteria from 84 percent to 59 percent [48].
As curcumin has antibacterial properties, including antioxidant and anti-inflammatory properties, we selected 70 curcumin analogues as ligands in this study. The schematic representation of the methodology applied in the present study is displayed in Figure 2.
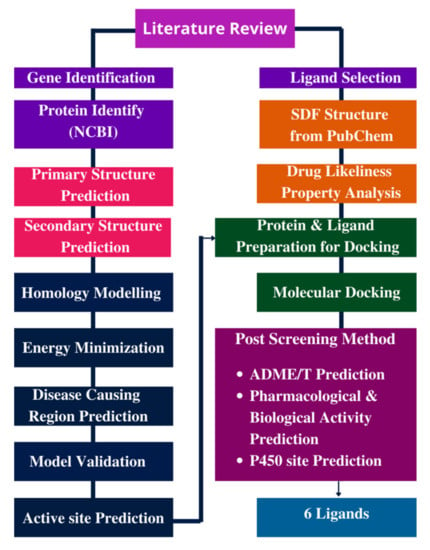
Figure 2.
Methodology/Overall study.
2. Materials and Methods
2.1. Drug-Likeness Features Interpretation
About 70 curcumin molecules were selected as ligands from literature to target the proteins β-lactamase and L, D-Transpeptidases of S. typhi. Each of the molecules was assessed for Lipinski’s rule of five or not [49,50]. Molinspiration Cheminformatics server (https://www.molinspi-ration.com/cgi-bin/properties accessed on 8 October 2020) was applied to experiment various drug-like parameters of the ligand molecules [51,52] (Table S2). Molinspiration allows for the prediction of significant molecular parameters (logP, polar surface area, number of hydrogen bond donors and acceptors, and so on), as well as the prediction of bioactivity scores for the most relevant therapeutic targets (GPCR ligands, kinase inhibitors, ion channel modulators, nuclear receptors). Compounds that did not comply with the rule were excluded from further study.
2.1.1. Protein Preparation for Docking
Sequence Retrieval
The complete protein sequences of β-lactamase and L, D-Transpeptidases were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/ accessed on 12 October 2020) in the standard FASTA format.
Physiochemical Property Identification
The physical and chemical parameters of the proteins, including molecular weight (MW), theoretical pI, amino acid composition, estimated half-life, instability index, aliphatic index, etc., were computed using the ProtParam tool of ExPasy server as it evaluates physicochemical data (molecular weight, theoretical pI, amino acid composition, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index, and grand average of hydropathicity (GRAVY)) from a protein sequence. (http://web.expasy.org/program/ accessed on 12 October 2020) [53].
Secondary and Tertiary Structure Prediction
The online tool SOPMA [54] (https://npsa-prabi.ibcp.fr/NPSA/npsa_sopma.html accessed on 15 October 2020) was applied for the secondary structure prediction of the proteins. Homology Modeling was performed in Swiss-Model, as its workspace is a Web-based modeling expert system that is integrated. We search a library of experimental protein architectures for acceptable templates for a specified target protein. A three-dimensional model of the target protein is constructed based on a sequence alignment between the target protein and the template structure [55,56,57,58,59] (https://swissmodel.expasy.org/, accessed on 18 October 2020) to construct the tertiary structures of the target proteins using the three-dimensional structure of a related protein as a template. Homology modeling was also done by Phyre2. It is a web-based collection of tools for predicting and analyzing protein structure, function, and mutations. Phyre2’s goal is to provide biologists with a simple and intuitive interface to cutting-edge protein bioinformatics tools. Phyre2, which builds 3D models, predicts ligand binding sites and analyzes the influence of amino acid changes (e.g., no synonymous SNPs (nsSNPs)) for a user’s protein sequence using advanced distant homology detection algorithms [60].
Disordered Regions Prediction
Disordered regions present in protein molecules remain unstable in the native state. To find out the disordered regions in proteins for which they lack a fixed tertiary structure, the Protein Disorder prediction System (PrDOS) server [61] (http://prdos.hgc.jp/cgi-bin/top.cgi, accessed on 18 October 2020) was exploited. This server predicts the disordered regions based on both local amino acid sequence and the template or homologous proteins through the SVM algorithm and PSI-BLAST, respectively. The prediction method consists of two predictors: one based on local amino acid sequence information and the other on template proteins. For each residue, the server aggregates the findings of the two predictors and delivers a two-state prediction (order/disorder) and a disorder probability.
Validation of Tertiary Protein Model
Model validation was carried out in PROCHECK [62,63] (http://www.ebi.ac.uk/thornton-srv/software/PROCHECK/, accessed on 20 December 2021), which assesses the stereo-chemical quality of a protein structure, i.e., how normal or unusual the pattern of the protein residues is, compared with a fined-tuned, high-resolution structure of a protein. PROCHECK evaluates the stereochemical quality of a protein structure by generating a series of PostScript graphs that analyze its overall and residue-by-residue geometry. It contains PROCHECK-NMR, which is used to check the quality of structures solved by NMR.
Active Site Prediction
For the prediction of active sites in the proteins where ligands will likely bind, two servers CASTP (Computed Atlas of Surface Topography of proteins) [64] (http://sts.bioe.uic.edu/castp/ accessed on 20 October 2020) and COACH [65,66] (http://zhanglab.ccmb.med.umich.edu/COACH/ accessed on 22 October 2020) were utilized. CASTP implements the theoretical and algorithmic results of computational geometry to predict the ligand-binding sites. It has several advantages: (1) pockets and cavities are recognized analytically, (2) the boundary between the bulk solvent and the pocket is accurately specified, and (3) all derived parameters are rotationally invariant, do not need discretization, and do not make use of dot surface or grid points. On the contrary, the COACH server applies two comparative methods, TM-SITE and S-SITE, to identify active sites in the protein.
2.1.2. Ligand and Protein Preparation for Docking
The ligand molecules were minimized in the Avogadro software using the mmff94 force field. Then, the protein structure was minimized in YASARA software using the AMBER14 force field. The docking program was carried out in the AutoDock Vina program. The ligands that will give the best results will be docked again using the SwissDock server. We used two servers to check the validity and to build a strong hypothesis. AutoDock Vina, a novel molecular docking and virtual screening application, has been introduced. Vina’s local optimization process employs a powerful gradient optimization algorithm. The gradient computation essentially provides the optimization algorithm with a “feeling of direction” from a single evaluation. Vina may speed up processing by making use of many CPUs or CPU cores by employing multithreading. SwissDock is a webserver dedicated to doing protein-ligand docking simulations in an easy and beautiful manner. SwissDock is protein-ligand docking software with a simple and integrated interface that is based on EADock DSS.
AutoDock Vina PDB file of protein was converted into a pdbqt file using the Auto Dock Vina tool. Then, every selected ligand file was converted to a PDB file using pymol because the AutoDock tool can only recognize the PDB format. Then, the PDB file of the ligand was converted into a pdbqt file, which is the criterion for the AutoDock run. After docking, we visualized the docked file using pymol.
SwissDock: At first, we set up the protein PDB file manipulating PDB code, which was retrieved from Uniprot or RCSB PDB. We specified chain A in β-lactamase. Then, ligands were selected from the ZINC database, but those that were not present in this database were uploaded in Mol2 format. The server provides very fast, fast, and accurate results.
2.2. ADME/T Prediction
ADME/T describes the Absorption, Distribution, Metabolism, Excretion and Toxicity of a drug-like substance. These properties account for the success of a drug in clinical trials. Therefore, in silico ADME/T profile examination of the candidate drugs is a prerequisite for the fruitful measure of drug designing expenditure [67,68]. The best 8 ligands (based on the docking score) were utilized to speculate their drug-like potential by observing pharmacokinetic and pharmacodynamics features. ADME/T profile of all the chosen ligands were calculated utilizing admetSAR 2.0 (http://lmmd.ecust.edu.cn/admetsar2/ accessed on 23 October 2020) and pkCSM (http://biosig.unimelb.edu.au/pkcsm/ accessed on 23 October 2020) server [69,70]. admetSAR presents an easy-to-use interface for searching for ADME/T (Absorption, Distribution, Metabolism, Excretion, and Toxicity) attributes profiling by name, CASRN, and similarity search. With QSAR models, admetSAR can predict around 50 ADMET endpoints. The pkCSM signatures were effectively employed to create predictive regression and classification models across five different pharmacokinetic property classes.
Validation of Tertiary Protein Model
ERRAT software was used to validate the protein model. ERRAT is a software that verifies crystallographically determined protein structures.
2.3. Pharmacological and Biological Activity Prediction
The elected ligands were employed to determine their pharmacological and biological activities accurately by using Prediction of Activity Spectra of Substances (PASS) Online (http://www.pharmaexpert.ru/passonline/ accessed on 26 October 2020) and Molinspiration Cheminformatics server [71]. These methods are used in conjunction with recognized compounds present in the database, depending on the structure-activity relationship (SAR). PASS Online predicts about 4000 different types of biological activity, such as pharmacological effects, mechanisms of action, toxic and unfavorable effects, interactions with metabolic enzymes and transporters, gene expression influence, and so on. Molinspiration Cheminformatics is also useful software to predict pharmacological and biological activities.
2.4. Pred. P450 Site of Metabolism Iction
In silico methods can contribute significantly to the prediction of drug metabolism sites focusing on the experimental view of the drug designing process. By conducting bioassay, these sites impart the knowledge of the molecules’ metabolic susceptibility and their fate inside the body [72]. RS-WebPredictor (http://reccr.chem.rpi.edu/Software/RS-WebPredictor/ accessed on 1 November 2020), an online server, was used to predict the best sites of drug metabolism mediated by CYP2C9, CYP2D6, and CYP3A4, three promiscuous isoforms of Cytochrome P450 (CYP) family. Predictions may be made for the promiscuous CYP isozymes 2C9, 2D6, and 3A4, as well as CYPs 1A2, 2A6, 2B6, 2C8, 2C19, and 2E1. The RS-WebPredictor service is the first publicly available server that predicts the regioselectivity of the last six isozymes.
3. Results
3.1. Drug-Likeness Features Interpretation
Among 70 types of ligands, all the compounds except DM1 were compatible with the drug-likeness features. Most of the selected compounds did not display any violations to Lipinski’s rules, which indicated the pharmacokinetic conformity of these compounds. Hence, all these compounds were taken into account for the next phases of the study (Tables S1 and S2).
3.2. Protein Preparation for Docking
3.2.1. Sequence Salvation
Two protein sequences of β-lactamase (EC 3.5.2.6) (Uniprot ID P62593; PDB ID 1ZG4) and L, D-Transpeptidases (Uniprot ID P22525; PDB ID 6NTW) were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/ accessed on 12 October 2020) in standard FASTA format.
3.2.2. Physiochemical Property Identification
Using the ProtParam tool of the ExPasy server, the physical and chemical parameters of the proteins were analyzed. β-lactamases is 286 amino acids long, weighted at 31515.20 grams, and has an instability index of 40.74. L, D-Transpeptidases is 615 amino acids long, 67812.49 (gm) weighted, with an instability index value of 43.79. The Instability Index is a metric for determining whether a protein will remain stable in a test tube. Table 1 shows the physicochemical properties of these two proteins.

Table 1.
The physiochemical properties of β-lactamase and L, D-Transpeptidases.
3.2.3. Secondary and Tertiary Structure Prediction
The SOPMA tool was used to predict the secondary structures of these two proteins. The values of alpha helix were 49.30% and 39.35% for β-lactamase and L, D-Transpeptidases, respectfully. The value of the extended strand was also greater for β-lactamase (12.94%) compared with the L, D-Transpeptidases (11.54%). L, D-Transpeptidases (43.58%) exceeds the β-lactamase (29.37%) in the case of a random coil. The values of 310 helix, pi helix, beta bridge, bend region, and ambiguous status were 0.00% for both proteins (Table 2).

Table 2.
The secondary structures of the proteins: β-lactamase and L, D-Transpeptidases.
The three-dimensional structures of β-lactamase and L, D-Transpeptidases were predicted in Swiss-Model web tools. The biounit oligo state of both proteins was a monomer. The template displayed 0.61 sequence similarities, coverage score of 1.0, and 24–286 range for β-lactamase, whereas L, D-transpeptidases showed 0.62 sequence similarity, coverage score of 0.95, and 37–615 range (Table 3). In Swiss-Model, two models for β-lactamase and three models for L, D-Transpeptidases were predicted based on the top 31 and 50 templates, respectively. On the contrary, the top 20 models were predicted by Phyre 2 for both proteins each. The best fit built by the two servers for β-lactamase had a confidence score of 100 when modeling 263 amino acid residues at positions 24–286, and for L, D-transpeptidases, it modelled 505 residues at positions 37–615 with a confidence score of 100.

Table 3.
The homology modeling parameters for β-lactamase and L, D-Transpeptidases.
3.2.4. Validation of Tertiary Protein Model
In ERRAT, the overall quality factor of β-lactamase and L, D-Transpeptidases was is 97.2549 and 84.1141, respectively. The two proteins also passed the verified 3D in their respective prediction results. The β-lactamase (EC 3.5.2.6) has an 11% disease-causing region, and its active sites are acyl ester intermediate (position 70) and proton acceptor (position 168). The Ramachandran plot analysis showed that both proteins delineated more than 90% of the amino acid residues in the most favored regions. The number of non-glycine and non-proline residues is 228 among 263 residues. Twenty-one glycine residues and 12 proline residues are present (Ramachandran plot), and 93.4% of residues are in the favored region. The L, D-Transpeptidases is 615 amino acids long, and its molecular weight is 67812.49. The active site of this protein stays in the 528 position. Here, 91.8% residues are in the favored region with 427 residues excluding glycine and proline, 8 terminal residues other than Gly and Pro, 31 glycine (represented as a triangle), and 3 proline residues (Table 4 and Figure 3 and Figure 4).

Table 4.
The quality of the hypothetical model protein.
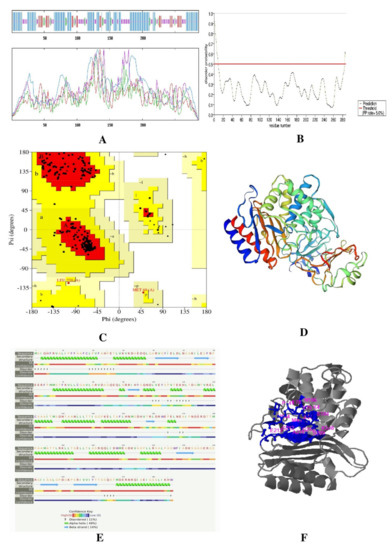
Figure 3.
Result of β-lactamase protein (A) secondary structure prediction result using SOPMA software, (B) disease-causing region prediction using PrDOS software, (C) Ramachandran plot using PROCHECK, (D) 3D structure predicted by Swiss-Model, (E) secondary structure and disordered region predicted by Phyre2, and (F) active site predicted by COACH Software.
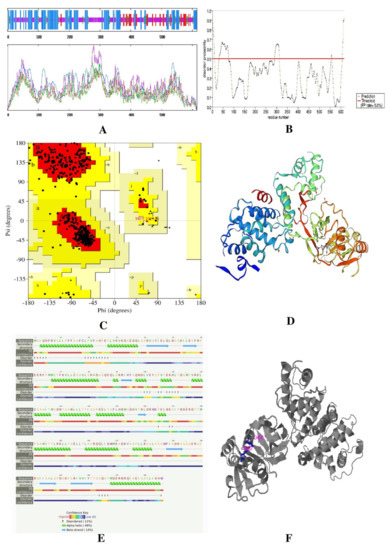
Figure 4.
Result of protein L, D-Transpeptidases (A) secondary structure prediction result using SOPMA software, (B) disease-causing region prediction using PrDOS software, (C) Ramachandran plot using PROCHECK, (D) 3D structure predicted by Swiss-Model, (E) secondary structure and disordered region predicted by Phyre2, and (F) active site predicted by COACH Software.
3.2.5. Active Site Prediction
3.2.6. Molecular Docking
The best five ligands were selected based on docking experiments among 70 ligands. Go-Y032, NSC-43319, Cyclovalone, Salsalate, and Cyclocurcumin showed the best docking results against the β-lactamase (1ZG4) enzyme (Table 5 and Table 6 and Figure 5), and NSC-43319, Cyclovalone, Cyclocurcumin, Difluorinated curcumin, and Go-Y032 showed the best docking results against the L, D-Transpeptidases (6NTW) enzyme (Table 5, Table 6 and Figure 5). Two controls were selected for each of the enzymes. Clavulanic acid and Tazobactum were docked against β-lactamase as they are existing drugs. These existing drugs showed low affinity to other ligands. For L, D-Transpeptidases, Carbapenem and Cephalosporin were selected as controls. These controls were also showed lower affinity.

Table 5.
Docking results of β-lactamase.

Table 6.
Docking Results of L, D-Transpeptidases.
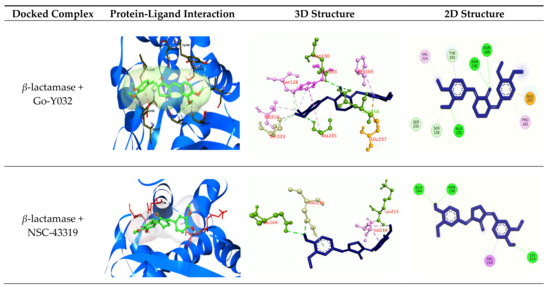
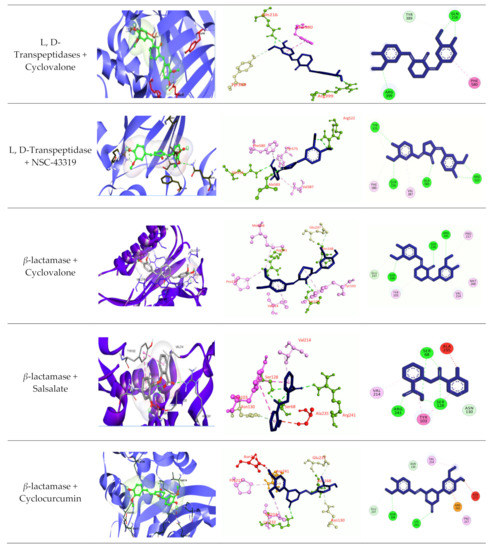
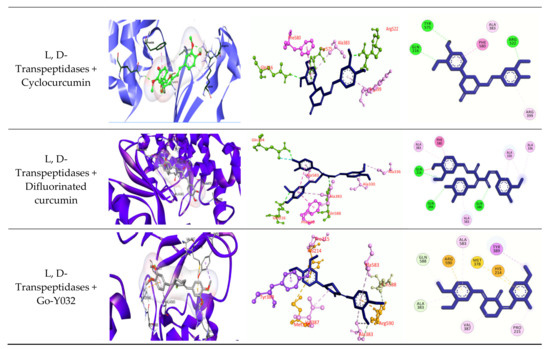
Figure 5.
Molecular docking experiments by targeting β-lactamase and L, D-Transpeptidases. Hydrogen bonds are displayed as green balls and sticks, hydrophobic bonds (Pi-Pi/Pi-sigma/amide-Pi interaction) are displayed as violet balls and sticks, hydrophobic bonds (Pi-alkyl/alkyl interaction stacking) are displayed as pink balls and sticks, hydrophobic (Pi-sulfur) are displayed as gold balls and sticks, and carbon–hydrogen bonds are displayed as white balls and sticks.
3.2.7. ADME/T Prognosis
ADME/T profiling was carried out for the ligands that gave the best docking scores in the molecular docking study (Table S6) and control group (Tables S3 and S4). Intestinal absorption and oral bioavailability were high for all of the ligands. Caco-2 permeability was high for the ligands Go-Y032 and NSC-43319, while the remaining ligand showed low Caco-2 permeability. Furthermore, NSC-43319 and Cyclovalone were predicted as substrates of P-glycoproteins, while the remaining was non-substrates of membrane P-glycoproteins. All of the ligands were able to enter the blood–brain barrier, excluding Cyclocurcumin. All of them exhibited substrate specificity of CYP3A4 except Cyclovalone. The NSC-43319, Salsalate, and Cyclovalone showed no specificity as substrates of CYP3C9. All of them were negative for AMES toxicity, and compounds demonstrated the inhibitions of the Human ether-a-go-go related gene (hERG) channel and also a less toxic profile.
3.3. Prediction of Pharmacological and Biological Activity
The screened ligands were inspected for the pharmacology-related study (Table S5) involving their accordance with Antibacterial, Bacterial Efflux Pump Inhibitor, Antibiotic Anthracycline-like activity, β-lactamase Inhibitor, Anti-mycobacterial and antibiotic activities. Here, Cyclovalone showed all of these pharmacological activities, whereas Go-Y032 was observed to have all of these properties except Antibiotic activity. The NSC-43319 showed antibacterial, bacterial efflux pump inhibitor, β-lactamase inhibitor, and anti-mycobacterial activities. In this study, Go-Y032, NSC-43319, and Cyclovalone were found as the best-performing ligands (Table 7). Thereafter, these five ligands were analyzed to observe whether they function against G protein-coupled receptor (GPCR) ligand, protein kinase, ion channels, enzyme protease, nuclear receptor ligand, etc., (Table S6).

Table 7.
Best-performing ligands after ADME/T, pharmacological, and biological activities prediction.
3.4. Prediction of P450-Mediated Sites of Metabolism (SOMs)
The most potential ligands were investigated for the prediction of their possible sites of metabolism (SOMs) by three main Cytochrome P450 (CYP) isozymes, i.e., 3A4, 2C9, and 2D6, respectively (Table 8). For CYP2D6 and CYP2C9 isoforms, Go-Y032, and for CYP3A4 and CYP2C9 isoforms, NSC-43319 have shown identical metabolism sites. However, for all of the isozymes, cyclovalone exhibited different metabolism sites.

Table 8.
Results of P450 sites of metabolism prediction. (Best three vulnerable atoms are marked in encircled number.)
4. Discussion
Molecular docking is the most significant approach in in silico drug designing. It assesses the binding affinity of a protein–ligand complex in the form of binding energy using computer algorithms. The lower the binding energy, the higher the affinity of the ligand bound to the target [73]. Five ligands gave the best free binding energies in the docking experiment conducted by AutoDock Vina and SwissDock, which included Go-Y032, NSC-43319 with β-lactamase enzyme, and NSC-43319, Cyclovalone with L, D-Transpeptidases (Table 5 and Table 6). The docking results were compared with controls. For the β-lactamase enzyme, the docking results with other ligands were compared with Clavulanic acid and Tazobactum (Table 9). Clavulanic acid and tazobactam are all plasmid-mediated β-lactamase inhibitors. Several studies have concluded that Clavulanic acid inhibits extended-spectrum TEM and SHV β-lactamases. They expressed lower affinity in our study compared to other ligands. Several studies reported that β-lactam antibiotics could work against L, D-Transpeptidases. Carbapenem and Cephalosporin are antibiotics in the beta-lactam class that kill bacteria by attaching to penicillin-binding proteins and blocking bacterial cell wall formation. As these are existing drugs, we compared their activities with our selected ligands. These controls were showed very low binding affinities in docking. Therefore, the ligands we selected have a better chance of working against those enzymes or as antibacterial drugs.

Table 9.
Docking result of the best-performing ligands.
After docking, the ligands were introduced to ADME/T prediction tools admetSAR 2.0 and pkCSM software for in silico prediction of ADME/T properties. This time-saving and cost-effective approach helps establish a candidate molecule as a promising drug through in vitro experiments [74,75]. A leading concern for drugs that mainly target the central nervous system (CNS) is that they need to be able to permeate across the blood–brain barrier. The most common route for drug delivery is the oral delivery system, through which the delivered drug enters into the intestine; hence, an investigated drug needs to be highly absorbed in the intestine. Cell membrane proteins, such as P-glycoproteins, facilitate the movement of many drugs through the cell membrane. Caco2 permeability of a drug reflects the permeability across the intestinal lining of humans, as this tissue is widely used in in vitro drug permeability studies due to its small intestinal mucosa-like behavior when cultured [75,76,77]. The Cytochrome P450 enzyme family focuses on the regulation of drug interaction, biotransformation, and their elimination outside the body. Acute toxicity, delayed removal, and eventual drug compound failure within the human body result from these enzymes’ inhibitory activity of the drugs [78,79,80]. The purpose of in silico AMES toxicity test is to determine the toxicity and mutagenicity of the chemicals [81,82]. Voltage-gated potassium ion channels, namely hERG channels, are involved in the transport of potassium ions through the cell membrane, which may be subjected to off-target drug interaction resulting in inhibition. Therefore, proper screening is necessary to see whether the investigated drugs have inhibitory activity on these transporters [83]. Renal organic cation transporter 2 (OCT2) plays a major role in the removal of drugs and xenobiotics via the kidney. It is considered that the substrates of this transporter protein are quickly excreted by urine [84]. In the ADME/T test, all the selected ligands showed almost similar properties (Table S6).
Afterwards, the pharmacological and biological activities of the ligands were carried out in the PASS online server and Molinspiration Cheminformatics server, respectively. Pharmacological activity (PASS prediction) is determined in terms of the likelihood of activity (Pa) and the likelihood of inactivity (Pi) of a drug, and the result of the prediction ranges between 0 and 1. The pharmacological activity of the drug is deemed possible if Pa > Pi [85]. The probability of anti-mycobacterial activity (Pa) for NSC-43319 and Cyclovalone was between 0.5–0.7, while for all ligands, Pa of all activities was <0.5, implying the unlikeliness of their activities [86]. However, Cyclovalone, Go-Y032, and NSC-43319 showed more satisfactory outcomes than others (Tables S5 and S6). Assessing biological activities against the most influential drug targets in the human body, such as G protein-coupled receptors (GPCRs), ion channels, enzymes, nuclear receptors, etc., is crucial because, when coupled with them, a drug mediates its therapeutic activity inside the body [87]. Probability scores for Petasiphenol representing activity against the targets were comparatively significant (Table S5).
Finally, the ligands were assessed in the RS-WebPredictor server to predict the probable sites where their metabolism will be likely to occur. Almost similar metabolism sites were reported for Go-Y032, NSC-43319, Cyclovalone, Difluorinated curcumin, which exhibited multiple sites of metabolism except for Cyclocurcumin and Salsalate. These two compounds showed few sites of metabolism compared to others (Table 8). After a successive screening of all potent ligands, this study suggests Go-Y032, Cyclovalone, NSC-43319, Difluorinated curcumin, Salsalate, and Cyclocurcumin as the best inhibitors of the enzymes β-lactamase and L, D-Transpeptidases, respectively, while NSC-43319, Cyclovalone, Cyclocurcumin, and Go-Y032 are recommended for both. These compounds may not express better performances at all screening tests. As our study was based on a computational approach, further in vitro analysis is needed to prove their activities. These compounds may give better performance in an in vitro approach than the controls we chose.
5. Conclusions
This study aimed to develop curcumin derivatives as potential drug candidates against S. typhi causing typhoid fever. The target proteins included two crucial proteins of this bacterium, β-lactamase and L, D-Transpeptidases, which are involved in acquiring antibiotic-resistance properties and toxin secretion, respectively. Although several antibiotics and β-lactam inhibitors are available to treat this fatal illness, none is effective enough to avoid side effects that eventually result in adverse complications. After surpassing multiple stages in assessing drug-like properties, among 70 ligands, three, including Go-Y032, Cyclovalone, and NSC-43319, were reported as the best-performing ligands. Further investigations based on in vivo and in vitro experiments are needed to ascertain the use of these ligands as drugs to treat S. typhi infection. Additionally, the other four ligands providing a satisfactory docking performance are also recommended for further wet laboratory investigation.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biomedinformatics2010005/s1, Table S1: The selected ligands; Table S2: Drug likeliness properties of the ligands; Table S3: ADME/T result of controls; Table S4: ADME/T prediction result; Table S5: Pharmacological activities; Table S6: Biological activities.
Author Contributions
Conceptualization, T.A., M.C., A.Y.T., S.M. and T.B.E.; methodology, T.A., M.C., A.Y.T., M.H.R., M.S.S.S., S.M., M.A.S., S.A.S. and T.B.E.; software, M.S.S.S., S.M., M.A.S., S.A.S. and T.B.E.; validation, M.H.R., S.M., M.A.S. and T.B.E.; formal analysis, S.M., M.A.S., S.A.S. and T.B.E.; investigation, T.A., M.C., A.Y.T., M.H.R., M.S.S.S., S.M., M.A.S., S.A.S. and T.B.E.; resources, M.A.S. and T.B.E.; data curation, S.M., M.A.S., S.A.S. and T.B.E.; writing—original draft preparation, T.A., M.C., A.Y.T., M.H.R., M.S.S.S., S.M. and T.B.E.; writing—review and editing, M.S.S.S., S.M., M.A.S., S.A.S. and T.B.E.; visualization, M.S.S.S., S.M., M.A.S., S.A.S. and T.B.E.; supervision, M.A.S. and T.B.E.; project administration, M.A.S. and T.B.E.; funding acquisition, S.M., M.A.S., S.A.S. and T.B.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Available data are presented in the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mogasale, V.; Maskery, B.; Ochiai, R.L.; Lee, J.S.; Mogasale, V.V.; Ramani, E.; Kim, Y.E.; Park, J.K.; Wierzba, T.F. Burden of typhoid fever in low-income and middle-income countries: A systematic, literature-based update with risk-factor adjustment. Lancet Glob. Health 2014, 2, e570–e580. [Google Scholar] [CrossRef] [Green Version]
- Crump, J.A.; Mintz, E.D. Global trends in typhoid and paratyphoid fever. Clin. Infect. Dis. 2010, 50, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Naheed, A.; Ram, P.K.; Brooks, W.A.; Hossain, M.A.; Parsons, M.B.; Talukder, K.A.; Mintz, E.; Luby, S.; Breiman, R.F. Burden of typhoid and paratyphoid fever in a densely populated urban community, Dhaka, Bangladesh. Int. J. Infect. Dis. 2010, 14, e93–e99. [Google Scholar] [CrossRef] [Green Version]
- Theiss-Nyland, K.; Qadri, F.; Colin-Jones, R.; Zaman, K.; Khanam, F.; Liu, X.; Voysey, M.; Khan, A.; Hasan, N.; Ashher, F. Assessing the impact of a Vi-polysaccharide conjugate vaccine in preventing typhoid infection among Bangladeshi children: A protocol for a phase IIIb trial. Clin. Infect. Dis. 2019, 68, S74–S82. [Google Scholar] [CrossRef]
- Stanaway, J.D.; Reiner, R.C.; Blacker, B.F.; Goldberg, E.M.; Khalil, I.A.; Troeger, C.E.; Andrews, J.R.; Bhutta, Z.A.; Crump, J.A.; Im, J. The global burden of typhoid and paratyphoid fevers: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019, 19, 369–381. [Google Scholar] [CrossRef] [Green Version]
- Kariuki, S.; Revathi, G.; Kiiru, J.; Mengo, D.M.; Mwituria, J.; Muyodi, J.; Munyalo, A.; Teo, Y.Y.; Holt, K.E.; Kingsley, R.A. Typhoid in Kenya is associated with a dominant multidrug-resistant Salmonella enterica serovar Typhi haplotype that is also widespread in Southeast Asia. J. Clin. Microbiol. 2010, 48, 2171–2176. [Google Scholar] [CrossRef] [Green Version]
- Geiger, T.; Lara-Tejero, M.; Xiong, Y.; Galán, J.E. Mechanisms of substrate recognition by a typhoid toxin secretion-associated muramidase. eLife 2020, 9, e53473. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; van der Bij, A.K.; Freeman, J.L.; Poirel, L.; Nordmann, P.; Costello, M.; Tchesnokova, V.L.; Pitout, J.D. Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum β-lactamases: Global distribution of the H 30-Rx sublineage. Antimicrob. Agents Chemother. 2014, 58, 3762–3767. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.M.; Palumbo, P.E.; Edelson, P.J. Salmonella strains resistant to multiple antibiotics: Therapeutic implications. Pediatric Infect. Dis. 1984, 3, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Samajpati, S.; Ray, U.; Roy, I.; Dutta, S. Antimicrobial resistance and molecular subtypes of Salmonella enterica serovar Typhi isolates from Kolkata, India over a 15 years period 1998–2012. Int. J. Med. Microbiol. 2017, 307, 28–36. [Google Scholar] [CrossRef]
- Melchiorre, M.G.; Chiatti, C.; Lamura, G.; Torres-Gonzales, F.; Stankunas, M.; Lindert, J.; Ioannidi-Kapolou, E.; Barros, H.; Macassa, G.; Soares, J.F. Social support, socio-economic status, health and abuse among older people in seven European countries. PLoS ONE 2013, 8, e54856. [Google Scholar]
- Iyer, R.N.; Jangam, R.R.; Jacinth, A.; Venkatalakshmi, A.; Nahdi, F.B. Prevalence and trends in the antimicrobial susceptibility pattern of Salmonella enterica serovars Typhi and Paratyphi A among children in a pediatric tertiary care hospital in South India over a period of ten years: A retrospective study. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2399–2404. [Google Scholar] [CrossRef] [PubMed]
- Tanmoy, A.M.; Westeel, E.; De Bruyne, K.; Goris, J.; Rajoharison, A.; Sajib, M.S.; van Belkum, A.; Saha, S.K.; Komurian-Pradel, F.; Endtz, H.P. Salmonella enterica serovar Typhi in Bangladesh: Exploration of genomic diversity and antimicrobial resistance. MBio 2018, 9, e02112-18. [Google Scholar] [CrossRef] [Green Version]
- Parvin, M.S.; Hasan, M.M.; Ali, M.Y.; Chowdhury, E.H.; Rahman, M.T.; Islam, M.T. Prevalence and Multidrug Resistance Pattern of Salmonella Carrying Extended-Spectrum β-Lactamase in Frozen Chicken Meat in Bangladesh. J. Food Prot. 2020, 83, 2107–2121. [Google Scholar] [CrossRef]
- Alam, S.B.; Mahmud, M.; Akter, R.; Hasan, M.; Sobur, A.; Nazir, K.; Noreddin, A.; Rahman, T.; El Zowalaty, M.E.; Rahman, M. Molecular detection of multidrug resistant Salmonella species isolated from broiler farm in Bangladesh. Pathogens 2020, 9, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhary, A.; Gopalakrishnan, R.; Senthur, N.P.; Ramasubramanian, V.; Ghafur, K.A.; Thirunarayan, M. Antimicrobial susceptibility of Salmonella enterica serovars in a tertiary care hospital in southern India. Indian J. Med. Res. 2013, 137, 800. [Google Scholar] [PubMed]
- Zhang, J.; Jin, H.; Hu, J.; Yuan, Z.; Shi, W.; Ran, L.; Zhao, S.; Yang, X.; Meng, J.; Xu, X. Serovars and antimicrobial resistance of non-typhoidal Salmonella from human patients in Shanghai, China, 2006–2010. Epidemiol. Infect. 2014, 142, 826–832. [Google Scholar] [CrossRef]
- Kuki, Á.; Nagy, L.; Zsuga, M.; Kéki, S. Fast identification of phthalic acid esters in poly (vinyl chloride) samples by direct analysis in real time (DART) tandem mass spectrometry. Int. J. Mass Spectrom. 2011, 303, 225–228. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Eggleston, K.; Rotimi, V.; Zeckhauser, R. Antibiotic resistance as a global threat: Evidence from China, Kuwait and the United States. Glob. Health 2006, 2, 2–6. [Google Scholar] [CrossRef] [Green Version]
- Lunguya, O.; Lejon, V.; Phoba, M.-F.; Bertrand, S.; Vanhoof, R.; Glupczynski, Y.; Verhaegen, J.; Muyembe-Tamfum, J.-J.; Jacobs, J. Antimicrobial resistance in invasive non-typhoid Salmonella from the Democratic Republic of the Congo: Emergence of decreased fluoroquinolone susceptibility and extended-spectrum beta lactamases. PLoS Negl. Trop. Dis. 2013, 7, e2103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulvey, M.R.; Boyd, D.A.; Finley, R.; Fakharuddin, K.; Langner, S.; Allen, V.; Ang, L.; Bekal, S.; El Bailey, S.; Haldane, D. Ciprofloxacin-resistant Salmonella enterica serovar Kentucky in Canada. Emerg. Infect. Dis. 2013, 19, 999. [Google Scholar] [CrossRef] [PubMed]
- Threlfall, E.; Ward, L. Ciprofloxacin-resistant Salmonella typhi and treatment failure. Lancet 1999, 353, 1590–1591. [Google Scholar] [CrossRef]
- Rushdy, A.A.; Mabrouk, M.I.; Abu-Sef, F.A.-H.; Kheiralla, Z.H.; Abdel-All, S.M.; Saleh, N.M. Contribution of different mechanisms to the resistance to fluoroquinolones in clinical isolates of Salmonella enterica. Braz. J. Infect. Dis. 2013, 17, 431–437. [Google Scholar] [CrossRef] [Green Version]
- Chandrasiri, P.; Elwitigala, J.; Nanayakkara, G. A multi centre laboratory study of Gram negative bacterial blood stream infections in Sri Lanka. Ceylon Med. J. 2013, 58, 56–61. [Google Scholar] [CrossRef]
- Chen, S.; Cui, S.; McDermott, P.F.; Zhao, S.; White, D.G.; Paulsen, I.; Meng, J. Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar Typhimurium to fluoroquinolones and other antimicrobials. Antimicrob. Agents Chemother. 2007, 51, 535–542. [Google Scholar] [CrossRef] [Green Version]
- Gaind, R.; Paglietti, B.; Murgia, M.; Dawar, R.; Uzzau, S.; Cappuccinelli, P.; Deb, M.; Aggarwal, P.; Rubino, S. Molecular characterization of ciprofloxacin-resistant Salmonella enterica serovar Typhi and Paratyphi A causing enteric fever in India. J. Antimicrob. Chemother. 2006, 58, 1139–1144. [Google Scholar] [CrossRef] [PubMed]
- Hirose, K.; Hashimoto, A.; Tamura, K.; Kawamura, Y.; Ezaki, T.; Sagara, H.; Watanabe, H. DNA sequence analysis of DNA gyrase and DNA topoisomerase IV quinolone resistance-determining regions of Salmonella enterica serovar Typhi and serovar Paratyphi A. Antimicrob. Agents Chemother. 2002, 46, 3249–3252. [Google Scholar] [CrossRef] [Green Version]
- Menezes, G.A.; Harish, B.N.; Khan, M.A.; Goessens, W.; Hays, J. Antimicrobial resistance trends in blood culture positive Salmonella Paratyphi A isolates from Pondicherry, India. Indian J. Med. Microbiol. 2016, 34, 222–227. [Google Scholar] [CrossRef]
- Wilke, M.S.; Lovering, A.L.; Strynadka, N.C. β-Lactam antibiotic resistance: A current structural perspective. Curr. Opin. Microbiol. 2005, 8, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Diamond, D.M.; Campbell, A.M.; Park, C.R.; Halonen, J.; Zoladz, P.R. The temporal dynamics model of emotional memory processing: A synthesis on the neurobiological basis of stress-induced amnesia, flashbulb and traumatic memories, and the Yerkes-Dodson law. Neural Plast. 2007, 2007, 060803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, A.; Lee, S.; Yang, Y.-A.; Song, J. Focus: Infectious diseases: The role of typhoid toxin in salmonella typhi virulence. Yale J. Biol. Med. 2017, 90, 283. [Google Scholar]
- Geiger, T.; Pazos, M.; Lara-Tejero, M.; Vollmer, W.; Galán, J.E. Peptidoglycan editing by a specific LD-transpeptidase controls the muramidase-dependent secretion of typhoid toxin. Nat. Microbiol. 2018, 3, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.D.; Vollmer, W.; Foster, S.J. Different walls for rods and balls: The diversity of peptidoglycan. Mol. Microbiol. 2014, 91, 862–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egan, A.J.; Biboy, J.; van’t Veer, I.; Breukink, E.; Vollmer, W. Activities and regulation of peptidoglycan synthases. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20150031. [Google Scholar] [CrossRef]
- Vollmer, W.; Blanot, D.; de Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [Green Version]
- Knapp, K.M.; English, B.K. Carbapenems. In Seminars in Pediatric Infectious Diseases; WB Saunders: Philadelphia, PA, USA, 2001; pp. 175–185. [Google Scholar]
- Khanna, N.R.; Gerriets, V. Beta Lactamase Inhibitors. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2021. [Google Scholar]
- Sienkiewicz, N.; Członka, S.; Kairyte, A.; Vaitkus, S. Curcumin as a natural compound in the synthesis of rigid polyurethane foams with enhanced mechanical, antibacterial and anti-ageing properties. Polym. Test. 2019, 79, 106046. [Google Scholar] [CrossRef]
- Alizadeh, N.; Malakzadeh, S. Antioxidant, antibacterial and anti-cancer activities of β-and γ-CDs/curcumin loaded in chitosan nanoparticles. Int. J. Biol. Macromol. 2020, 147, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Bhavsar, A.; Damani, A. A Comparative Study of the Performance of Selected Mutual Fund Growth Schemes from the Private Sector and Public Sector Schemes in India. Anvesha 2014, 7, 1–9. [Google Scholar]
- Rahayu, S.I.; Nurdiana, N.; Santoso, S. The effect of curcumin and cotrimoxazole in Salmonella typhimurium infection in vivo. Int. Sch. Res. Not. 2013, 2013, 601076. [Google Scholar] [CrossRef] [Green Version]
- Mohammed, N.A.; Habil, N.Y. Evaluation of antimicrobial activity of curcumin against two oral bacteria. Autom. Control Intell. Syst. 2015, 3, 18. [Google Scholar] [CrossRef]
- Sarkar, A.; De, R.; Mukhopadhyay, A.K. Curcumin as a potential therapeutic candidate for Helicobacter pylori associated diseases. World J. Gastroenterol. 2016, 22, 2736. [Google Scholar] [CrossRef] [PubMed]
- Adamczak, A.; Ożarowski, M.; Karpiński, T.M. Curcumin, a natural antimicrobial agent with strain-specific activity. Pharmaceuticals 2020, 13, 153. [Google Scholar] [CrossRef]
- Teow, S.-Y.; Liew, K.; Ali, S.A.; Khoo, A.S.-B.; Peh, S.-C. Antibacterial action of curcumin against Staphylococcus aureus: A brief review. J. Trop. Med. 2016, 2016, 2853045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, D.; Huang, C.; Huang, H.; Zhao, Y.; Khan, M.R.U.; Zhao, H.; Huang, L. Antibacterial mechanism of curcumin: A review. Chem. Biodivers. 2020, 17, e2000171. [Google Scholar] [CrossRef] [PubMed]
- Zorofchian Moghadamtousi, S.; Abdul Kadir, H.; Hassandarvish, P.; Tajik, H.; Abubakar, S.; Zandi, K. A review on antibacterial, antiviral, and antifungal activity of curcumin. BioMed Res. Int. 2014, 2014, 186864. [Google Scholar] [CrossRef]
- Marathe, S.A.; Balakrishnan, A.; Negi, V.D.; Sakorey, D.; Chandra, N.; Chakravortty, D. Curcumin reduces the motility of Salmonella enterica serovar Typhimurium by binding to the flagella, thereby leading to flagellar fragility and shedding. J. Bacteriol. 2016, 198, 1798–1811. [Google Scholar] [CrossRef] [Green Version]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Ullah, A.; Prottoy, N.I.; Araf, Y.; Hossain, S.; Sarkar, B.; Saha, A. Molecular docking and pharmacological property analysis of phytochemicals from Clitoria ternatea as potent inhibitors of cell cycle checkpoint proteins in the cyclin/CDK pathway in cancer cells. Comput. Mol. Biosci. 2019, 9, 81. [Google Scholar] [CrossRef] [Green Version]
- Bolton, E.E.; Wang, Y.; Thiessen, P.A.; Bryant, S.H. PubChem: Integrated platform of small molecules and biological activities. In Annual Reports in Computational Chemistry; Elsevier: Amsterdam, The Netherlands, 2008; Volume 4, pp. 217–241. [Google Scholar]
- Molinspiration Cheminformatics, Nova ulica, SK-900 26, Slovensky Grob, Slovak Republic. 2018. Available online: https://www.molinspiration.com/ (accessed on 20 October 2020).
- Walker, J.M. The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvement in protein secondary structure prediction by c prediction from alignments and joint prediction. CABIOS 1995, 11, 681–684. [Google Scholar] [PubMed]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo-and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 1–15. [Google Scholar]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo—distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository—New features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, M.; Kaptein, R.; Thornton, J. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar]
- Laskowski, R.; MacArthur, M.; Moss, D.; Thornton, J. SFCHECK: A unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Liu, D.; Liu, F.; Wu, J.; Zou, J.; Xiao, X.; Zhao, F.; Zhu, B. HTQC: A fast quality control toolkit for Illumina sequencing data. BMC Bioinform. 2013, 14, 33. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Roy, A.; Zhang, Y. BioLiP: A semi-manually curated database for biologically relevant ligand–protein interactions. Nucleic Acids Res. 2012, 41, D1096–D1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, S.; Sarkar, B.; Prottoy, M.N.I.; Araf, Y.; Taniya, M.A.; Ullah, M.A. Thrombolytic activity, drug likeness property and ADME/T analysis of isolated phytochemicals from ginger (Zingiber officinale) using in silico approaches. Mod. Res. Inflamm. 2019, 8, 29–43. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Adedoyin, A. ADME–Tox in drug discovery: Integration of experimental and computational technologies. Drug Discov. Today 2003, 8, 852–861. [Google Scholar] [CrossRef]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Filimonov, D.; Lagunin, A.; Gloriozova, T.; Rudik, A.; Druzhilovskii, D.; Pogodin, P.; Poroikov, V. Prediction of the biological activity spectra of organic compounds using the PASS online web resource. Chem. Heterocycl. Compd. 2014, 50, 444–457. [Google Scholar] [CrossRef]
- Tarcsay, Á.; Keserű, G.M. In silico site of metabolism prediction of cytochrome P450-mediated biotransformations. Expert Opin. Drug Metab. Toxicol. 2011, 7, 299–312. [Google Scholar] [CrossRef]
- Gschwend, D.A.; Good, A.C.; Kuntz, I.D. Molecular docking towards drug discovery. J. Mol. Recognit. Interdiscip. J. 1996, 9, 175–186. [Google Scholar] [CrossRef]
- Tian, S.; Wang, J.; Li, Y.; Li, D.; Xu, L.; Hou, T. The application of in silico drug-likeness predictions in pharmaceutical research. Adv. Drug Deliv. Rev. 2015, 86, 2–10. [Google Scholar] [CrossRef]
- Wang, Y.; Xing, J.; Xu, Y.; Zhou, N.; Peng, J.; Xiong, Z.; Liu, X.; Luo, X.; Luo, C.; Chen, K. In silico ADME/T modelling for rational drug design. Q. Rev. Biophys. 2015, 48, 488–515. [Google Scholar] [CrossRef] [Green Version]
- Li, A.P. Screening for human ADME/Tox drug properties in drug discovery. Drug Discov. Today 2001, 6, 357–366. [Google Scholar] [CrossRef]
- Paul Gleeson, M.; Hersey, A.; Hannongbua, S. In-silico ADME models: A general assessment of their utility in drug discovery applications. Curr. Top. Med. Chem. 2011, 11, 358–381. [Google Scholar] [CrossRef] [PubMed]
- De Graaf, C.; Vermeulen, N.P.; Feenstra, K.A. Cytochrome P450 in silico: An integrative modeling approach. J. Med. Chem. 2005, 48, 2725–2755. [Google Scholar] [CrossRef] [PubMed]
- Lamb, D.C.; Waterman, M.R.; Kelly, S.L.; Guengerich, F.P. Cytochromes P450 and drug discovery. Curr. Opin. Biotechnol. 2007, 18, 504–512. [Google Scholar] [CrossRef]
- Anzenbacher, P.; Anzenbacherova, E. Cytochromes P450 and metabolism of xenobiotics. Cell. Mol. Life Sci. CMLS 2001, 58, 737–747. [Google Scholar] [CrossRef]
- Xu, C.; Cheng, F.; Chen, L.; Du, Z.; Li, W.; Liu, G.; Lee, P.W.; Tang, Y. In silico prediction of chemical Ames mutagenicity. J. Chem. Inf. Model. 2012, 52, 2840–2847. [Google Scholar] [CrossRef]
- Ames, B.N.; Gurney, E.G.; Miller, J.A.; Bartsch, H. Carcinogens as frameshift mutagens: Metabolites and derivatives of 2-acetylaminofluorene and other aromatic amine carcinogens. Proc. Natl. Acad. Sci. USA 1972, 69, 3128–3132. [Google Scholar] [CrossRef] [Green Version]
- Priest, B.; Bell, I.M.; Garcia, M. Role of hERG potassium channel assays in drug development. Channels 2008, 2, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Hacker, K.; Maas, R.; Kornhuber, J.; Fromm, M.F.; Zolk, O. Substrate-dependent inhibition of the human organic cation transporter OCT2: A comparison of metformin with experimental substrates. PLoS ONE 2015, 10, e0136451. [Google Scholar] [CrossRef] [Green Version]
- Stepanchikova, A.; Lagunin, A.; Filimonov, D.; Poroikov, V. Prediction of biological activity spectra for substances: Evaluation on the diverse sets of drug-like structures. Curr. Med. Chem. 2003, 10, 225–233. [Google Scholar] [CrossRef] [Green Version]
- Lagunin, A.; Stepanchikova, A.; Filimonov, D.; Poroikov, V. PASS: Prediction of activity spectra for biologically active substances. Bioinformatics 2000, 16, 747–748. [Google Scholar] [CrossRef] [PubMed]
- Overington, J.P.; Al-Lazikani, B.; Hopkins, A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006, 5, 993–996. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).