Mapping Invasive Herbaceous Plant Species with Sentinel-2 Satellite Imagery: Echium plantagineum in a Mediterranean Shrubland as a Case Study
Abstract
1. Introduction
- (1)
- Can Sentinel-2 imagery be used to discriminate the invasive alien plant species, E. plantagineum?
- (2)
- What variables are important in detecting E. plantagineum in satellite imagery?
2. Materials and Methods
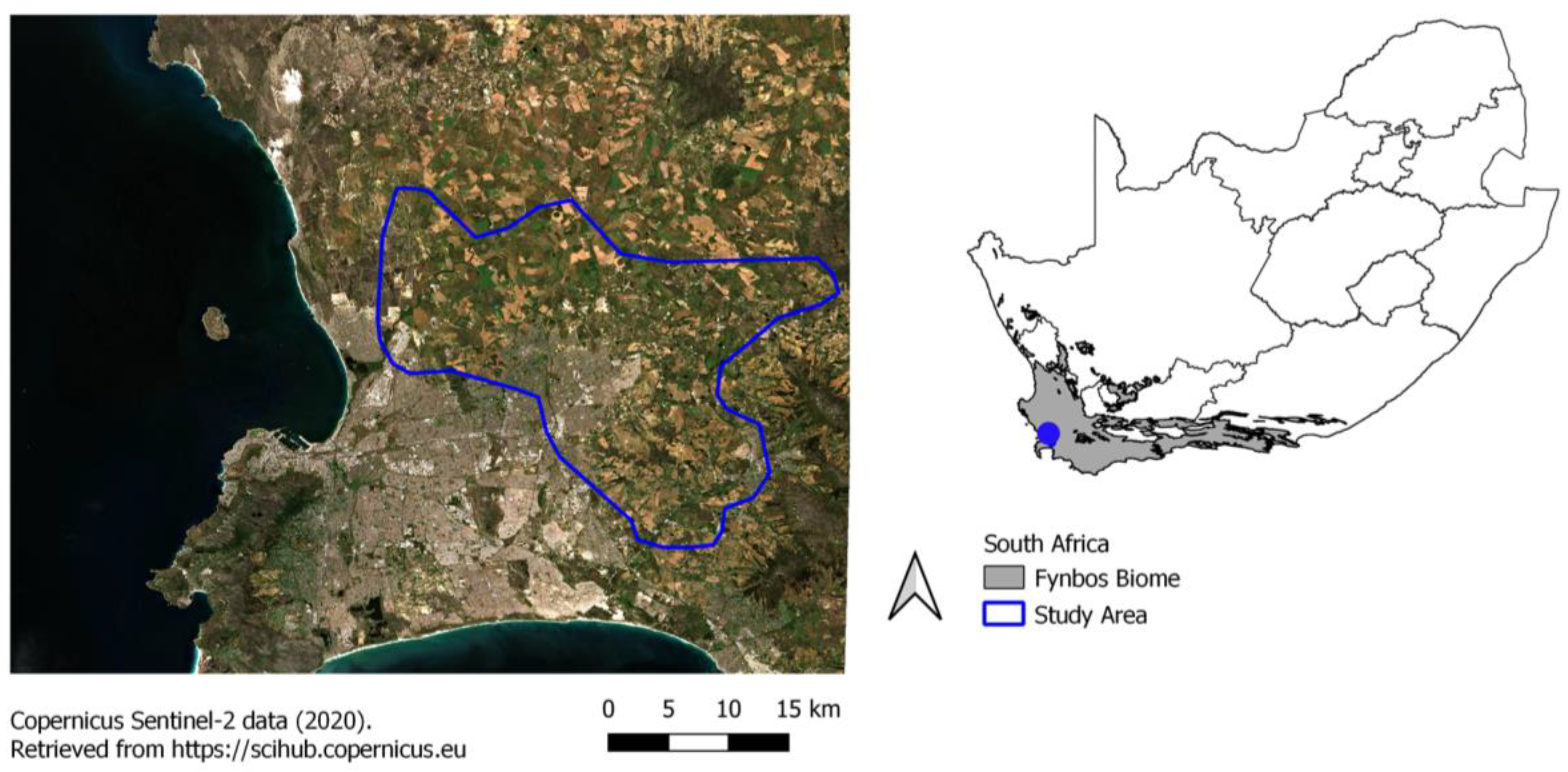
2.1. Study Area
2.2. Study Species
2.3. Training and Validation Data
2.4. Image Acquisition and Processing
| Sentinel-2 Image Band or Index | Description of Image Band or Equation of Index | Reference | Bands + VIs | Most Important Bands + VIs | Bands |
|---|---|---|---|---|---|
| B2 | Blue | x | x | x | |
| B3 | Green | x | x | x | |
| B4 | Red | x | x | ||
| B5 | Vegetation Red Edge 1 | x | x | x | |
| B6 | Vegetation Red Edge 2 | x | x | x | |
| B7 | Vegetation Red Edge 3 | x | x | x | |
| B8 | Near Infrared (NIR) wide | x | x | x | |
| B8A | Near Infrared (NIR) narrow | x | x | x | |
| B11 | Short Wave Infrared (SWIR) 1 | x | x | x | |
| B12 | Short Wave Infrared (SWIR) 2 | x | x | x | |
| ARVI | (NIR - (2 × Red) + Blue)/(NIR + (2 × Red) + Blue) | [93] | x | ||
| DVI | NIR - Red | [94] | x | ||
| EVI | 2.5 × ((NIR - Red)/((NIR) + (6 × Red) - (7.5 × Blue) + 1)) | [95] | x | ||
| EVI2 | 2.5 × ((NIR - Red)/(NIR + 2.4 × Red + 1) | [96] | x | ||
| GDVI | NIR - Green | [94] | x | x | |
| GNDVI | (NIR - Green)/(NIR + Green) | [97] | x | ||
| IPVI | NIR/(NIR + Red) | [98] | x | ||
| NDGI | (Green - Red)/(Green + Red) | [94] | x | ||
| NDRE | (NIR - Red Edge)/(NIR + Red Edge) | [99] | x | x | |
| NDVI | (NIR - Red)/(NIR + Red) | [89] | x | ||
| NFRE | (Red Edge - Red)/(Red Edge + Red) | [60] | x | x | |
| RI | (Red - Green)/(Red + Green) | [100] | x | x | |
| SAVI | ((NIR - Red)/(NIR + Red + L)) × (1 + L). Where L = 0.5 | [101] | x | ||
| SR | NIR/Red | [102] | x | ||
| VARI | (Green - Red)/(Green + Red - Blue) | [103] | x | ||
| VDVI | (2 × Green - Red - Blue)/(2 × Green + Red + Blue) | [104] | x |
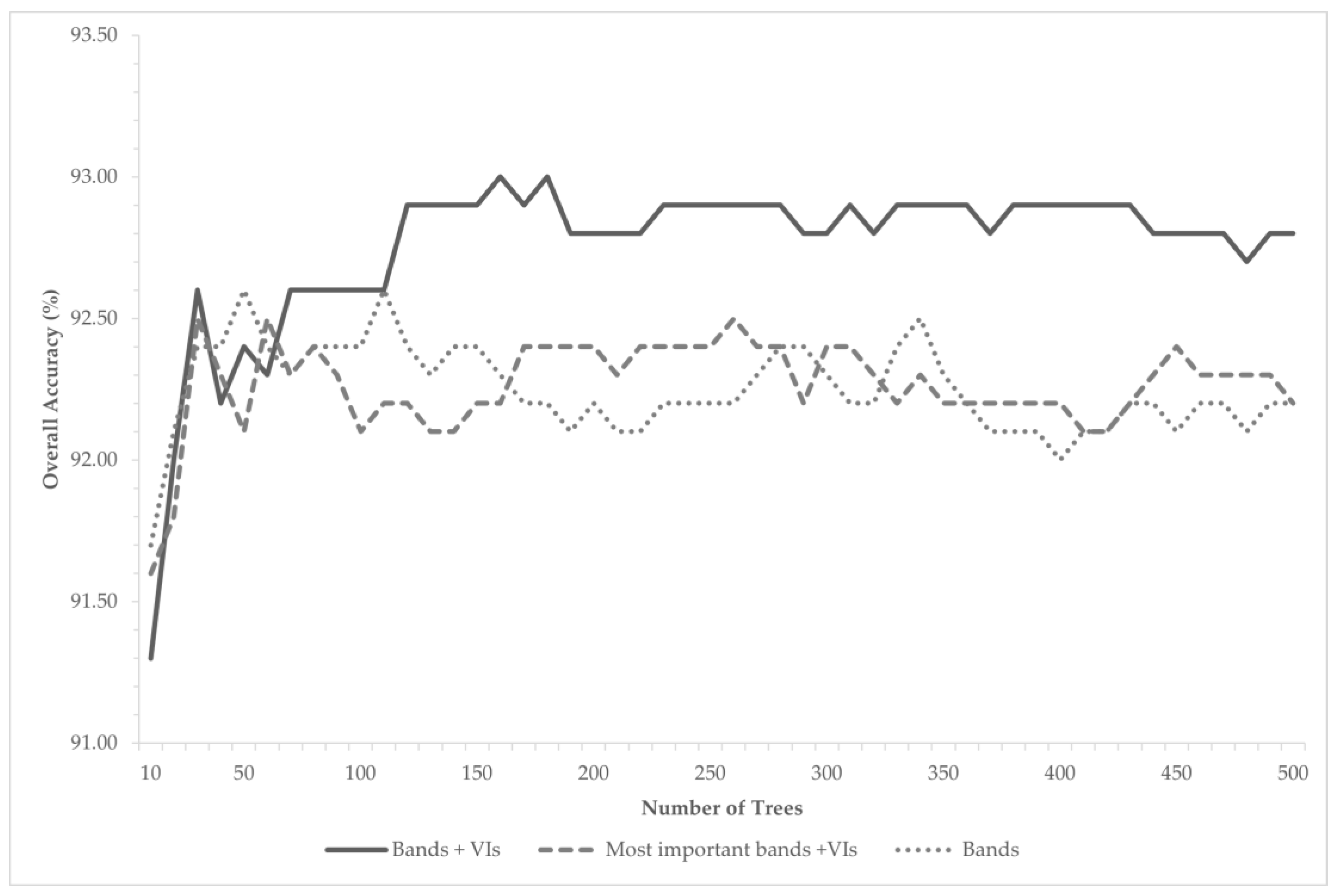
2.5. Image Classification—Random Forest
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pyšek, P.; Jarošík, V.; Hulme, P.E.; Pergl, J.; Hejda, M.; Schaffner, U.; Vilà, M. A global assessment of invasive plant impacts on resident species, communities and ecosystems: The interaction of impact measures, invading species’ traits and environment. Glob. Change Biol. 2012, 18, 1725–1737. [Google Scholar] [CrossRef]
- Paini, D.R.; Sheppard, A.W.; Cook, D.C.; De Barro, P.J.; Worner, S.P.; Thomas, M.B. Global threat to agriculture from invasive species. Proc. Natl. Acad. Sci. USA 2016, 113, 7575–7579. [Google Scholar] [CrossRef]
- Richardson, D.M.; van Wilgen, B.W. Invasive alien plants in South Africa: How well do we understand the ecological impacts? S. Afr. J. Sci. 2004, 100, 45–52. [Google Scholar]
- de Lange, W.J.; van Wilgen, B.W. An economic assessment of the contribution of biological control to the management of invasive alien plants and to the protection of ecosystem services in South Africa. Biol. Invasions 2010, 12, 4113–4124. [Google Scholar] [CrossRef]
- van Wilgen, B.W.; de Lange, W.J. The Costs and Benefits of Biological Control of Invasive Alien Plants in South Africa. Afr. Entomol. 2011, 19, 504–514. [Google Scholar] [CrossRef]
- van Wilgen, B.W.; Fill, J.M.; Baard, J.; Cheney, C.; Forsyth, A.T.; Kraaij, T. Historical costs and projected future scenarios for the management of invasive alien plants in protected areas in the Cape Floristic Region. Biol. Conserv. 2016, 200, 168–177. [Google Scholar] [CrossRef]
- Sheppard, A.W.; Smyth, M. Echium plantagineum L.—Paterson’s curse. In Biological Control of Weeds in Australia; Julien, M.H., McFadyen, R.E., Cullen, J., Eds.; CSIRO Publishing: Clayton, VIC, Australia, 2012; ISBN 9780643099937. [Google Scholar]
- Pimentel, D.; McNair, S.; Janecka, J.; Wightman, J.; Simmonds, C.; O’Connell, C.; Wong, E.; Russel, L.; Zern, J.; Aquino, T.; et al. Economic and environmental threats of alien plant, animal, and microbe invasions. Agric. Ecosyst. Environ. 2001, 84, 1–20. [Google Scholar] [CrossRef]
- Monaco, T.J.; Weller, S.C.; Ashton, F.M. WEED SCIENCE Principles and Practices; John Wiley & Sons, Inc.: New York, NY, USA, 2002; ISBN 0471370517. [Google Scholar]
- Pyšek, P.; Richardson, D.M. Invasive Species, Environmental Change and Management, and Health. Annu. Rev. Environ. Resour. 2010, 35, 25–55. [Google Scholar] [CrossRef]
- Erckie, L.; Adedoja, O.; Geerts, S.; van Wyk, E.; Boatwright, J.S. Impacts of an invasive alien Proteaceae on native plant species richness and vegetation structure. S. Afr. J. Bot. 2022, 144, 332–338. [Google Scholar] [CrossRef]
- Bradley, B.A. Remote detection of invasive plants: A review of spectral, textural and phenological approaches. Biol. Invasions 2014, 16, 1411–1425. [Google Scholar] [CrossRef]
- Van den Berg, E.C.; Kotze, I.; Beukes, H. Detection, Quantification and Monitoring of Prosopis in the Northern Cape Province of South Africa using Remote Sensing and GIS. S. Afr. J. Geomat. 2013, 2, 68–81. [Google Scholar]
- Geerts, S.; Rossenrode, T.; Irlich, U.M.; Visser, V. Emerging Ornamental Plant Invaders in Urban Areas—Centranthus ruber in Cape Town, South Africa as a Case Study. Invasive Plant Sci. Manag. 2017, 10, 322–331. [Google Scholar] [CrossRef]
- Afonso, L.; Esler, K.; Gaertner, M.; Geerts, S. The invasive alien Hypericum canariense in South Africa: Management, cost, and eradication feasibility. S. Afr. J. Bot. 2022, 146, 685–694. [Google Scholar] [CrossRef]
- Matthys, C.; Jubase, N.; Visser, V.; Geerts, S. Distribution of Melaleuca rugulosa (Schlechtendal ex Link) Craven (Myrtaceae) in South Africa: Assessment of invasiveness and feasibility of eradication. S. Afr. J. Bot. 2022, 148, 228–237. [Google Scholar] [CrossRef]
- du Plessis, N.S.; Rebelo, A.J.; Richardson, D.M.; Esler, K.J. Guiding restoration of riparian ecosystems degraded by plant invasions: Insights from a complex social-ecological system in the Global South. Ambio 2021, 51, 1552–1568. [Google Scholar] [CrossRef] [PubMed]
- Holmes, P.M.; Esler, K.J.; Gaertner, M.; Geerts, S.; Hall, S.A.; Nsikani, M.M.; Richardson, D.M.; Ruwanza, S. Biological Invasions and Ecological Restoration in South Africa. In Biological Invasions in South Africa; van Wilgen, B.W., Measey, J., Richardson, D.M., Wilson, J.R., Zengeya, T.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; pp. 665–700. [Google Scholar]
- Lopez, R.D.; Frohn, R.C. Remote Sensing for Landscape Ecology: Monitoring, Modeling, and Assessment of Ecosystems, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2017; ISBN 9781315152714. [Google Scholar]
- Zwiggelaar, R. A review of spectral properties of plants and their potential use for crop/weed discrimination in row-crops. Crop. Prot. 1998, 17, 189–206. [Google Scholar] [CrossRef]
- Lamb, D.W.; Brown, R.B. Remote-sensing and mapping of weeds in crops. J. Agric. Eng. Res. 2001, 78, 117–125. [Google Scholar] [CrossRef]
- Thorp, K.R.; Tian, L.F. A review on remote sensing of weeds in agriculture. Precis. Agric. 2004, 5, 477–508. [Google Scholar] [CrossRef]
- Royimani, L.; Mutanga, O.; Odindi, J.; Dube, T.; Matongera, T.N. Advancements in satellite remote sensing for mapping and monitoring of alien invasive plant species (AIPs). Phys. Chem. Earth 2019, 1, 237–245. [Google Scholar] [CrossRef]
- Vaz, A.S.; Alcaraz-Segura, D.; Vicente, J.R.; Honrado, J.P. The Many Roles of Remote Sensing in Invasion Science. Front. Ecol. Evol. 2019, 7, 370. [Google Scholar] [CrossRef]
- Huang, C.; Asner, G.P. Applications of remote sensing to alien invasive plant studies. Sensors 2009, 9, 4869–4889. [Google Scholar] [CrossRef]
- Campbell, J.B.; Wyynne, R.H. Introduction to Remote Sensing; Guilford Press: New York, NY, USA, 2011; ISBN 9781609181765. [Google Scholar]
- Vidhya, R.; Vijayasekaran, D.; Ramakrishnan, S.S. Mapping invasive plant Prosopis juliflora in arid land using high resolution remote sensing data and biophysical parameters. Indian J. Geo-Mar. Sci. 2017, 46, 1135–1144. [Google Scholar]
- Peerbhay, K.; Mutanga, O.; Lottering, R.; Bangamwabo, V.; Ismail, R. Detecting bugweed (Solanum mauritianum) abundance in plantation forestry using multisource remote sensing. ISPRS J. Photogramm. Remote Sens. 2016, 121, 167–176. [Google Scholar] [CrossRef]
- Evangelista, P.H.; Stohlgren, T.J.; Morisette, J.T.; Kumar, S. Mapping invasive tamarisk (Tamarix): A comparison of single-scene and time-series analyses of remotely sensed data. Remote Sens. 2009, 1, 519–533. [Google Scholar] [CrossRef]
- Kimothi, M.M.; Anitha, D.; Vasistha, H.B.; Soni, P.; Chandola, S.K. Remote sensing to map the invasive weed, Lantana camara in forests. Trop. Ecol. 2010, 51, 67–74. [Google Scholar]
- Oumar, Z. Assessing the utility of the SPOT 6 sensor in detecting and mapping Lantana camara for a community clearing project in KwaZulu-Natal, South Africa. S. Afr. J. Geomat. 2016, 5, 214–226. [Google Scholar] [CrossRef]
- Masemola, C.; Cho, M.A.; Ramoelo, A. Sentinel-2 time series based optimal features and time window for mapping invasive Australian native Acacia species in KwaZulu Natal, South Africa. Int. J. Appl. Earth Obs. Geoinf. 2020, 93, 102207. [Google Scholar] [CrossRef]
- Matongera, T.N.; Mutanga, O.; Dube, T.; Sibanda, M. Detection and mapping the spatial distribution of bracken fern weeds using the Landsat 8 OLI new generation sensor. Int. J. Appl. Earth Obs. Geoinf. 2017, 57, 93–103. [Google Scholar] [CrossRef]
- Kiala, Z.; Mutanga, O.; Odindi, J.; Masemola, C. Optimal window period for mapping Parthenium weed in South Africa, using high temporal resolution imagery and the ExtraTrees classifier. Biol. Invasions 2021, 23, 2881–2892. [Google Scholar] [CrossRef]
- Müllerová, J.; Brůna, J.; Bartaloš, T.; Dvořák, P.; Vítková, M.; Pyšek, P. Timing Is Important: Unmanned Aircraft vs. Satellite Imagery in Plant Invasion Monitoring. Front. Plant Sci. 2017, 8, 887. [Google Scholar] [CrossRef]
- Vaz, A.S.; Alcaraz-Segura, D.; Campos, J.C.; Vicente, J.R.; Honrado, J.P. Managing plant invasions through the lens of remote sensing: A review of progress and the way forward. Sci. Total Environ. 2018, 642, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Müllerová, J.; Pergl, J.; Pyšek, P. Remote sensing as a tool for monitoring plant invasions: Testing the effects of data resolution and image classification approach on the detection of a model plant species Heracleum mantegazzianum (giant hogweed). Int. J. Appl. Earth Obs. Geoinf. 2013, 25, 55–65. [Google Scholar] [CrossRef]
- Martin, F.-M.M.; Müllerová, J.; Borgniet, L.; Dommanget, F.; Breton, V.; Evette, A. Using Single- and Multi-Date UAV and Satellite Imagery to Accurately Monitor Invasive Knotweed Species. Remote Sens. 2018, 10, 1662. [Google Scholar] [CrossRef]
- Mudereri, B.T.; Dube, T.; Adel-Rahman, E.M.; Niassy, S.; Kimathi, E.; Khan, Z.; Landmann, T. A comparative analysis of PlanetScope and Sentinel-2 space-borne sensors in mapping Striga weed using Guided Regularised Random Forest classification ensemble. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, 42, 701–708. [Google Scholar] [CrossRef]
- Royimani, L.; Mutanga, O.; Odindi, J.; Zolo, K.S.; Sibanda, M.; Dube, T. Distribution of Parthenium hysterophoru L. with variation in rainfall using multi-year SPOT data and random forest classification. Remote Sens. Appl. Soc. Environ. 2019, 13, 215–223. [Google Scholar] [CrossRef]
- Henderson, L. Invasive, naturalized and casual alien plants in southern Africa: A summary based on the Southern African Plant Invaders Atlas (SAPIA). Bothalia 2007, 37, 215–248. [Google Scholar] [CrossRef]
- Rebelo, A.G.; Boucher, C.; Helme, N.; Mucina, L.; Rutherford, M.C. Fynbos Biome. In Vegetation Map of South Africa, Lesotho and Swaziland; Mucina, L., Rutherford, M.C., Eds.; South African National Biodiversity Institute: Pretoria, South Africa, 2006; pp. 52–219. [Google Scholar]
- van Wilgen, B.W.; Measey, J.; Richardson, D.M.; Wilson, J.R.; Zengeya, T.A. (Eds.) Biological Invasions in South Africa; Springer International Publishing: Cham, Switzerland, 2020; ISBN 978-3-030-32393-6. [Google Scholar]
- Holden, P.B.; Rebelo, A.J.; New, M.G. Mapping invasive alien trees in water towers: A combined approach using satellite data fusion, drone technology and expert engagement. Remote Sens. Appl. Soc. Environ. 2021, 21, 100448. [Google Scholar] [CrossRef]
- Mtengwana, B.; Dube, T.; Mkunyana, Y.P.; Mazvimavi, D. Use of multispectral satellite datasets to improve ecological understanding of the distribution of Invasive Alien Plants in a water-limited catchment, South Africa. Afr. J. Ecol. 2020, 58, 709–718. [Google Scholar] [CrossRef]
- Nel, J.L.; Richardson, D.M.; Rouget, M.; Mgidi, T.N.; Mdzeke, N.; Le Maitre, D.C.; van Wilgen, B.W.; Schonegevel, L.; Henderson, L.; Neser, S. A proposed classification of invasive alien plant species in South Africa: Towards prioritizing species and areas for management action. S. Afr. J. Sci. 2004, 100, 53–64. [Google Scholar]
- Sharma, G.P.; Esler, K.J. Phenotypic plasticity among Echium plantagineum populations in different habitats of Western Cape, South Africa. S. Afr. J. Bot. 2008, 74, 746–749. [Google Scholar] [CrossRef]
- Henderson, L. Mapping of Invasive Alien Plants: The Contribution of the Southern African Plant Invaders Atlas (SAPIA) to Biological Weed Control. Afr. Entomol. 2011, 19, 498–503. [Google Scholar] [CrossRef]
- Henderson, L. Alien Weeds and Invasive Plants: A Complete Guide to Declared Weed Invaders of South Africa; Plant Protection Research Institute: Pretoria, South Africa, 2001.
- Henderson, L. The Southern African Plant Invaders Atlas (SAPIA) and Its Contribution to Biological Weed Control. 1999, pp. 159–163. Available online: https://www.arc.agric.za/arc-ppri/News%20Articles%20Library/Henderson.pdf (accessed on 1 December 2022).
- Henderson, L.; Wilson, J.R.U. Changes in the composition and distribution of alien plants in South Africa: An update from the Southern African Plant. Bothalia 2017, 47, 1–26. [Google Scholar] [CrossRef]
- Grigulis, K.; Sheppard, A.W.; Ash, J.E.; Groves, R.H. The comparative demography of the pasture weed Echium plantagineum between its native and invaded ranges. J. Appl. Ecol. 2001, 38, 281–290. [Google Scholar] [CrossRef]
- Government of Western Australia. Paterson’s Curse: What You Should Know. 2022. Available online: https://www.agric.wa.gov.au/biological-control/patersons-curse-what-you-should-know (accessed on 16 September 2022).
- Hulting, A.; Krenz, J.; Parker, R. Paterson’s Curse in the Pacific Northwest. A Pacific Northwest Extension; Oregon State University: Corvallis, OR, USA, 2007. [Google Scholar]
- Piggin, C.M. The nutritive value of Echium plantagineum L. and Trifolium subterraneum L. Weed Res. 1977, 17, 361–365. [Google Scholar] [CrossRef]
- Smyth, M.J.; Sheppard, A.W.; Swirepik, A. The effect of grazing on seed production in Echium plantagineum. Weed Res. 1997, 37, 63–70. [Google Scholar] [CrossRef]
- Piggin, C.M. Flowering and seed production of Echium plantagineum L. Weed Res. 1978, 18, 83–87. [Google Scholar] [CrossRef]
- Piggin, C.M.; Sheppard, A.W. Echium plantagineum L. In The Biology of Australian Weeds Vol 1; Groves, R.H., Shepherd, R.C.H., Richardson, R.G., Eds.; R.G. and F.J. Richardson: Melbourne, VIC, Australia, 1995. [Google Scholar]
- Ullah, E.; Shepherd, R.C.H.; Baxter, J.T.; Peterson, J.A. Mapping flowering Paterson’s curse (Echium plantagineum) around Lake Hume, north eastern Victoria, using Landsat TM data. Plant Prot. Q. 1989, 4, 155–157. [Google Scholar]
- McIntyre, D.L. Application of High Resolution Remote Sensing to Detect and Map the Pasture Weed Paterson’s Curse (Echium plantagineum) in Western Australia; Curtin University: Bentley, WA, Australia, 2015. [Google Scholar]
- McIntyre, D.L.; Corner, R.J. Using EO-1 Hyperion satellite hyperspectral imagery to detect the pasture weed Paterson’s curse (Echium plantagineum L.) in southern Western Australia. In Proceedings of the Twentieth Australasian Weeds Conference, Perth, WA, Australia, 11–15 September 2016; pp. 196–199. [Google Scholar]
- Mirik, M.; Ansley, R.J.; Steddom, K.; Jones, D.C.; Rush, C.M.; Michels, G.J.; Elliott, N.C. Remote distinction of a noxious weed (Musk Thistle: Carduus Nutans) using airborne hyperspectral imagery and the support vector machine classifier. Remote Sens. 2013, 5, 612–630. [Google Scholar] [CrossRef]
- Mundt, J.T.; Glenn, N.F.; Weber, K.T.; Prather, T.S.; Lass, L.W.; Pettingill, J. Discrimination of hoary cress and determination of its detection limits via hyperspectral image processing and accuracy assessment techniques. Remote Sens. Environ. 2005, 96, 509–517. [Google Scholar] [CrossRef]
- Paz-Kagan, T.; Silver, M.; Panov, N.; Karnieli, A. Multispectral Approach for Identifying Invasive Plant Species Based on Flowering Phenology Characteristics. Remote Sens. 2019, 11, 953. [Google Scholar] [CrossRef]
- Singh, K.K.; Chen, Y.H.; Smart, L.; Gray, J.; Meentemeyer, R.K. Intra-annual phenology for detecting understory plant invasion in urban forests. ISPRS J. Photogramm. Remote Sens. 2018, 142, 151–161. [Google Scholar] [CrossRef]
- Houborg, R.; McCabe, M.F. Daily retrieval of NDVI and LAI at 3 m resolution via the fusion of CubeSat, Landsat, and MODIS data. Remote Sens. 2018, 10, 890. [Google Scholar] [CrossRef]
- Matongera, T.N.; Mutanga, O.; Dube, T.; Lottering, R.T. Detection and mapping of bracken fern weeds using multispectral remotely sensed data: A review of progress and challenges. Geocarto Int. 2018, 33, 209–224. [Google Scholar] [CrossRef]
- Hamada, Y.; Stow, D.A.; Coulter, L.L.; Jafolla, J.C.; Hendricks, L.W. Detecting Tamarisk species (Tamarix spp.) in riparian habitats of Southern California using high spatial resolution hyperspectral imagery. Remote Sens. Environ. 2007, 109, 237–248. [Google Scholar] [CrossRef]
- Lourenço, P.; Teodoro, A.C.; Gonçalves, J.A.; Honrado, J.P.; Cunha, M.; Sillero, N. Assessing the performance of different OBIA software approaches for mapping invasive alien plants along roads with remote sensing data. Int. J. Appl. Earth Obs. Geoinf. 2021, 95, 102263. [Google Scholar] [CrossRef]
- Große-Stoltenberg, A.; Hellmann, C.; Thiele, J.; Werner, C.; Oldeland, J. Early detection of GPP-related regime shifts after plant invasion by integrating imaging spectroscopy with airborne LiDAR. Remote Sens. Environ. 2018, 1, 780–792. [Google Scholar] [CrossRef]
- Kattenborn, T.; Lopatin, J.; Förster, M.; Braun, A.C.; Fassnacht, F.E. UAV data as alternative to field sampling to map woody invasive species based on combined Sentinel-1 and Sentinel-2 data. Remote Sens. Environ. 2019, 227, 61–73. [Google Scholar] [CrossRef]
- Große-Stoltenberg, A.; Hellmann, C.; Werner, C.; Oldeland, J.; Thiele, J. Evaluation of continuous VNIR-SWIR spectra versus narrowband hyperspectral indices to discriminate the invasive Acacia longifolia within a mediterranean dune ecosystem. Remote Sens. 2016, 8, 334. [Google Scholar] [CrossRef]
- Andrew, M.E.; Ustin, S.L. The role of environmental context in mapping invasive plants with hyperspectral image data. Remote Sens. Environ. 2008, 112, 4301–4317. [Google Scholar] [CrossRef]
- Holmes, P.M.; Rebelo, A.G.; Dorse, C.; Wood, J. Can Cape Town’s unique biodiversity be saved? Balancing conservation imperatives and development needs. Ecol. Soc. 2012, 17, 2. [Google Scholar] [CrossRef]
- Afonso, L.; Esler, K.J.; Gaertner, M.; Geerts, S. Comparing invasive alien plant community composition between urban, peri-urban and rural areas; the city of Cape Town as a case study. In Urban Ecology: Emerging Patterns and Social-Ecological Systems; Verma, P., Singh, P., Singh, R., Rashubanshi, A.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; ISBN 978-0-12-820730-7. [Google Scholar]
- Davison, A.; Marshak, M. State of Environment Report; Communication Department, City of Cape Town: Cape Town, South Africa, 2012. [Google Scholar]
- Moran, V.C.; Hoffmann, J.H. Conservation of the fynbos biome in the Cape Floral Region: The role of biological control in the management of invasive alien trees. BioControl 2012, 57, 139–149. [Google Scholar] [CrossRef]
- Goldblatt, P.; Manning, J.C. Plant Diversity of the Cape Region of Southern Africa. Ann. Mo. Bot. Gard. Press 2002, 89, 281–302. [Google Scholar] [CrossRef]
- Mucina, L.; Rutherford, M.C. (Eds.) The Vegetation of South Africa, Lesotho and Swaziland; Strelitzia 19, South African National Biodiversity Institute: Pretoria, South Africa, 2006. [Google Scholar]
- Manning, J.C.; Paterson-Jones, C. Field Guide to Fynbos, 2nd ed.; Struik Nature: Cape Town, South Africa, 2018. [Google Scholar]
- Piggin, C.M. The herbaceous species of Echium (Boraginaceae) naturalized in Australia. Muelleria 1977, 3, 215–244. [Google Scholar] [CrossRef]
- IAC. Biological Control of Echium Species (Including Paterson’s Curse/Salvation Jane); Industries Assistance Commission Report No. 371; Australian Government Publishing Service: Canberra, ACT, Australia, 1985.
- Holloway, J.; Mengersen, K. Statistical machine learning methods and remote sensing for sustainable development goals: A review. Remote Sens. 2018, 10, 1365. [Google Scholar] [CrossRef]
- Adam, E.; Mureriwa, N.; Newete, S. Mapping Prosopis glandulosa (mesquite) in the semi-arid environment of South Africa using high-resolution WorldView-2 imagery and machine learning classifiers. J. Arid Environ. 2017, 145, 43–51. [Google Scholar] [CrossRef]
- Abdel-Rahman, E.M.; Mutanga, O.; Adam, E.; Ismail, R. Detecting Sirex noctilio grey-attacked and lightning-struck pine trees using airborne hyperspectral data, random forest and support vector machines classifiers. ISPRS J. Photogramm. Remote Sens. 2014, 88, 48–59. [Google Scholar] [CrossRef]
- Congalton, R.G. Accuracy assessment and validation of remotely sensed and other spatial information. Int. J. Wildl. Fire 2001, 10, 321–328. [Google Scholar] [CrossRef]
- ESA. Sentinel Online. 2021. Available online: https://sentinel.esa.int/web/sentinel/home (accessed on 31 January 2021).
- Xie, Y.; Sha, Z.; Yu, M. Remote sensing imagery in vegetation mapping: A review. J. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- Rouse, J.W., Jr.; Hass, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the great plains with ERTS. Third Earth Resour. Technol. Satell. Symp. 1973, 1, 309–317. [Google Scholar]
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 2017, 1353691. [Google Scholar] [CrossRef]
- Barnes, E.M.; Clarke, T.R.; Richards, S.E.; Colaizzi, P.D.; Haberland, J.; Kostrzewski, M.; Waller, P.; Choi, C.; Riley, E.; Thompson, T.; et al. Coincident detection of crop water status, nitrogen status, and canopy density using ground-based multispectral data. In Proceedings of the Fifth International Conference on Precision Agriculture, American Society of Agronomy, Bloomington, MN, USA, 16–19 July 2000. [Google Scholar]
- Chuvieco, E.; Huete, A. Fundamentals of Satellite Remote Sensing; CRC Press: Boca Raton, FL, USA, 2009; ISBN 9781420021516. [Google Scholar]
- Kaufman, Y.; Tanre, D. Atmospherically resistant vegetation index. IEEE Trans. Geosci. Remote Sens. 1992, 30, 260–271. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Liu, H.Q.; Huete, A. Feedback based modification of the NDVI to minimize canopy background and atmospheric noise. IEEE Trans. Geosci. Remote Sens. 1995, 33, 457–465. [Google Scholar] [CrossRef]
- Jiang, Z.; Huete, A.R.; Kim, Y.; Didan, K. 2-band enhanced vegetation index without a blue band and its application to AVHRR data. Remote Sens. Model. Ecosyst. Sustain. IV 2007, 6679, 667905. [Google Scholar]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Crippen, R.E. Calculating the vegetation index faster. Remote Sens. Environ. 1990, 34, 71–73. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Quantitative estimation of chlorophyll–A using reflectance spectra: Experiments with autumn chestnut and maple leaves. J. Photochem. Photobiol. B Biol. 1994, 22, 247–252. [Google Scholar] [CrossRef]
- Escadafal, R.; Huete, A. Improvement in remote sensing of low vegetation cover in arid regions by correcting vegetation indices for soil noise. CR Académie Des Sci. Paris 1991, 312, 1385–1391. [Google Scholar]
- Huete, A.R. A Soil-Adjusted Vegetation Index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of leaf-area index from quality of light on the forest floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel Algorithms for Remote Estimation of Vegetation Fraction. Remote Sens. Environ. 2002, 1, 76–87. [Google Scholar] [CrossRef]
- Wang, X.; Wang, M.; Wang, S.; Wu, Y. Extraction of Vegetation Information from Visible Unmanned Aerial Vehicle Images. Trans. Chin. Soc. Agric. Eng. 2015, 31, 152–159. [Google Scholar]
- Bannari, A.; Morin, D.; Bonn, F.; Huete, A.R. A review of vegetation indices. Remote Sens. Rev. 1995, 13, 95–120. [Google Scholar] [CrossRef]
- Ndlovu, H.S.; Sibanda, M.; Odindi, J.; Buthelezi, S.; Mutanga, O. Detecting and mapping the spatial distribution of Chromoleana odorata invasions in communal areas of South Africa using Sentinel-2 multispectral remotely sensed data. Phys. Chem. Earth 2022, 126, 103081. [Google Scholar] [CrossRef]
- Makori, D.; Abdel-Rahman, E.M.; Landmann, T.; Mutanga, O.; Odindi, J.; Nguku, E.; Tonnang, H.; Raina, S. Suitability of resampled multispectral datasets for mapping flowering plants in the Kenyan savannah. PLoS ONE 2020, 15, e0232313. [Google Scholar] [CrossRef]
- Genuer, R.; Poggi, J.; Tuleau-malot, C.; Genuer, R.; Poggi, J.; Variable, C.T.; Forests, R.; Genuer, R.; Poggi, J.; Tuleau-malot, C. Variable selection using Random Forests. Pattern Recognit. Lett. 2010, 31, 2225–2236. [Google Scholar] [CrossRef]
- Google. API Reference. 2022. Available online: https://developers.google.com/earth-engine/apidocs/ee-classifier-smilerandomforest (accessed on 26 August 2022).
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liu, H.; Motoda, H. Feature Extraction, Construction and Selection: A Data Mining Perspective; Kluwer Academic Publishers: Vancouver, BC, Canada, 1998; ISBN 0792381963. [Google Scholar]
- Pal, M.; Foody, G.M. Feature selection for classification of hyperspectral data by SVM. IEEE Trans. Geosci. Remote Sens. 2010, 48, 2297–2307. [Google Scholar] [CrossRef]
- Poona, N.K.; Van Niekerk, A.; Nadel, R.L.; Ismail, R. Random Forest (RF) Wrappers for Waveband Selection and Classification of Hyperspectral Data. Appl. Spectrosc. 2016, 70, 322–333. [Google Scholar] [CrossRef]
- Han, H.; Guo, X.; Yu, H. Variable selection using Mean Decrease Accuracy and Mean Decrease Gini based on Random Forest. In Proceedings of the 2016 7th IEEE International Conference on Software Engineering and Service Science (ICSESS), Beijing, China, 26–28 August 2016; Volume 1, pp. 219–224. [Google Scholar]
- Belgiu, M.; Drăgu, L.; Drăgut, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Probst, P.; Wright, M.N.; Boulesteix, A.L. Hyperparameters and tuning strategies for random forest. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Nygren, R.; Petkov, A. Evaluation of Hyperparameter Optimization Methods for Random Forest Classifiers; Kth Royal Institute of Technology School of Electrical Engineering and Computer Science: Stockholm, Sweden, 2021. [Google Scholar]
- Rebelo, A.J.; Gokool, S.; Holden, P.B.; New, M.G. Can Sentinel-2 be used to detect invasive alien trees and shrubs in Savanna and Grassland Biomes? Remote Sens. Appl. Soc. Environ. 2021, 23, 100600. [Google Scholar] [CrossRef]
- Shoko, C.; Mutanga, O. Examining the strength of the newly-launched Sentinel 2 MSI sensor in detecting and discriminating subtle differences between C3 and C4 grass species. ISPRS J. Photogramm. Remote Sens. 2017, 129, 32–40. [Google Scholar] [CrossRef]
- Cho, M.A.; Mathieu, R.; Asner, G.P.; Naidoo, L.; van Aardt, J.; Ramoelo, A.; Debba, P.; Wessels, K.; Main, R.; Smit, I.P.J.; et al. Mapping tree species composition in South African savannas using an integrated airborne spectral and LiDAR system. Remote Sens. Environ. 2012, 125, 214–226. [Google Scholar] [CrossRef]
- Odindi, J.; Mutanga, O.; Rouget, M.; Hlanguza, N. Mapping alien and indigenous vegetation in the KwaZulu-Natal Sandstone Sourveld using remotely sensed data. Bothalia 2016, 46, 1–9. [Google Scholar] [CrossRef]
- Otunga, C.; Odindi, J.; Mutanga, O.; Adjorlolo, C. Evaluating the potential of the red edge channel for C3 (Festuca spp.) grass discrimination using Sentinel-2 and Rapid Eye satellite image data. Geocarto Int. 2019, 34, 1123–1143. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Lichtenthaler, H.K. Detection of red edge position and chlorophyll content by reflectance measurements near 700 nm. J. Plant Physiol. 1996, 148, 501–508. [Google Scholar] [CrossRef]
- Rajah, P.; Odindi, J.; Mutanga, O.; Kiala, Z. The utility of Sentinel-2 Vegetation Indices (VIs) and Sentinel-1 Synthetic Aperture Radar (SAR) for invasive alien species detection and mapping. Nat. Conserv. 2019, 35, 41–61. [Google Scholar] [CrossRef]
- César De Sá, N.; Carvalho, S.; Castro, P.; Marchante, E.; Marchante, H. Using Landsat Time Series to Understand How Management and Disturbances Influence the Expansion of an Invasive Tree. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 3243–3253. [Google Scholar] [CrossRef]
- Ottosen, T.B.; Lommen, S.T.E.; Skjøth, C.A. Remote sensing of cropping practice in Northern Italy using time-series from Sentinel-2. Comput. Electron. Agric. 2019, 157, 232–238. [Google Scholar] [CrossRef]
- Lass, L.W.; Shafii Thill, B.; Prather, T.S. Detecting Spotted Knapweed (Centaurea maculosa) with Hyperspectral Remote Sensing. Weed Technol. 2002, 16, 426–432. [Google Scholar] [CrossRef]
- Pastick, N.J.; Dahal, D.; Wylie, B.K.; Parajuli, S.; Boyte, S.P.; Wu, Z. Characterizing land surface phenology and exotic annual grasses in dryland ecosystems using landsat and sentinel-2 data in harmony. Remote Sens. 2020, 12, 725. [Google Scholar] [CrossRef]
- Sage, A.J.; Genschel, U.; Nettleton, D. Tree aggregation for random forest class probability estimation. Stat. Anal. Data Min. 2020, 13, 134–150. [Google Scholar] [CrossRef]
- Chen, M.; Ke, Y.; Bai, J.; Li, P.; Lyu, M.; Gong, Z.; Zhou, D. Monitoring early stage invasion of exotic Spartina alterniflora using deep-learning super-resolution techniques based on multisource high-resolution satellite imagery: A case study in the Yellow River Delta, China. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102180. [Google Scholar] [CrossRef]


| Number of Samples | |||
|---|---|---|---|
| Land Cover Class | Training | Validation | Total |
| Bare ground | 682 | 323 | 1005 |
| Built-up | 907 | 371 | 1278 |
| Agriculture | 725 | 279 | 1004 |
| Shrubland | 721 | 317 | 1038 |
| Water | 1006 | 361 | 1367 |
| Echium plantagineum | 256 | 119 | 375 |
| Total number of samples | 4297 | 1170 | 6067 |
| Area of samples | 0.43 km2 (43 ha) | 0.18 km2 (18 ha) | 0.61 km2 (61 ha) |
| Parameter | Default Value | Description |
| Number of Trees | No default value. Must be specified | The number of decision trees to create. |
| Variables Per Split | Square root of the number of variables | The number of variables per split. |
| Min. Leaf Population | 1 | Only create nodes whose training set contains at least this number of points. |
| Bag Fraction | 0.5 | The fraction of input to bag per tree. |
| Max. Nodes | No limit | The max. number of leaf nodes in each tree. |
| Seed | 0 | The randomization seed. |
| Bands + VIs | |||||||
| Class | B | BU | AG | SH | W | EP | UA (%) |
| B | 304 | 19 | 0 | 0 | 0 | 0 | 94.12 |
| BU | 8 | 357 | 5 | 0 | 0 | 1 | 96.23 |
| AG | 0 | 3 | 251 | 16 | 0 | 9 | 89.96 |
| SH | 0 | 3 | 1 | 300 | 5 | 8 | 94.64 |
| W | 0 | 0 | 0 | 2 | 359 | 0 | 99.45 |
| EP | 2 | 1 | 20 | 19 | 0 | 77 | 64.71 |
| PA (%) | 96.82 | 93.21 | 90.61 | 89.02 | 98.63 | 81.05 | |
| OA (%) | 93.11 | ||||||
| K | 0.92 | ||||||
| Most Important Bands + VIs | |||||||
| Class | B | BU | AG | SH | W | EP | UA (%) |
| B | 299 | 21 | 0 | 3 | 0 | 0 | 92.57 |
| BU | 7 | 358 | 5 | 0 | 0 | 1 | 96.50 |
| AG | 0 | 6 | 250 | 14 | 0 | 9 | 89.61 |
| SH | 0 | 4 | 5 | 299 | 5 | 4 | 94.32 |
| W | 0 | 0 | 0 | 2 | 359 | 0 | 99.45 |
| EP | 2 | 2 | 22 | 18 | 0 | 75 | 63.03 |
| PA (%) | 97.08 | 91.56 | 88.65 | 88.99 | 98.63 | 84.27 | |
| OA (%) | 92.66 | ||||||
| K | 0.91 | ||||||
| Bands | |||||||
| Class | B | BU | AG | SH | W | EP | UA (%) |
| B | 306 | 17 | 0 | 0 | 0 | 0 | 94.74 |
| BU | 12 | 353 | 4 | 1 | 0 | 1 | 95.15 |
| AG | 0 | 2 | 258 | 13 | 0 | 6 | 92.47 |
| SH | 0 | 3 | 5 | 300 | 5 | 4 | 94.64 |
| W | 0 | 0 | 0 | 3 | 358 | 0 | 99.17 |
| EP | 2 | 1 | 32 | 17 | 0 | 67 | 56.30 |
| PA (%) | 95.63 | 93.88 | 86.29 | 89.82 | 98.62 | 85.90 | |
| OA (%) | 92.77 | ||||||
| K | 0.91 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duncan, P.; Podest, E.; Esler, K.J.; Geerts, S.; Lyons, C. Mapping Invasive Herbaceous Plant Species with Sentinel-2 Satellite Imagery: Echium plantagineum in a Mediterranean Shrubland as a Case Study. Geomatics 2023, 3, 328-344. https://doi.org/10.3390/geomatics3020018
Duncan P, Podest E, Esler KJ, Geerts S, Lyons C. Mapping Invasive Herbaceous Plant Species with Sentinel-2 Satellite Imagery: Echium plantagineum in a Mediterranean Shrubland as a Case Study. Geomatics. 2023; 3(2):328-344. https://doi.org/10.3390/geomatics3020018
Chicago/Turabian StyleDuncan, Patricia, Erika Podest, Karen J. Esler, Sjirk Geerts, and Candice Lyons. 2023. "Mapping Invasive Herbaceous Plant Species with Sentinel-2 Satellite Imagery: Echium plantagineum in a Mediterranean Shrubland as a Case Study" Geomatics 3, no. 2: 328-344. https://doi.org/10.3390/geomatics3020018
APA StyleDuncan, P., Podest, E., Esler, K. J., Geerts, S., & Lyons, C. (2023). Mapping Invasive Herbaceous Plant Species with Sentinel-2 Satellite Imagery: Echium plantagineum in a Mediterranean Shrubland as a Case Study. Geomatics, 3(2), 328-344. https://doi.org/10.3390/geomatics3020018






