Abstract
The microalga Phaeodactylum tricornutum is considered a model diatom. It is the second diatom whose genome was sequenced and the first one genetically engineered. This permits its use as a cell factory for the production of high-value compounds for nutraceutical, cosmeceutical, pharmaceutical, biodiesel, and bioplastic applications. This study is focused on analyzing expression levels of enzymes involved in the synthesis of sulfoglycolipids and monogalactosyldiacylglycerols, compounds known to have anticancer and immunomodulatory activities, and genes coding antioxidant, heat shock and stress-responsive proteins, in various culturing conditions. Our data showed that both nutrient starvation and senescence induced the down-regulation of both sulfoglycolipid and monogalactosyldiacylglycerol synthesis-related genes and stress-responsive genes (compared to the replete condition), suggesting that the control condition, consisting of cells in the exponential phase in replete medium, is the condition with the highest expression of the genes of interest and worth of further bioactivity screening and chemical analyses for drug discovery and biotechnological applications.
1. Introduction
Phaeodactylum tricornutum is a marine diatom and the only species of the genus Phaeodactylum. It is widely studied and represents a model diatom. It is not widely distributed in nature, but found mainly in coastal unstable environments (denoting its great adaptation capability) [1]. It is the first diatom genetically engineered [2] and the second diatom of which the genome has been completely sequenced [3]. This allows the use of the most advanced genome editing techniques, such as TALEN and CRISPR/Cas9, in order to study algal physiology [4,5,6,7,8,9] and biotechnological applications [4]. Information and tools available for P. tricornutum have allowed its use as producer of non-native products such as monoclonal antibodies [10] and plants triterpenoids [11] or for high-added value compounds which are generally produced at low levels, such as docosahexaenoic acid (DHA; [12]). Its sequenced genome, genetic engineered tools, and biological properties make P. tricornutum an important candidate as diatom cell factory. In addition, several studies report P. tricornutum as a source of high-value natural compounds with interesting bioactivities, such as (1) the compound nonyl 8-acetoxy-6-methyloctanoate which suppressed the growth of human promyelocytic leukemia cells triggering apoptotic pathway [13]; (2) two monogalactosyldiacylglycerols (MGDGs) which induced apoptosis in immortal mouse cell lines isolated from mouse embryo [14]; (3) fucoxantin [15], able to decrease the production of pro-inflammatory cytokines such as Interleukin 1-beta (IL-1β), Interleukin 6 (IL-6), and Tumor Necrosis Factor-alpha (TNF-α) [16]; and (4) sulfated polysaccharides, which inhibited hepatocellular carcinoma cell lines (HepG2) growth in vitro [17].
Considering the need for new drugs in order to expand our weapons against cancer and other human pathologies, such as emerging viral and antibiotic-resistant infections, studies on metabolic pathways responsible for high-value compounds synthesis are increasing [18,19,20,21,22,23]. In addition, several papers showed that depending on the culturing conditions, microalgae may activate specific metabolic pathways and produce key metabolites with potential applications for human health. This experimental approach is also known as “one strain many compounds” (OSMAC; [24]) and has been used to improve the production of compounds of interest [24,25,26]. In fact, lipids and triacylglycerols (TAG) accumulation have been often related to stressful exposure, growth phase, and nutrient starvation in various microalgae including P. tricornutum [27,28,29,30,31].
In this study, considering that the enzymes involved in the synthesis of MGDGs and sulfoglycolipids, compounds known to have anticancer and immunomodulatory activities, have been shown to be present in P. tricornutum [32], our aim was to evaluate the expression levels of genes involved in their synthesis: UDP-sulfoquinovose synthase [EC:3.13.1.1] (SQD1) and sulfoquinovosyltransferase [EC:2.4.1.-] (SQD2) involved in sulfolipids synthesis and 1,2-diacylglycerol 3-beta-galactosyltransferase [EC:2.4.1.46] (MGD) involved in MDGDs synthesis. In addition, we also analyze the expression of the stress responsive genes involved in detoxification pathways, antioxidant processes, cell death/division, and general stress-response (i.e., heat shock proteins) [33,34,35,36,37,38]. Transcripts were related to enzymes involved in the first and second line of defense [39]. The first includes the multixenobiotic resistance system and the related multidrug resistance protein (MDR). The second line of defense consists in two phases, catalyzed by Phase I and Phase II enzymes. Cytochrome P450 (CYP450) belongs to Phase I enzymes involved in oxidation, reduction, hydrolysis, hydration and dehalogenation reactions. Phase II consists in conjugation reactions, such as with the antioxidant compound glutathione. Glutathione synthase (GSH) and glutathione peroxidase (GPX) are involved in glutathione metabolism [35,40]. In addition, we studied heat shock proteins (HSPs), known to be activated in response to various environmental stress factors [34], superoxide dismutase (SOD) responsible for reactive oxygen species detoxification [39], proteins related to cell death/division (i.e., mitotic spindle assembly checkpoint proteins (MAD1 and MAD2) and metacaspase (CASP) [38,41]), and other stress responsive genes, such as ascorbate peroxidase (APX), glycolate oxidase (GOX), tocopherol cyclase (TOC) and hystone deacetylase (HYS) [36,37,42,43]. In particular, we analyzed expression levels of our genes of interest (GOI) in different culturing conditions, including a replete condition and two nutrient starvation conditions (silica and nitrogen starvation), as well as different growth phases (exponential, early and late stationery phase). Nutrient starvation and different growth phases were selected since previous studies have shown that diatoms may produce bioactive compounds in these conditions [44,45,46]. Regarding P. tricornutum, for example, fucoxantin production was shown to be regulated by lower light intensity and nitrogen supplementation [47], while nitrogen depletion caused thylakoid senescence and growth arrest [29] and nitrogen starvation induced the up-regulation of 86 up to 212 transcription factors (in particular, two specific motifs named 6 and 17) [48]. These stressful conditions (nutrient starvation and senescence) were selected in order to identify the culturing condition with the highest expression of sulfoglycolipid and MGDG synthesis-related genes and to propose them for future bioactivity screening and bioactive metabolite identification.
2. Results
2.1. Reference Gene Assessment
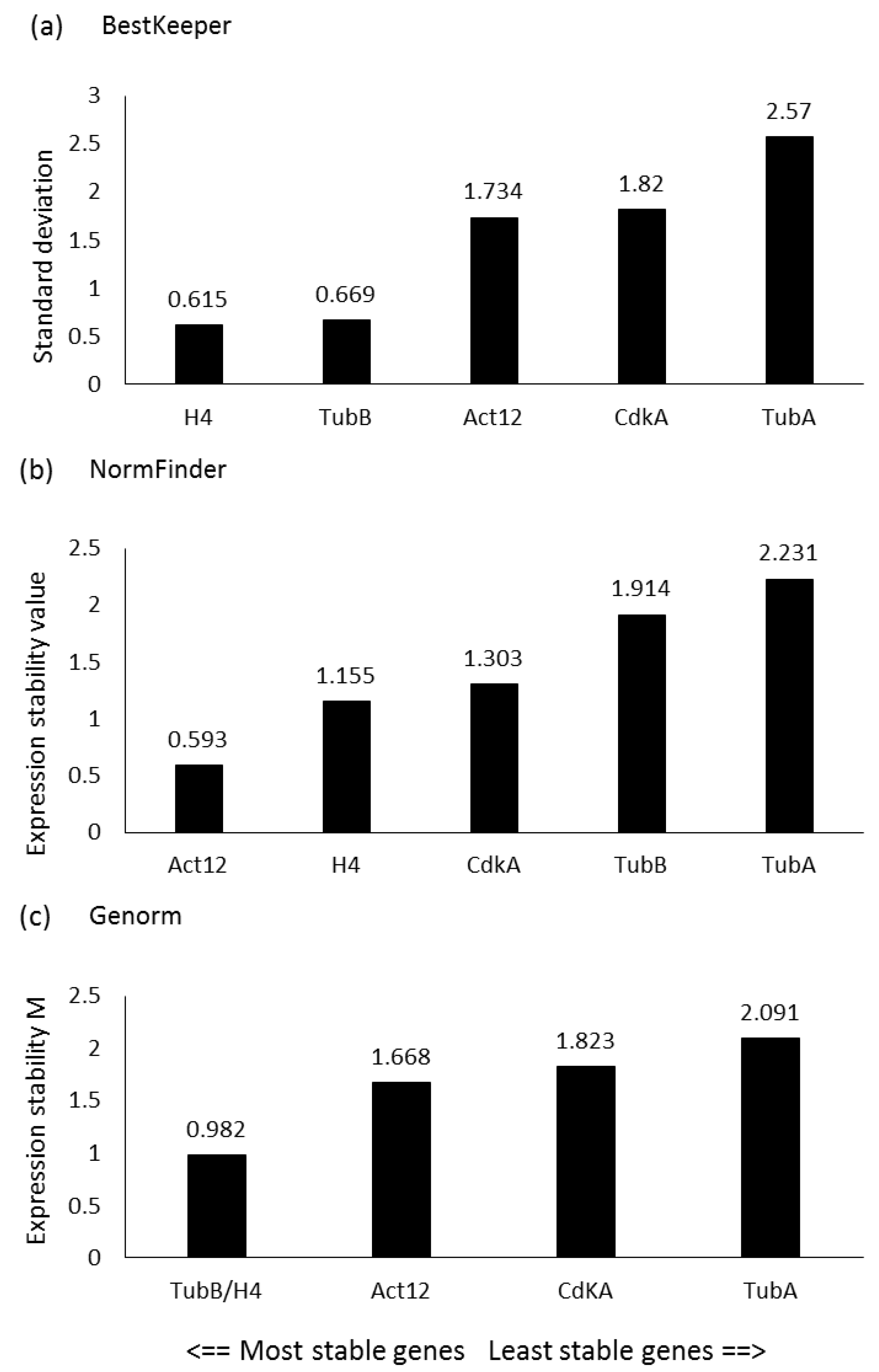
In order to study the expression levels of the genes of interest (GOIs), a panel of putative reference genes (RGs) was first screened in the experimental conditions presented in the current study, since RG stability can vary depending on the studied species, growth phases, nutrient culturing conditions and/or stressor exposures. Genes that were selected included those that had already been tested as RGs in the diatom P. tricornutum and/or used in similar studies for other diatoms [19,38,49,50]. In particular, according to the results obtained by BestKeeper, the lowest standard deviation (SD) was obtained for Histone 4 (H4), followed by β-Tubulin (TubB) and Actin 12 (Act12) (Figure 1a). According to NormFinder, the lowest stability values were observed for Act12, followed by H4 and cyclin-dependent kinase A (CdKA) (Figure 1b), while according to the statistical approach of geNorm, the two most stable genes (i.e., with the lowest expression stability, M) were TubB and H4 (Figure 1c). Considering the best RGs assigned by each software, Act12, TubB, and H4 were selected as RGs for the RT-qPCR analyses.
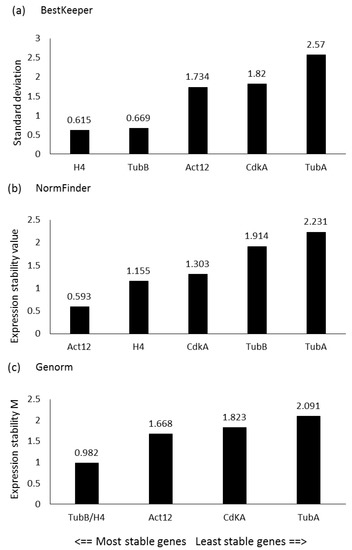
Figure 1.
Reference gene assessment for P. tricornutum. Arrows indicate the most stable reference genes according to (a) BestKeeper (lowest standard deviation), (b) NormFinder (lowest expression stability value), and (c) geNorm (lowest average expression stability M) softwares. The selected genes were: Actin 12 (Act12), cyclin-dependent kinase (Cdk), histone 4 (H4), α-tubulin (TubA) and β-tubulin (TubB).
2.2. Expression Levels of Genes Involved in the Synthesis of Sulfoquinovosyldiacylglycerols (SQDGs) and Monogalactosyldiacylglycerols (MGDGs)
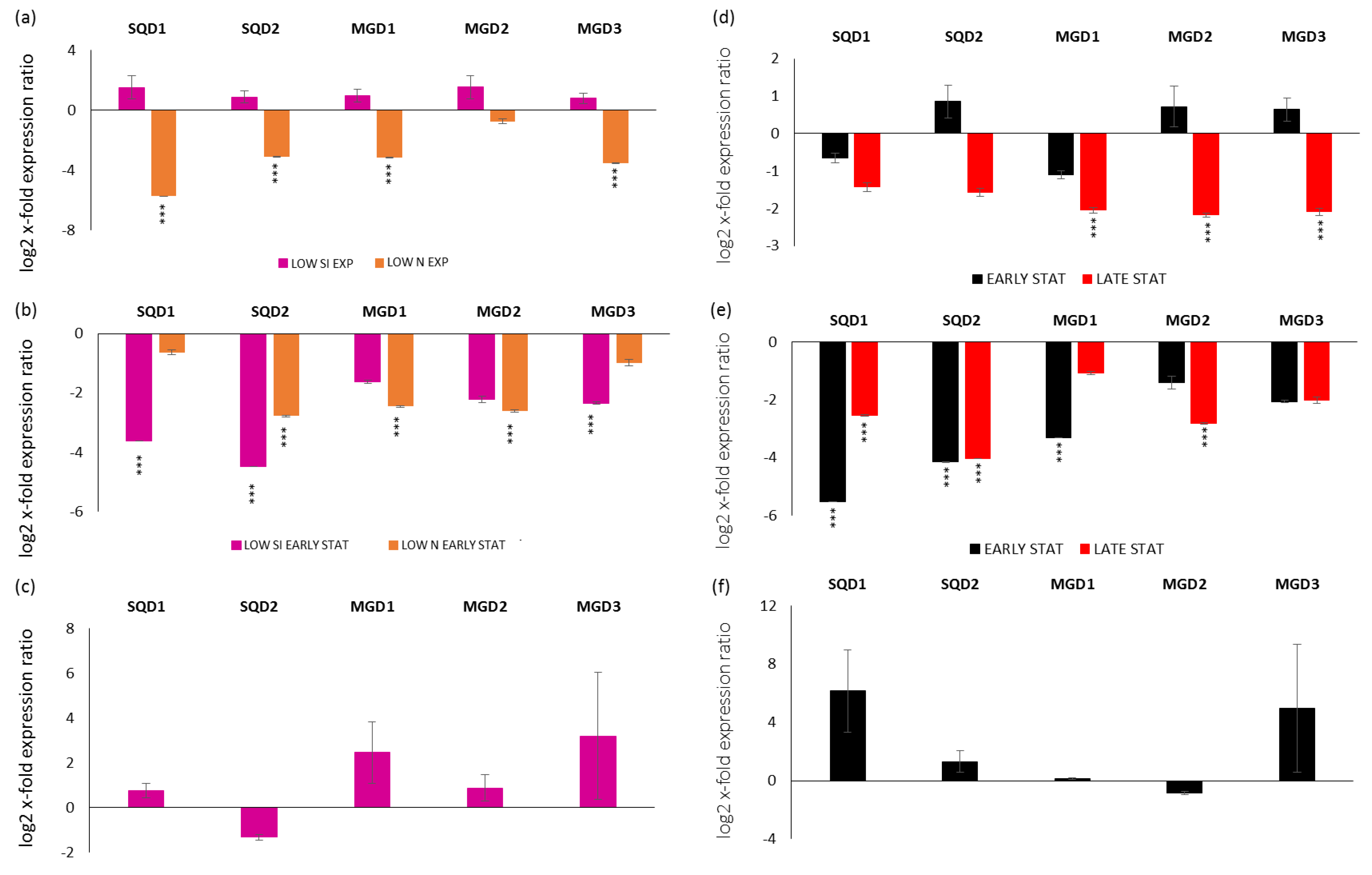
Gene expression analyses were performed also for transcripts coding enzymes involved in the synthesis of SQDGs (transcripts were named SQD1 and SQD2) and MGDGs (transcripts were named MGD1, MGD2 and MGD3). In particular, looking at the exponential phase in LOW SI and LOW N compared to the replete condition (x-axis), there was a down-regulation of SQD1, SQD2, MGD1 and MGD3 in the LOW N exponential condition (p < 0.001 for all; Figure 2a). Regarding the stationery phase, in the early case there was the down-regulation of SQD1 and MGD3 for the LOW SI condition, the down-regulation of MGD1 and MGD2 for the LOW N condition and the down-regulation of SQD2 for both LOW SI and LOW N conditions (p < 0.001 for all; Figure 2b). There were no significant changes for the late stationery condition (Figure 2c).
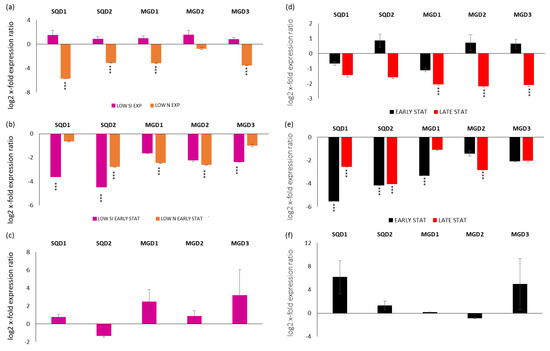
Figure 2.
Expression levels of genes involved in sulfoquinovosyldiacylglycerol and monogalactosyldiacylglycerol synthesis in P. tricornutum cells in (a) LOW SI and LOW N exponential phase compared to the control condition (Replete condition exponential phase; x-axis); (b) LOW SI and LOW N early stationery phase compared to the control condition (Replete condition early stationery phase); (c) LOW SI late stationery phase compared to the control condition (Replete condition late stationery phase); (d) Replete condition early stationery and late stationery compared to the control condition (Replete; x-axis) exponential phase; (e) LOW SI condition early stationery and late stationery compared to the LOW SI condition exponential phase; (f) LOW N early stationery phase compared to LOW N exponential phase. Data are represented as log2 x-fold expression ratio ± SD (n = 4; *** p < 0.001, Student’s t-test). Abbreviations stands for: UDP-sulfoquinovose synthase (SQD1), sulfoquinovosyldiacylglycerol synthase (SQD2), monogalactosyldiacylglycerol synthases 1, 2 and 3 (MGD1, MGD2 and MGD3, respectively).
Regarding their expression along the growth curves, for the replete condition, there was the down-regulation of the three MGD in the late stationery phase (Figure 2d; p < 0.001 for all). For the LOW SI culturing condition, there was the down-regulation of SQD1 and SQD2 in both early and late stationery growth phases, the down-regulation of MGD1 in the early stationery phase and MGD2 in the late stationery phase (p < 0.001 for all; Figure 2e). On the contrary, there were no significant differences in gene expression in the LOW N condition (p > 0.05 for all; Figure 2f).
2.3. Expression Levels of Stress-Responsive Genes in Silica and Nitrogen Starvation Versus Replete Condition
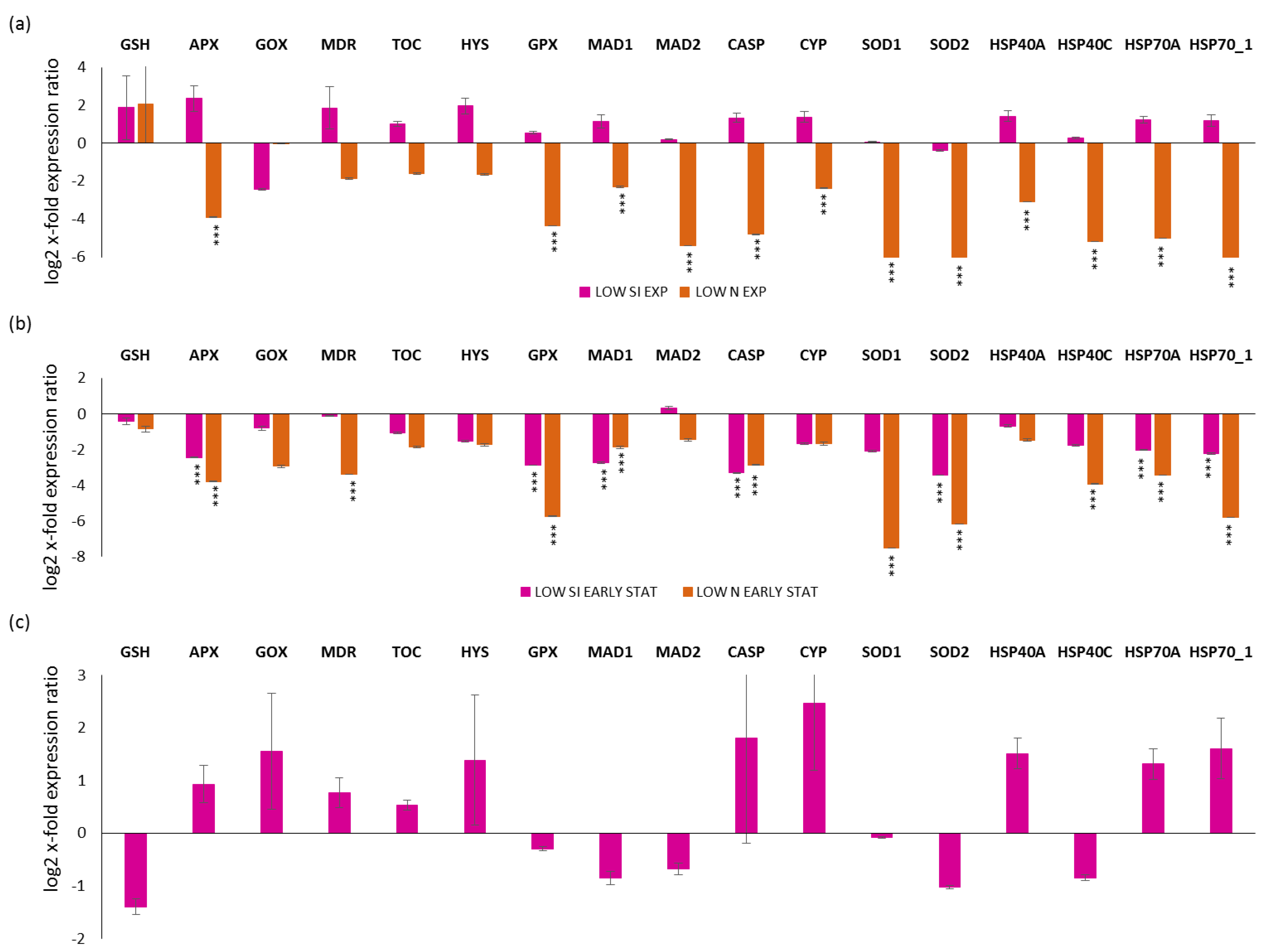
Regarding the selected stress-responsive genes, a first analysis was performed by comparing the exponential, early and late stationery growth phases in replete, LOW SI and LOW N conditions (for silica and nitrogen-starvation conditions, respectively; Figure 3). For the LOW N culturing condition, it was not possible to collect the late stationery condition, since the culture rapidly went into the decline phase. Looking at the exponential phase in LOW SI and LOW N compared to the replete condition (x-axis; Figure 3a), there was no statistical significant changes for LOW SI, while there was a significant down-regulation for APX, GPX, MAD1, MAD2, CASP, CYP, SOD1, SOD2, HSP40A, HSP40C, HSP70A, and HSP70_1 (p < 0.001 for all) for LOW N. Regarding the early stationery condition, there was the down-regulation of APX, GPX, MAD1, CASP, SOD2, HSP70A, and HSP70_1 for both LOW SI and LOW N (p < 0.001 for all) and the down-regulation of MDR, SOD1 and HSP40C only in the LOW N condition (p < 0.001 for all; Figure 3b). Regarding the late stationery analysis, there were no significant gene expression changes (p > 0.05 for all; Figure 3c).
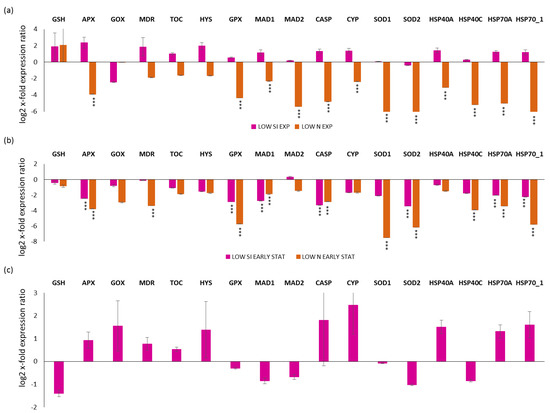
Figure 3.
Expression levels of selected stress responsive genes in P. tricornutum cells in (a) LOW SI and LOW N exponential phase compared to the control condition (Replete condition exponential phase; x-axis); (b) LOW SI and LOW N early stationery phase compared to the control condition (Replete condition early stationery phase); (c) LOW SI late stationery phase compared to the control condition (Replete condition late stationery phase). Data are represented as log2 x-fold expression ratio ± SD (n = 4; *** p < 0.001, Student’s t-test). Abbreviations stands for glutathione synthase (GSH), ascorbate peroxidase (APX), glycolate oxidase (GOX), multidrug resistance protein (MDR), tocopherol cyclase (TOC), hystone deacetylase (HYS), glutathione peroxidase (GPX), mitotic spindle assembly checkpoint proteins (MAD1 and MAD2), metacaspase (CASP), cytochrome P450 (CYP450), superoxide dismutase 1 and 2 (SOD1 and SOD2, respectively) and heat shock proteins 40A, 40C, 70.1, and 70A (HSP40A, HSP40C, HSP70_1, HSP70A).
2.4. Expression Levels of Stress-Responsive Genes along the Growth Curve
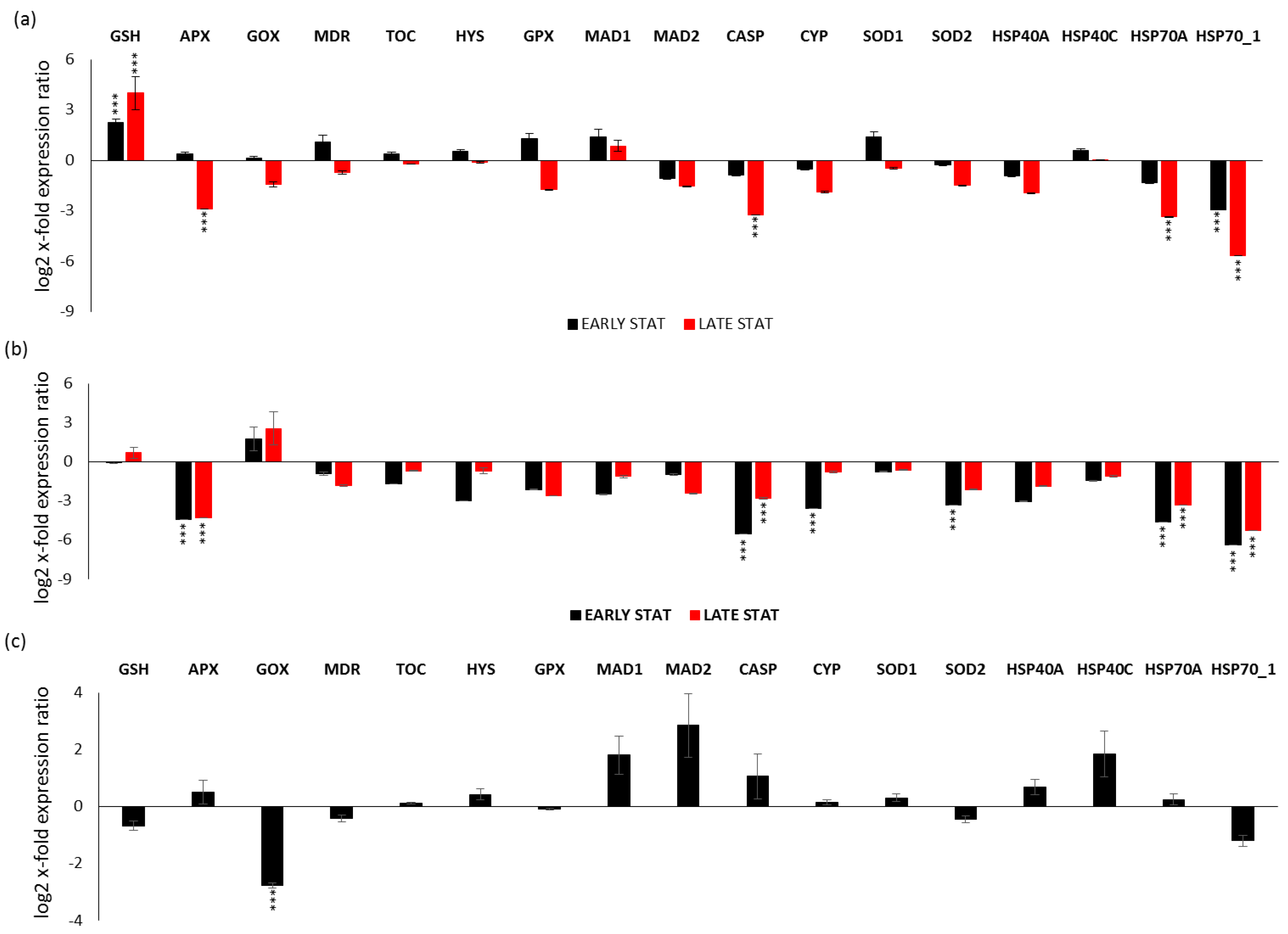
Gene expression analyses were also performed for each studied condition (i.e., replete, LOW SI, and LOW N during the growth curve) by comparing early stationery and late stationery (except for LOW N) versus the exponential phase (x-axis; Figure 4). Regarding the replete condition (Figure 4a), there was the up-regulation of the antioxidant enzyme GSH in both early and late stationery phase (p < 0.001 for both conditions), while the down-regulation of HSP70_1 in the early stationery phase and APX, CASP, HSP70A and HSP70_1 in the late stationery phase (p < 0.001 for all; Figure 4a). In the LOW SI condition (Figure 4b), there was the down-regulation of CYP and SOD2 only in the early stationery phase, while there was down-regulation of APX, CASP, HSP70A, and HSP70_1 both in the early and late stationery phase (p < 0.001 for all). Regarding the LOW N condition (Figure 4c), there was down-regulation only in the case of GOX (p < 0.001).
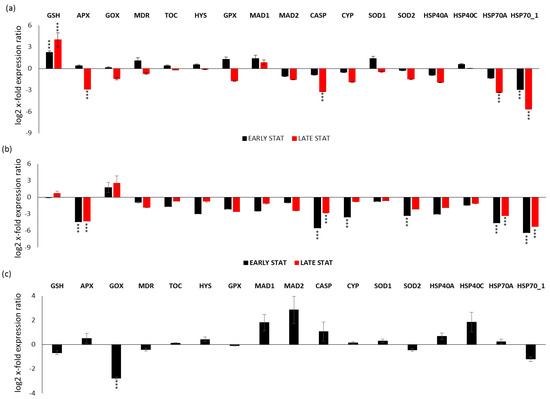
Figure 4.
Expression levels of selected stress responsive genes in P. tricornutum cells in: (a) Replete condition early stationery and late stationery compared to the control condition (Replete; x-axis) exponential phase; (b) LOW SI condition early stationery and late stationery compared to the LOW SI condition exponential phase; (c) LOW N early stationery phase compared to LOW N exponential phase. Data are represented as log2 x-fold expression ratio ± SD (n = 4; ** p < 0.01 and *** p < 0.001, Student’s t-test). Abbreviations stands for glutathione synthase (GSH), ascorbate peroxidase (APX), glycolate oxidase (GOX), multidrug resistance protein (MDR), tocopherol cyclase (TOC), hystone deacetylase (HYS), glutathione peroxidase (GPX), mitotic spindle assembly checkpoint proteins (MAD1 and MAD2), metacaspase (CASP), cytochrome P450 (CYP450), superoxide dismutase 1 and 2 (SOD1 and SOD2, respectively) and heat shock proteins 40A, 40C, 70.1, and 70A (HSP40A, HSP40C, HSP70_1, HSP70A).
3. Discussion
Microalgae are continuously exposed to various stressors in the natural environment, including both biotic and abiotic stressors. Upon exposure, cells react in order to cope with the stressor and survive, such as by activating antioxidant and xenobiotic-degrading enzymes, defense systems, and remodeling cellular metabolism [51,52]. In addition, in certain stressful conditions, it has been shown that microalgae may activate enzymatic pathways responsible of the synthesis of secondary metabolites with potential bioactivities for human health. This is the case of polyunsaturated aldehydes, shown to have anticancer properties [53], which are produced by the diatom Skeletonema marinoi at the end of the stationery phase (depending on nutrient stress exposure) [44]. However, this is not a rule. In fact, it was shown that the production of bioactive compounds is different depending on the species, clones, growth phases, and exposure to biotic and abiotic interferences [46,54,55], including predator exposure [26]. This phenomenon is known as the OSMAC approach [24]. Lauritano et al. [45] showed that specific microalgal clones were active for anti-inflammatory applications only when cultivated in control condition (replete medium (i.e., for the diatoms Cylindrotheca closterium, Odontella mobiliensis, Pseudonitzschia pseudodelicatissima)), while others were active only in stressful conditions (i.e., nutrient starvation), such as the diatom S. marinoi (which is active against human melanoma cells or pathogenic bacteria). Our data show that nutrient deficiencies (silica and nitrogen nutrient starvation or late stationery growth phase) induce the down-regulation of genes involved in SQDG and MGDG synthesis, as well as genes coding stress-responsive proteins (a schematic summary is reported in Figure 5). The regulation of SQDG and MGDG biosynthetic pathways may be of industrial interest since the final products possess anticancer and immunomodulatory activity. Abida end co-workers [29] analyzed the glycerolipid composition of P. tricornutum after five days of nitrogen starvation. The authors reported a decreased content of MGDG, while the content of SQDG remained almost unchanged. With regards to MGDG, our results are perfectly in line with Abida et al. On the contrary, we reported a down-regulation of the genes involved in SQDG synthesis in nitrogen starvation condition. Overall, these data suggest that for higher gene expression, and especially monogalactosyldiacylglycerol production, the best culturing condition to consider for possible biotechnological exploitation is the exponential phase of the replete culturing condition.
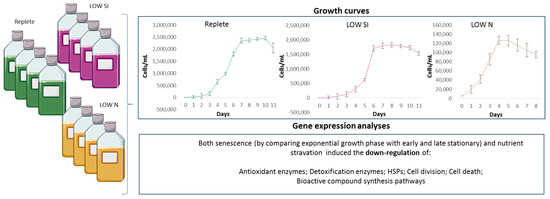
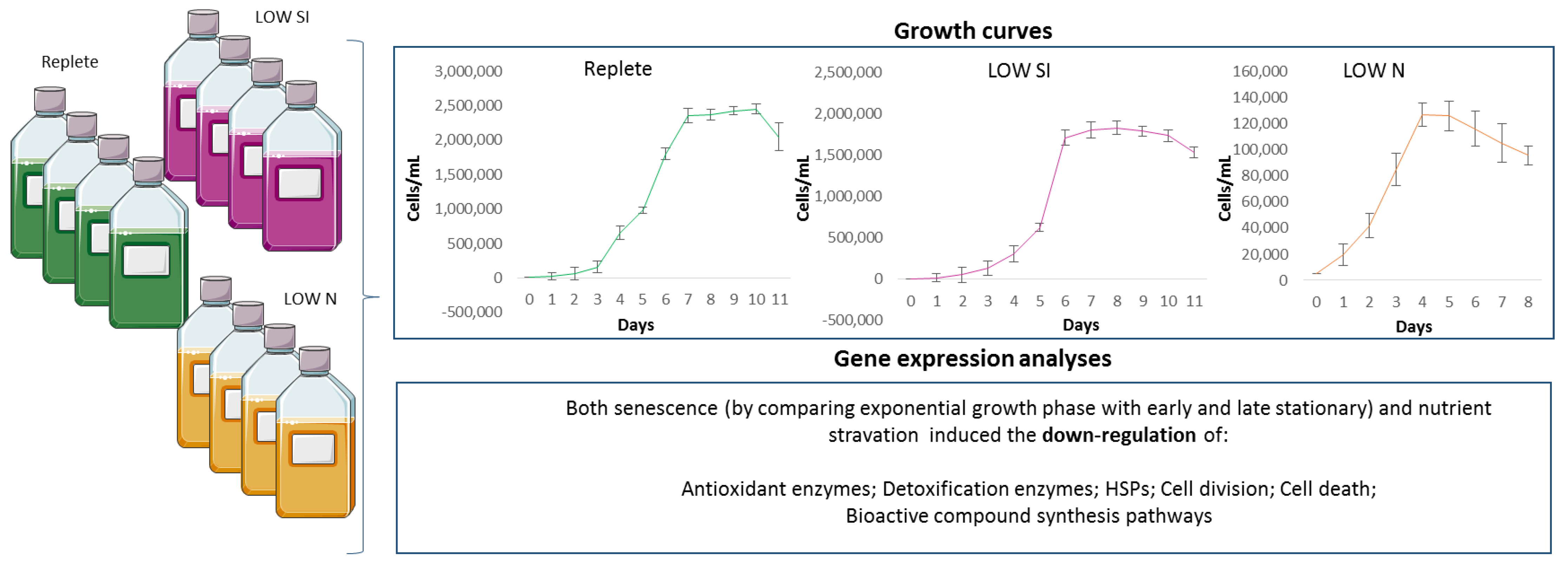
Figure 5.
Schematic representation of the main results. In the reported growth curves of the three culturing conditions (replete, LOW SI and LOW N conditions) error bars represent the standard deviation of biological triplicates.
Regarding already available molecular information for P. tricornutum, several studies reported transcriptomes, expressed sequence tags, arrays and gene-editing approaches. Recently, Ait-Mohamed et al. [56] studied 187 publicly available RNAseq datasets of P. tricornutum generated under various culturing conditions in terms of nitrogen, iron and phosphate. The aim was to understand the co-regulation underpinning transcription in P. tricornutum. They used WGCNA (Weighted Gene Correlation Network Analysis), identifying 28 modules of co-expressed genes. However, considering that the concentration of nitrogen/silica, culturing volumes, and general growth conditions are often different each other, it is difficult to compare them with our gene expression data.
Down-regulation has been observed upon senescence or nutrient starvation also for various genes related to cell death or stress response for other diatoms, such as Skeletonema marinoi [38,49]. Matthijs et al. [48] also analyzed P. tricornutum in nitrogen starvation finding a series of genes differentially expressed, but the experimental set up was different from the current study: the clone was different (P. tricornutum Pt1 Bohlin Strain 8.6), the exponential growth phase was studied, the culturing volume was 500 mL, and, for the stress experiment, the pre-cultured cells were diluted two-fold daily. Another study on P. tricornutum by Longworth and colleagues [30] was focused on the proteome in response to nitrogen depletion and, in particular, for the lipid accumulation response. For this experiment, the studied clone was P. tricornutum CCAP 1055/1, nitrogen was completely depleted, and our GOI were not investigated. Lipid accumulation is often studied for the economic potential of lipids for industrial applications and the most common finding from transcriptome analyses in nitrogen starvation is lipid accumulation [48,57,58]. In general, gene expression variations have been shown to be species/clones and conditions-dependent, and these variations have been suggested to explain the environmental response of diatoms to grow as blooms and to out-compete other classes of microalgae [59].
For instance, nitrogen deprivation was the more severe stress for P. tricornutum, inducing thylakoid senescence and growth arrest [29]. Di Dato et al. [60] showed how prostaglandin synthesis related gene expression varied depending on growth phases in the diatom Thalassiosira rotula or depending on different clones of the diatom Skeletonema marinoi [61]. Many of our GOI have been analyzed in several other marine algae and marine or terrestrial plants upon stress exposure [49,62,63,64,65]. Also, in this case regulation of gene expression is very species- and stress specific-dependent. Variations of expression of the same stress-responsive genes in different populations of the same species were also observed for the marine copepod Calanus helgolandicus and it was suggested that the population more used to cope to the stress exposure (i.e., in that experiment, it was the exposure to an alga producing compounds with teratogenic effects) were more promptly able to activate the stress-responsive genes [51].
Considering that also RGs may change depending on the species, clones, different conditions and stressful exposures, we had to find the best reference gene for our studied conditions. Transcripts coding genes with different functions were selected to avoid that they may be co-regulated. Our analyses identified RGs, Act12, TubB, and H4 as the best and most stable genes. TubB was the most stable gene, according to the geNorm software, also for the diatom Pseudo-nitzschia arenysensis along a growth curve experiment [66]. Similarly, H4 and Act were the most stable genes, according to the BestKeeper and geNorm softwares, along the growth curve of the diatom Skeletonema marinoi, and H4 was also between the best reference genes for silica depletion and starvation conditions, according to geNorm, while Act for a CO2-enriched experiment, according to geNorm and NormFinder softwares, in the same species [49].
The limitation of the current study is that, with gene expression analyses, it is only possible to observe what happens at transcription level. However, it is also important to highlight that other changes/regulations may occur at translational and post-translational levels. Considering that mRNA levels may not always reflect changes in protein levels/activities, further studies, such as protein and chemical analyses, can represent future directions for a better understanding of SQDG and MGDG production.
P. tricornutum is considered a model species since, in addition to the genome and various transcriptome sequenced [3,56,67,68], several engineering-modification techniques have been applied (e.g., TALEN (transcription activator-like effector nuclease) and CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats)) [6,69]. For instance, a study by McCharty et al. [8] generated a knockout in the nitrate reductase (NR) gene in P. tricornutum to monitor NR cellular localization in relation to N source and availability. As recently summarized by Butler et al. [70], P. tricornutum is routinely transformed by biolistic transformation, conjugation, and electroporation. In addition, it is considered a sustainable microalgal cell factory (for example, of eicosapentaenoic acid, fucoxanthin, neutral lipids, and crysolaminarin) by using green extraction processes and environment-friendly manufacturing solutions. Diatoms generally have faster growth compared to other systems, such as plants; they can be cultivated both in open ponds or controlled enclosed photobioreactors and can assure large biomass productivities. Considering that products obtained by P. tricornutum may find applications in different sectors, ranging from the pharmaceutical, nutraceutical, biodiesel, and bioplastic fields [70]. It represents a cell factory with potential for a wide spectrum of marketable products worth further investigations.
4. Materials and Methods
4.1. Cell culturing and Harvesting
The diatom P. tricornutum (RCC69) was cultured in Guillard’s F/2 medium [71]. Experimental culturing was performed in triplicate for both replete (complete medium), silica and nitrogen starvation (named LOW SI and LOW N, respectively). For nutrient starvation conditions, a low concentration of silicic acid (36 μM Si(OH)4) and nitrogen (30 μM of NO3−) were used. Culturing was performed in 2-litre polycarbonate bottles, constant bubbling with air filtered through 0.2 μm membrane filters in a climate chamber at 19 °C on a 12:12 h light:dark cycle and at 100 μmol photons m−2 s−1). Initial cell concentration in each bottle was 5000 cells/mL.
For each condition, cells were counted daily by sampling 2 mL of culture, fixing them with one drop of Lugol (final concentration of about 2%, v/v) and counting cell number in a Bürker counting chamber under an Axioskop 2 microscope (20×) (Carl Zeiss GmbH, Oberkochen, Germany) as previously described [72]. Growth curves for each condition are shown in Figure 5. Culture aliquots (50 mL) were sampled in quadruplicate in the exponential (Day 3 for LOW N, Day 4 for replete and LOW SI), early stationery (Day 4 for LOW N, Day 7 for replete and LOW SI) and late stationery (D10; only for replete and LOW SI) conditions from each bottle. Aliquots were centrifuged for 15 min at 4 °C at 1900× g (Eppendorf, 5810R, Hamburg, Germany). Pellets for each condition and growth stage were then re-suspended in 500 µL of TRIZOL© (Invitrogen, Carlsbad, CA, USA), incubated for 2–3 min at 60 °C until completely dissolved, and stored at −80 °C until RNA extraction.
4.2. Bioinformatic Search of Putative Enzymes of Interest Involved in SQDG and MGDG Synthesis
Genes encoding for the enzymes of interest were found on KEGG: Kyoto Encyclopedia of Genes and Genomes (accessed on 6 June 2022). P. tricornutum possesses a sequence encoding a UDP-sulfoquinovose synthase [EC:3.13.1.1] (SQD1) and a sequence encoding a sulfoquinovosyltransferase [EC:2.4.1.-] (SQD2), respectively at https://www.genome.jp/entry/pti:PHATR_21201 and https://www.genome.jp/entry/pti:PHATRDRAFT_50356 (accessed on 10 June 2022) These two genes are involved in sulfoquinovosyldiacylglycerol synthesis.
P. tricornutum has been found to possess 3 sequences encoding 1,2-diacylglycerol 3-beta-galactosyltransferases [EC:2.4.1.46]. These three sequences, reported as MGD1, MGD2 and MGD3, are respectively PHATRDRAFT_14125, PHATR_43938 and PHATRDRAFT_9619 on https://www.genome.jp/entry/pti:PHATRDRAFT_14125+pti:PHATRDRAFT_9619+pti:PHATR_43938 (accessed on 10 June 2022).
4.3. Selection and Characterization of Putative Reference Genes (RGs) and Other Genes of Interest (GOIs) and Primer Design
Reference genes (RGs) selection was carried out by testing a series of genes previously used as a reference for P. tricornutum [50] or other diatoms [38,66]. The selected genes were: Actin (Act12), cyclin-dependent kinase A (CdkA), histone 4 (H4), α-tubulin (TubA) and β-tubulin (TubB). The primers used for RGs are those reported in [50]. A series of genes related to stress/detoxification responses, cell division, cell death were selected as genes of interest, together to those involved in the synthesis of sulfoquinovosyldiacylglycerols and monogalactosyldiacylglycerols (reported in the paragraph 4.2). Primers were designed using the software Primer3 version 4.1.0 (http://primer3.ut.ee/; accessed on 10 January 2021). The size of the amplicons was kept in the range of 150 to 250 base pairs, GC content at 50%, the length of primers between 19 and 21 nucleotides, and melting temperature from 59 °C to 61 °C. Primers were synthesized by Sigma-Aldrich (St. Louis, MO, USA). PCR conditions were optimized on a C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA, USA). Supplementary Table S1 illustrates primer sequences for all of the selected genes as well as their abbreviation and full name, GenBank Accession Number, amplicon sizes and oligo efficiencies (E).
4.4. RNA Extraction and cDNA Synthesis
RNA extraction was performed as in [49]. Each sample was treated with DNase I (Invitrogen) using the instruction’s manual in order to remove residual DNA. The assessment of RNA quantity and quality was carried out monitoring the absorbance at 260 nm and the 260/280 nm and 260/230 nm ratios. Both ratios were approximately 2. RNA quality was also assessed on 1% agarose gels, showing intact 18S and 28S ribosomal bands. RNA samples (1 μg/each) were retrotranscribed into complementary DNA (cDNA) by using the iScript™ cDNA Synthesis Kit (Bio-Rad), as in [62], in the C1000 Touch Thermal Cycler (Bio-Rad).
4.5. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
The cDNA samples were then used for the RT-qPCR experiments performed in a Viia7 real-time PCR system (Applied Biosystems) at a 1:25 dilution. The PCR volume of each sample was 10 μL with 5 μL of 2× qPCR SYBR Green Master Mix, 0.7 pmol/μL for each primer and 1 μL of 1:10 diluted cDNA template. RT-qPCR experiments were performed as previously described [73]. The best reference genes were identified using three different algorithms, i.e., BestKeeper [74], geNorm [75] and NormFinder [76] by using RefFinder (http://blooge.cn/RefFinder/; accessed on 1 June 2022), a user-friendly web-based comprehensive tool developed for evaluating and screening reference genes [77]. To study expression levels for each gene of interest relative to the most stable RGs, we used the REST tool (Relative Expression Software Tool) [78].
4.6. Statistical Analysis
Statistical analysis (Student’s t-test) was performed by using GraphPad Prim Statistic Software, V4.00 (GraphPad Software; http://www.graphpad.com/quickcalcs/; accessed on 1 June 2022). Differences with p < 0.05 were regarded as significant.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/stresses2030022/s1. Table S1: Primer abbreviation, GenBank accession numbers, full names, sequences, amplicon sizes and oligo efficiencies (E) are provided for the genes of interest in P. tricornutum.
Author Contributions
Conceptualization, C.L.; methodology, G.R.; formal analysis, G.R. and C.L.; writing—original draft preparation, G.R. and C.L.; writing—review and editing, G.R. and C.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the project “Antitumor Drugs and Vaccines from the Sea (ADViSE)” (CUP B43D18000240007–SURF 17061BP000000011; PG/2018/0494374) funded by POR Campania FESR 2014–2020 “Technology Platform for Therapeutic Strategies against Cancer”-Action 1.1.2 and 1.2.2.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Gennaro Riccio was supported by a research grant within the project “Antitumor Drugs and Vaccines from the Sea (ADViSE)” (CUP B43D18000240007–SURF 17061BP000000011; PG/2018/0494374) funded by POR Campania FESR 2014–2020 “Technology Platform for Thera-peutic Strategies against Cancer”-Action 1.1.2 and 1.2.2. Authors thank Servier Medical Art (SMART) website (https://smart.servier.com/; accessed on 10 June 2022) by Servier for the bottle elements of Figure 5.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ovide, C.; Kiefer-Meyer, M.-C.; Bérard, C.; Vergne, N.; Lecroq, T.; Plasson, C.; Burel, C.; Bernard, S.; Driouich, A.; Lerouge, P.; et al. Comparative in Depth RNA Sequencing of P. tricornutum’s Morphotypes Reveals Specific Features of the Oval Morphotype. Sci. Rep. 2018, 8, 14340. [Google Scholar] [CrossRef] [PubMed]
- Zaslavskaia, L.A.; Lippmeier, J.C.; Shih, C.; Ehrhardt, D.; Grossman, A.R.; Apt, K.E. Trophic Conversion of an Obligate Photoautotrophic Organism through Metabolic Engineering. Science 2001, 292, 2073–2075. [Google Scholar] [CrossRef]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum Genome Reveals the Evolutionary History of Diatom Genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef]
- Daboussi, F.; Leduc, S.; Maréchal, A.; Dubois, G.; Guyot, V.; Perez-Michaut, C.; Amato, A.; Falciatore, A.; Juillerat, A.; Beurdeley, M.; et al. Genome Engineering Empowers the Diatom Phaeodactylum tricornutum for Biotechnology. Nat. Commun. 2014, 5, 3831. [Google Scholar] [CrossRef]
- Weyman, P.D.; Beeri, K.; Lefebvre, S.C.; Rivera, J.; McCarthy, J.K.; Heuberger, A.L.; Peers, G.; Allen, A.E.; Dupont, C.L. Inactivation of Phaeodactylum tricornutum Urease Gene Using Transcription Activator-like Effector Nuclease-Based Targeted Mutagenesis. Plant Biotechnol. J. 2015, 13, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Stukenberg, D.; Zauner, S.; Dell’Aquila, G.; Maier, U.G. Optimizing CRISPR/Cas9 for the Diatom Phaeodactylum tricornutum. Front. Plant Sci. 2018, 9, 740. [Google Scholar] [CrossRef]
- Serif, M.; Dubois, G.; Finoux, A.-L.; Teste, M.-A.; Jallet, D.; Daboussi, F. One-Step Generation of Multiple Gene Knock-Outs in the Diatom Phaeodactylum tricornutum by DNA-Free Genome Editing. Nat. Commun. 2018, 9, 3924. [Google Scholar] [CrossRef]
- McCarthy, J.K.; Smith, S.R.; McCrow, J.P.; Tan, M.; Zheng, H.; Beeri, K.; Roth, R.; Lichtle, C.; Goodenough, U.; Bowler, C.P.; et al. Nitrate Reductase Knockout Uncouples Nitrate Transport from Nitrate Assimilation and Drives Repartitioning of Carbon Flux in a Model Pennate Diatom. Plant Cell 2017, 29, 2047–2070. [Google Scholar] [CrossRef] [PubMed]
- Nymark, M.; Sharma, A.K.; Hafskjold, M.C.G.; Sparstad, T.; Bones, A.M.; Winge, P. CRISPR/Cas9 Gene Editing in the Marine Diatom Phaeodactylum tricornutum. Bio-Protoc. 2017, 7, e2442. [Google Scholar] [CrossRef]
- Hempel, F.; Maurer, M.; Brockmann, B.; Mayer, C.; Biedenkopf, N.; Kelterbaum, A.; Becker, S.; Maier, U.G. From Hybridomas to a Robust Microalgal-Based Production Platform: Molecular Design of a Diatom Secreting Monoclonal Antibodies Directed against the Marburg Virus Nucleoprotein. Microb. Cell Factories 2017, 16, 131. [Google Scholar] [CrossRef] [PubMed]
- D’Adamo, S.; di Visconte, G.S.; Lowe, G.; Szaub-Newton, J.; Beacham, T.; Landels, A.; Allen, M.J.; Spicer, A.; Matthijs, M. Engineering the Unicellular Alga Phaeodactylum tricornutum for High-Value Plant Triterpenoid Production. Plant Biotechnol. J. 2019, 17, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, M.L.; Warwick, J.; Terry, A.; Allen, M.J.; Napier, J.A.; Sayanova, O. Towards the Industrial Production of Omega-3 Long Chain Polyunsaturated Fatty Acids from a Genetically Modified Diatom Phaeodactylum tricornutum. PLoS ONE 2015, 10, e0144054. [Google Scholar] [CrossRef] [PubMed]
- Samarakoon, K.W.; Ko, J.-Y.; Lee, J.-H.; Kwon, O.-N.; Kim, S.-W.; Jeon, Y.-J. Apoptotic Anticancer Activity of a Novel Fatty Alcohol Ester Isolated from Cultured Marine Diatom, Phaeodactylum tricornutum. J. Funct. Foods 2014, 6, 231–240. [Google Scholar] [CrossRef]
- Andrianasolo, E.H.; Haramaty, L.; Vardi, A.; White, E.; Lutz, R.; Falkowski, P. Apoptosis-Inducing Galactolipids from a Cultured Marine Diatom, Phaeodactylum tricornutum. J. Nat. Prod. 2008, 71, 1197–1201. [Google Scholar] [CrossRef]
- Kim, S.M.; Jung, Y.-J.; Kwon, O.-N.; Cha, K.H.; Um, B.-H.; Chung, D.; Pan, C.-H. A Potential Commercial Source of Fucoxanthin Extracted from the Microalga Phaeodactylum tricornutum. Appl. Biochem. Biotechnol. 2012, 166, 1843–1855. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.-H.; Shin, H.-Y.; Park, J.-H.; Koo, S.Y.; Kim, S.M.; Yang, S.-H. Fucoxanthin from Microalgae Phaeodactylum tricornutum Inhibits Pro-Inflammatory Cytokines by Regulating Both NF-ΚB and NLRP3 Inflammasome Activation. Sci. Rep. 2021, 11, 543. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wan, H.; Wang, R.; Hao, D. Sulfated Polysaccharides from Phaeodactylum tricornutum: Isolation, Structural Characteristics, and Inhibiting HepG2 Growth Activity In Vitro. PeerJ 2019, 7, e6409. [Google Scholar] [CrossRef] [PubMed]
- Vingiani, G.M.; De Luca, P.; Ianora, A.; Dobson, A.D.W.; Lauritano, C. Microalgal Enzymes with Biotechnological Applications. Mar. Drugs 2019, 17, 459. [Google Scholar] [CrossRef] [PubMed]
- Elagoz, A.M.; Ambrosino, L.; Lauritano, C. De Novo Transcriptome of the Diatom Cylindrotheca closterium Identifies Genes Involved in the Metabolism of Anti-Inflammatory Compounds. Sci. Rep. 2020, 10, 4138. [Google Scholar] [CrossRef]
- Vingiani, G.M.; Štālberga, D.; De Luca, P.; Ianora, A.; De Luca, D.; Lauritano, C. De Novo Transcriptome of the Non-Saxitoxin Producing Alexandrium Tamutum Reveals New Insights on Harmful Dinoflagellates. Mar. Drugs 2020, 18, 386. [Google Scholar] [CrossRef]
- Di Dato, V.; Di Costanzo, F.; Barbarinaldi, R.; Perna, A.; Ianora, A.; Romano, G. Unveiling the Presence of Biosynthetic Pathways for Bioactive Compounds in the Thalassiosira rotula Transcriptome. Sci. Rep. 2019, 9, 9893. [Google Scholar] [CrossRef]
- Riccio, G.; Martinez, K.A.; Ianora, A.; Lauritano, C. De Novo Transcriptome of the Flagellate Isochrysis galbana Identifies Genes Involved in the Metabolism of Antiproliferative Metabolites. Biology 2022, 11, 771. [Google Scholar] [CrossRef] [PubMed]
- De Luca, D.; Lauritano, C. In Silico Identification of Type III PKS Chalcone and Stilbene Synthase Homologs in Marine Photosynthetic Organisms. Biology 2020, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Jackson, S.; Patry, S.; Dobson, A. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Hemphill, C.F.P.; Sureechatchaiyan, P.; Kassack, M.U.; Orfali, R.S.; Lin, W.; Daletos, G.; Proksch, P. OSMAC Approach Leads to New Fusarielin Metabolites from Fusarium tricinctum. J. Antibiot. 2017, 70, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Osvik, R.D.; Ingebrigtsen, R.A.; Norrbin, M.F.; Andersen, J.H.; Eilertsen, H.C.; Hansen, E.H. Adding Zooplankton to the OSMAC Toolkit: Effect of Grazing Stress on the Metabolic Profile and Bioactivity of a Diatom. Mar. Drugs 2021, 19, 87. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Song, H.; Lavoie, M.; Zhu, K.; Su, Y.; Ye, H.; Chen, S.; Fu, Z.; Qian, H. Proteomic Analyses Bring New Insights into the Effect of a Dark Stress on Lipid Biosynthesis in Phaeodactylum tricornutum. Sci. Rep. 2016, 6, 25494. [Google Scholar] [CrossRef]
- Levitan, O.; Dinamarca, J.; Zelzion, E.; Lun, D.S.; Guerra, L.T.; Kim, M.K.; Kim, J.; Van Mooy, B.A.S.; Bhattacharya, D.; Falkowski, P.G. Remodeling of Intermediate Metabolism in the Diatom Phaeodactylum tricornutum under Nitrogen Stress. Proc. Natl. Acad. Sci. USA 2015, 112, 412–417. [Google Scholar] [CrossRef]
- Abida, H.; Dolch, L.-J.; Meï, C.; Villanova, V.; Conte, M.; Block, M.A.; Finazzi, G.; Bastien, O.; Tirichine, L.; Bowler, C.; et al. Membrane Glycerolipid Remodeling Triggered by Nitrogen and Phosphorus Starvation in Phaeodactylum tricornutum. Plant Physiol. 2015, 167, 118–136. [Google Scholar] [CrossRef]
- Longworth, J.; Wu, D.; Huete-Ortega, M.; Wright, P.C.; Vaidyanathan, S. Proteome Response of Phaeodactylum tricornutum, during Lipid Accumulation Induced by Nitrogen Depletion. Algal Res. 2016, 18, 213–224. [Google Scholar] [CrossRef]
- Alipanah, L.; Rohloff, J.; Winge, P.; Bones, A.M.; Brembu, T. Whole-Cell Response to Nitrogen Deprivation in the Diatom Phaeodactylum tricornutum. J. Exp. Bot. 2015, 66, 6281–6296. [Google Scholar] [CrossRef] [PubMed]
- Riccio, G.; De Luca, D.; Lauritano, C. Monogalactosyldiacylglycerol and Sulfolipid Synthesis in Microalgae. Mar. Drugs 2020, 18, 237. [Google Scholar] [CrossRef] [PubMed]
- van Straalen, N.M.; Roelofs, D. An Introduction to Ecological Genomics; Oxford University Press: Oxford, NY, USA, 2011. [Google Scholar]
- Feder, M.E.; Hofmann, G.E. Heat-Shock Proteins, Molecular Chaperones, and the Stress Response: Evolutionary and Ecological Physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef]
- Forman, H.J.; Zhang, H.; Rinna, A. Glutathione: Overview of Its Protective Roles, Measurement, and Biosynthesis. Mol. Aspects Med. 2009, 30, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.-L.; Lu, Y.; Li, Y.; Yang, C.; Peng, X.-X. Overexpression of Glycolate Oxidase Confers Improved Photosynthesis under High Light and High Temperature in Rice. Front. Plant Sci. 2016, 7, 1165. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Cheng, K.; Xu, Y.; Yang, S.; Wu, K. Plant Responses to Abiotic Stress Regulated by Histone Deacetylases. Front. Plant Sci. 2017, 8, 2147. [Google Scholar] [CrossRef]
- Orefice, I.; Lauritano, C.; Procaccini, G.; Ianora, A.; Romano, G. Insights into Possible Cell-Death Markers in the Diatom Skeletonema marinoi in Response to Senescence and Silica Starvation. Mar. Genom. 2015, 24, 81–88. [Google Scholar] [CrossRef]
- Lauritano, C.; Procaccini, G.; Ianora, A. Gene Expression Patterns and Stress Response in Marine Copepods. Mar. Environ. Res. 2012, 76, 22–31. [Google Scholar] [CrossRef]
- Lauritano, C.; Carotenuto, Y.; Roncalli, V. Glutathione S-Transferases in Marine Copepods. J. Mar. Sci. Eng. 2021, 9, 1025. [Google Scholar] [CrossRef]
- Tunquist, B.J.; Eyers, P.A.; Chen, L.G.; Lewellyn, A.L.; Maller, J.L. Spindle Checkpoint Proteins Mad1 and Mad2 Are Required for Cytostatic Factor–Mediated Metaphase Arrest. J. Cell Biol. 2003, 163, 1231–1242. [Google Scholar] [CrossRef]
- Ouyang, S.; He, S.; Liu, P.; Zhang, W.; Zhang, J.; Chen, S. The Role of Tocopherol Cyclase in Salt Stress Tolerance of Rice (Oryza Sativa). Sci. China Life Sci. 2011, 54, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Caverzan, A.; Passaia, G.; Rosa, S.B.; Ribeiro, C.W.; Lazzarotto, F.; Margis-Pinheiro, M. Plant Responses to Stresses: Role of Ascorbate Peroxidase in the Antioxidant Protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Ribalet, F.; Wichard, T.; Pohnert, G.; Ianora, A.; Miralto, A.; Casotti, R. Age and Nutrient Limitation Enhance Polyunsaturated Aldehyde Production in Marine Diatoms. Phytochemistry 2007, 68, 2059–2067. [Google Scholar] [CrossRef] [PubMed]
- Lauritano, C.; Andersen, J.H.; Hansen, E.; Albrigtsen, M.; Escalera, L.; Esposito, F.; Helland, K.; Hanssen, K.Ø.; Romano, G.; Ianora, A. Bioactivity Screening of Microalgae for Antioxidant, Anti-Inflammatory, Anticancer, Anti-Diabetes, and Antibacterial Activities. Front. Mar. Sci. 2016, 3, 68. [Google Scholar] [CrossRef]
- Ingebrigtsen, R.A.; Hansen, E.; Andersen, J.H.; Eilertsen, H.C. Light and Temperature Effects on Bioactivity in Diatoms. J. Appl. Phycol. 2016, 28, 939–950. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Wei, D. Improving Fucoxanthin Production in Mixotrophic Culture of Marine Diatom Phaeodactylum tricornutum by LED Light Shift and Nitrogen Supplementation. Front. Bioeng. Biotechnol. 2020, 8, 820. [Google Scholar] [CrossRef]
- Matthijs, M.; Fabris, M.; Broos, S.; Vyverman, W.; Goossens, A. Profiling of the Early Nitrogen Stress Response in the Diatom Phaeodactylum tricornutum Reveals a Novel Family of RING-Domain Transcription Factors. Plant Physiol. 2016, 170, 489–498. [Google Scholar] [CrossRef]
- Lauritano, C.; Orefice, I.; Procaccini, G.; Romano, G.; Ianora, A. Key Genes as Stress Indicators in the Ubiquitous Diatom Skeletonema marinoi. BMC Genom. 2015, 16, 411. [Google Scholar] [CrossRef]
- Siaut, M.; Heijde, M.; Mangogna, M.; Montsant, A.; Coesel, S.; Allen, A.; Manfredonia, A.; Falciatore, A.; Bowler, C. Molecular Toolbox for Studying Diatom Biology in Phaeodactylum tricornutum. Gene 2008, 406, 23–35. [Google Scholar] [CrossRef]
- Lauritano, C.; Carotenuto, Y.; Miralto, A.; Procaccini, G.; Ianora, A. Copepod Population-Specific Response to a Toxic Diatom Diet. PLoS ONE 2012, 7, e47262. [Google Scholar] [CrossRef]
- Vingiani, G.M.; Leone, S.; De Luca, D.; Borra, M.; Dobson, A.D.W.; Ianora, A.; De Luca, P.; Lauritano, C. First Identification and Characterization of Detoxifying Plastic-Degrading DBP Hydrolases in the Marine Diatom Cylindrotheca closterium. Sci. Total Environ. 2022, 812, 152535. [Google Scholar] [CrossRef] [PubMed]
- Miralto, A.; Barone, G.; Romano, G.; Poulet, S.A.; Ianora, A.; Russo, G.L.; Buttino, I.; Mazzarella, G.; Laabir, M.; Cabrini, M.; et al. The Insidious Effect of Diatoms on Copepod Reproduction. Nature 1999, 402, 173–176. [Google Scholar] [CrossRef]
- Levasseur, W.; Perré, P.; Pozzobon, V. A Review of High Value-Added Molecules Production by Microalgae in Light of the Classification. Biotechnol. Adv. 2020, 41, 107545. [Google Scholar] [CrossRef]
- Gerecht, A.; Romano, G.; Ianora, A.; d’Ippolito, G.; Cutignano, A.; Fontana, A. Plasticity Of Oxylipin Metabolism Among Clones Of The Marine Diatom Skeletonema marinoi (Bacillariophyceae). J. Phycol. 2011, 47, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Ait-Mohamed, O.; Vanclová, A.M.G.N.; Joli, N.; Liang, Y.; Zhao, X.; Genovesio, A.; Tirichine, L.; Bowler, C.; Dorrell, R.G. PhaeoNet: A Holistic RNAseq-Based Portrait of Transcriptional Coordination in the Model Diatom Phaeodactylum tricornutum. Front. Plant Sci. 2020, 11, 590949. [Google Scholar] [CrossRef]
- Hu, Q.; Sommerfeld, M.; Jarvis, E.; Ghirardi, M.; Posewitz, M.; Seibert, M.; Darzins, A. Microalgal Triacylglycerols as Feedstocks for Biofuel Production: Perspectives and Advances. Plant J. Cell Mol. Biol. 2008, 54, 621–639. [Google Scholar] [CrossRef]
- Lv, H.; Qu, G.; Qi, X.; Lu, L.; Tian, C.; Ma, Y. Transcriptome Analysis of Chlamydomonas reinhardtii during the Process of Lipid Accumulation. Genomics 2013, 101, 229–237. [Google Scholar] [CrossRef]
- Shrestha, R.P.; Tesson, B.; Norden-Krichmar, T.; Federowicz, S.; Hildebrand, M.; Allen, A.E. Whole Transcriptome Analysis of the Silicon Response of the Diatom Thalassiosira pseudonana. BMC Genom. 2012, 13, 499. [Google Scholar] [CrossRef]
- Di Dato, V.; Barbarinaldi, R.; Amato, A.; Di Costanzo, F.; Fontanarosa, C.; Perna, A.; Amoresano, A.; Esposito, F.; Cutignano, A.; Ianora, A.; et al. Variation in Prostaglandin Metabolism during Growth of the Diatom Thalassiosira rotula. Sci. Rep. 2020, 10, 5374. [Google Scholar] [CrossRef]
- Di Dato, V.; Orefice, I.; Amato, A.; Fontanarosa, C.; Amoresano, A.; Cutignano, A.; Ianora, A.; Romano, G. Animal-like Prostaglandins in Marine Microalgae. Isme J. 2017, 11, 1722. [Google Scholar] [CrossRef]
- Lauritano, C.; Ruocco, M.; Dattolo, E.; Buia, M.C.; Silva, J.; Santos, R.; Olivé, I.; Costa, M.M.; Procaccini, G. Response of Key Stress-Related Genes of the Seagrass Posidonia oceanica in the Vicinity of Submarine Volcanic Vents. Biogeosciences 2015, 12, 4185–4194. [Google Scholar] [CrossRef]
- Tutar, O.; Marín-Guirao, L.; Ruiz, J.M.; Procaccini, G. Antioxidant Response to Heat Stress in Seagrasses. A Gene Expression Study. Mar. Environ. Res. 2017, 132, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Brumbarova, T.; Ivanov, R. The Nutrient Response Transcriptional Regulome of Arabidopsis. iScience 2019, 19, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Nazir, M.; Pandey, R.; Siddiqi, T.O.; Ibrahim, M.M.; Qureshi, M.I.; Abraham, G.; Vengavasi, K.; Ahmad, A. Nitrogen-Deficiency Stress Induces Protein Expression Differentially in Low-N Tolerant and Low-N Sensitive Maize Genotypes. Front. Plant Sci. 2016, 7, 298. [Google Scholar] [CrossRef] [PubMed]
- Adelfi, M.G.; Borra, M.; Sanges, R.; Montresor, M.; Fontana, A.; Ferrante, M.I. Selection and Validation of Reference Genes for QPCR Analysis in the Pennate Diatoms Pseudo-Nitzschia multistriata and P. arenysensis. J. Exp. Mar. Biol. Ecol. 2014, 451, 74–81. [Google Scholar] [CrossRef]
- Montsant, A.; Jabbari, K.; Maheswari, U.; Bowler, C. Comparative Genomics of the Pennate Diatom Phaeodactylum tricornutum. Plant Physiol. 2005, 137, 500–513. [Google Scholar] [CrossRef]
- Rastogi, A.; Maheswari, U.; Dorrell, R.G.; Vieira, F.R.J.; Maumus, F.; Kustka, A.; McCarthy, J.; Allen, A.E.; Kersey, P.; Bowler, C.; et al. Integrative Analysis of Large Scale Transcriptome Data Draws a Comprehensive Landscape of Phaeodactylum tricornutum Genome and Evolutionary Origin of Diatoms. Sci. Rep. 2018, 8, 4834. [Google Scholar] [CrossRef]
- Serif, M.; Lepetit, B.; Weißert, K.; Kroth, P.G.; Rio Bartulos, C. A Fast and Reliable Strategy to Generate TALEN-Mediated Gene Knockouts in the Diatom Phaeodactylum tricornutum. Algal Res. 2017, 23, 186–195. [Google Scholar] [CrossRef]
- Butler, T.; Kapoore, R.V.; Vaidyanathan, S. Phaeodactylum tricornutum: A Diatom Cell Factory. Trends Biotechnol. 2020, 38, 606–622. [Google Scholar] [CrossRef]
- Guillard, R.R.L. Culture of Phytoplankton for Feeding Marine Invertebrates. In Culture of Marine Invertebrate Animals; Smith, W.L., Chanley, M.H., Eds.; Plenum Press: New York, NY, USA, 1975; pp. 29–60. [Google Scholar]
- Lauritano, C.; De Luca, D.; Amoroso, M.; Benfatto, S.; Maestri, S.; Racioppi, C.; Esposito, F.; Ianora, A. New Molecular Insights on the Response of the Green Alga Tetraselmis Suecica to Nitrogen Starvation. Sci. Rep. 2019, 9, 3336. [Google Scholar] [CrossRef]
- Lauritano, C.; Carotenuto, Y.; Vitiello, V.; Buttino, I.; Romano, G.; Hwang, J.-S.; Ianora, A. Effects of the Oxylipin-Producing Diatom Skeletonema marinoi on Gene Expression Levels of the Calanoid Copepod Calanus sinicus. Mar. Genom. 2015, 24, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper—Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Roy, N.; De Paepe, A. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome. Biol. 2002, 3, research0034. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. MiRDeepFinder: A MiRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative Expression Software Tool (REST©) for Group-Wise Comparison and Statistical Analysis of Relative Expression Results in Real-Time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).