Physiological Responses to Drought, Salinity, and Heat Stress in Plants: A Review
Abstract
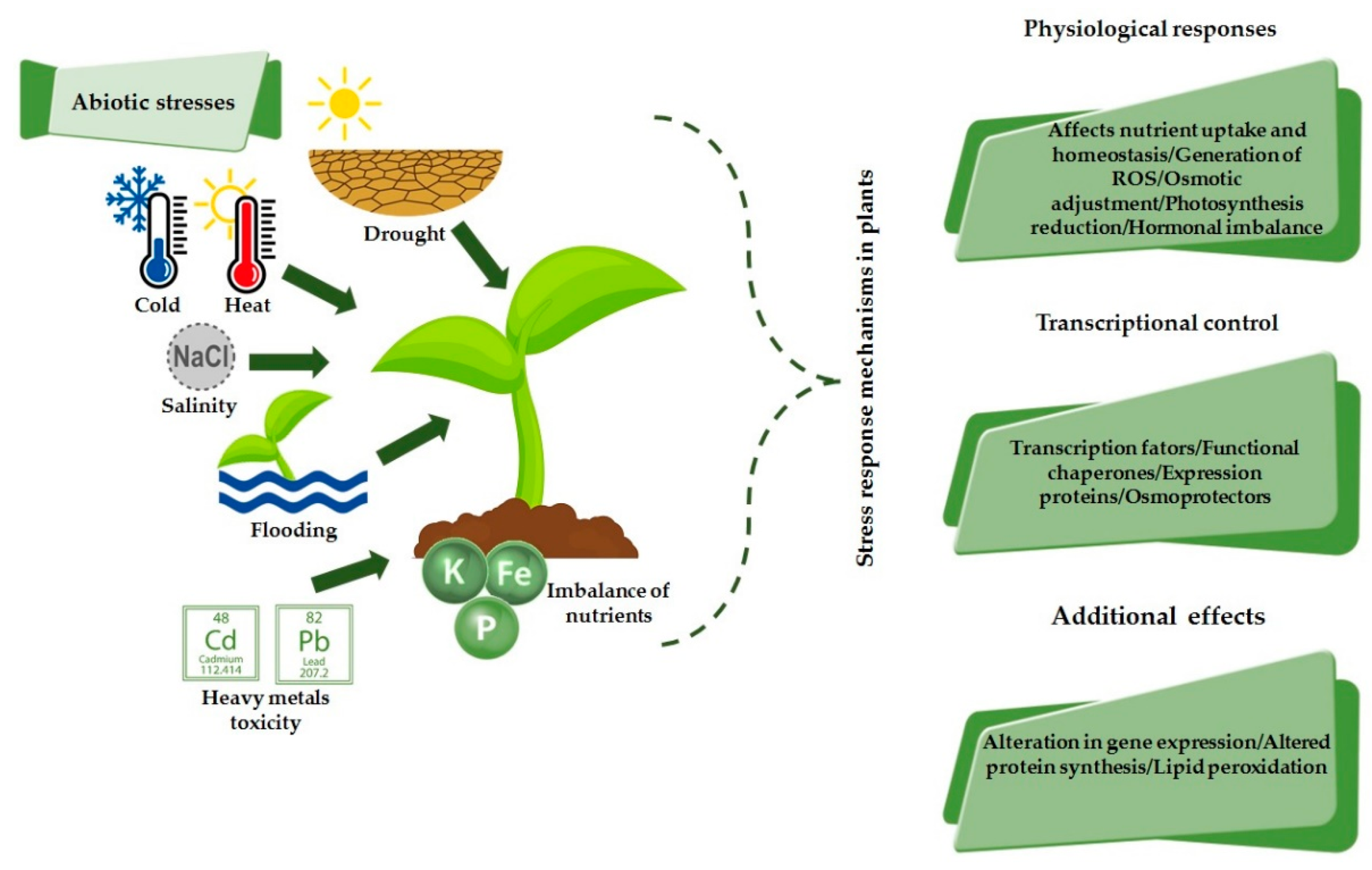
1. Introduction
2. Response Mechanism to Abiotic Stresses in Plants
3. Water Deficit in Plants
3.1. Strategies to Combat the Water Deficit
3.2. Physiological Strategies for Increasing Productivity under Water Deficit Conditions
3.3. Difficulties and Advances in the Development of Drought-Tolerant Cultivars
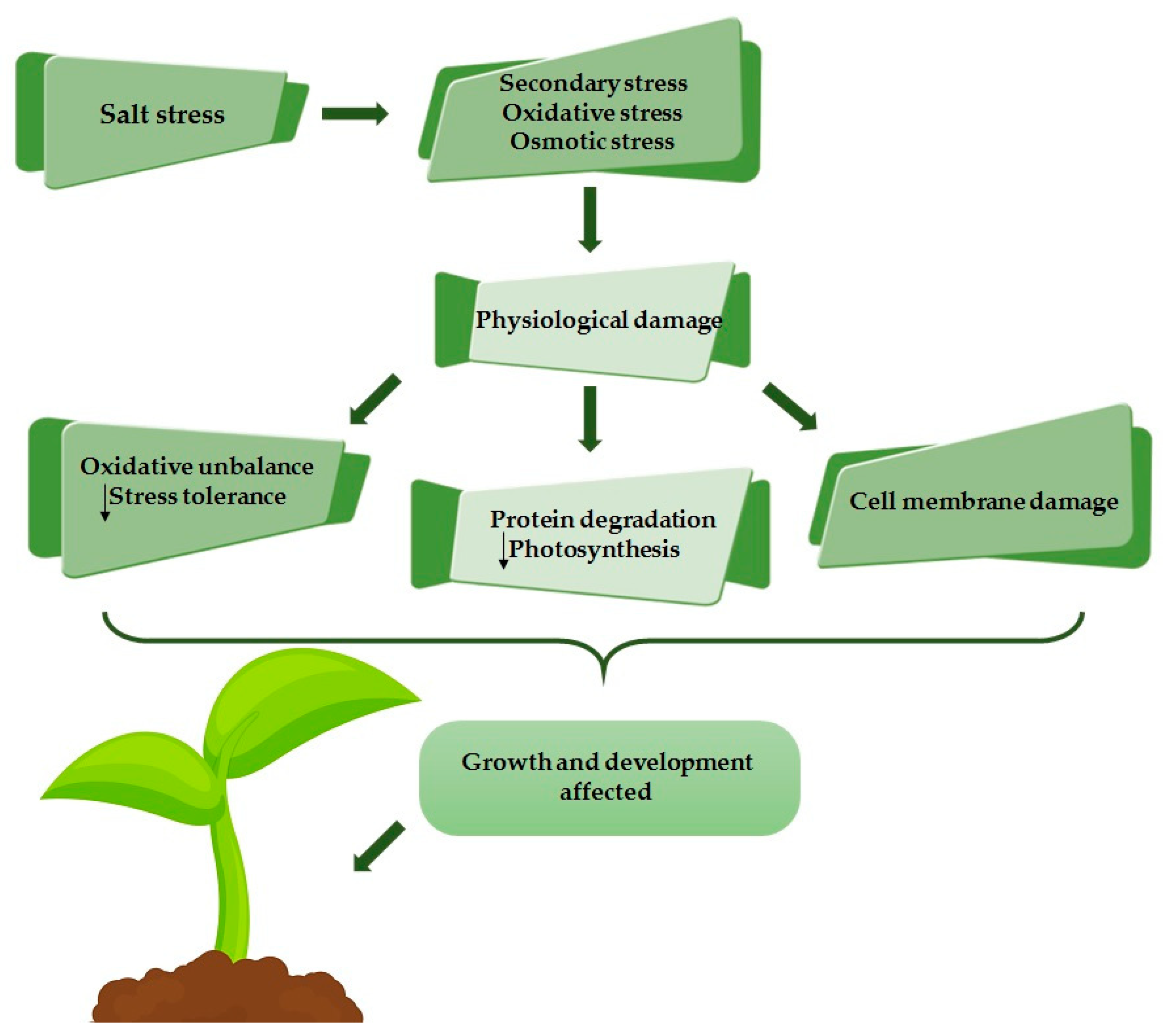
4. Salinity
4.1. Response Mechanisms to Saline Stress
4.2. Molecular Approachs to Salt Stress
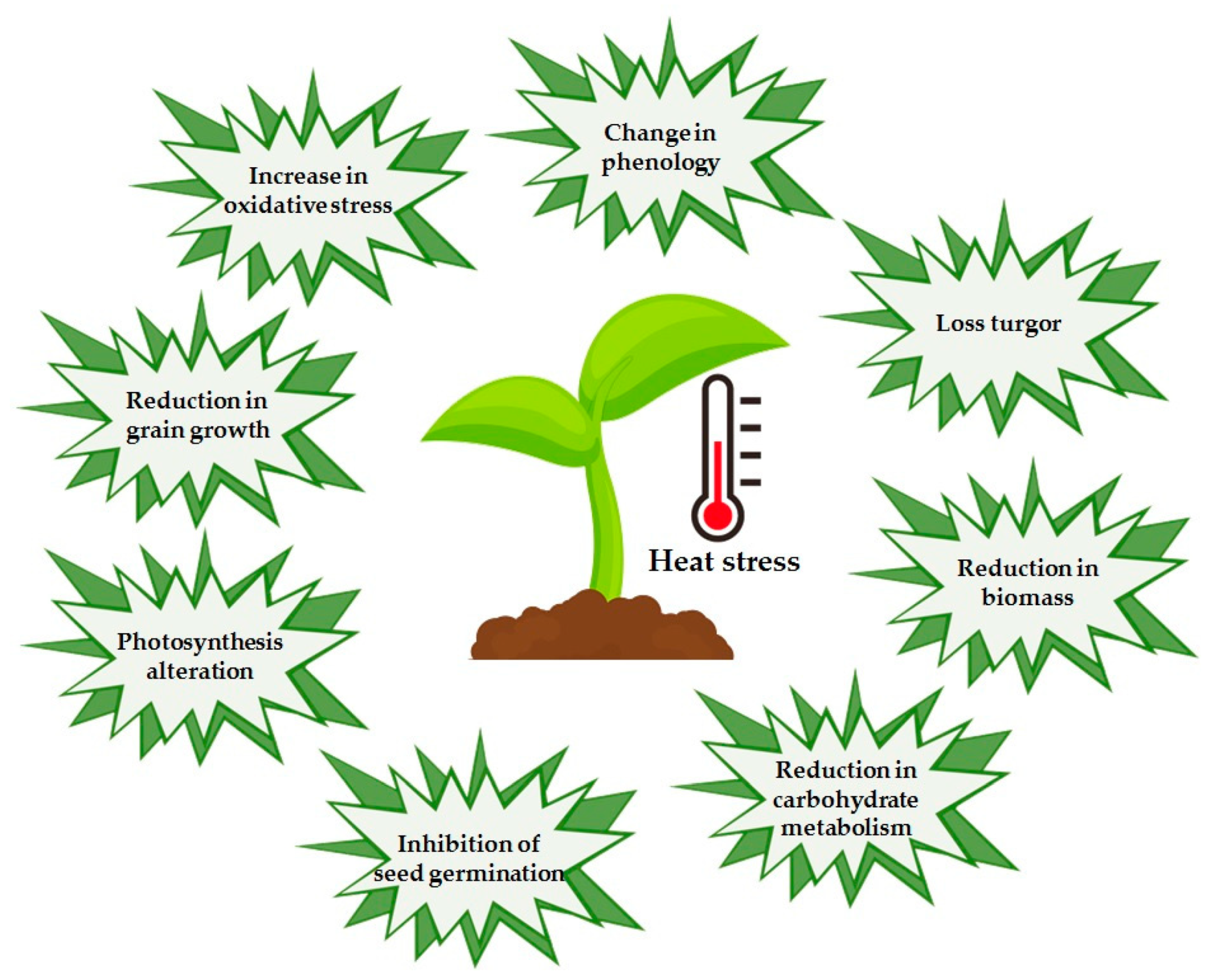
5. Heat Stress
5.1. Response Mechanisms to Heat Stress
5.2. Approaches to Generate Heat-Tolerant Plants
6. Conclusions and Future Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| WUE | water use efficiency |
| ABA | abscisic acid |
| ETC | electron transport chain |
| ROS | reactive oxygen species |
| ATP | adenosine triphosphate |
| NADH | nicotinamide adenine dinucleotide |
| TCA | tricarboxylic acid cycle |
| AOX | alternative oxidase |
| GABA | γ-aminobutyric acid |
| NO | nitric oxide |
| HR | hypersensitive response |
| PCD | programmed cell death |
| MAS | marker-assisted selection |
| GWAS | genome-wide selection |
| QTL | quantitative trait loci |
| PSII | photosystem II |
| RWC | relative water content |
| PGRs | plant growth regulator |
| OEC | oxygen-evolving complex |
| ASC | ascorbate |
| GSH | glutathione |
| SOD | superoxide dismutase |
| POX | peroxidase |
| CAT | catalase |
| GSH | glutathione |
| APX | ascorbate peroxidase |
| MDHAR | monodehydroascorbate dehydrogenase |
| DHAR | dehydroascorbate reductase |
| K | potassium |
| N | nitrogen |
| HSPs | heat shock proteins |
| GR | glutathione reductase |
| SA | salicilic acid |
| BRs | brassinosteroids |
| CK | cytokinin |
| JA | jasmonate |
| ET | ethylene |
| CuZn-SOD | copper-zinc superoxide dismutase |
| Ged | genome edition |
| RNA-seq | RNA sequencing |
| ChIP-seq | chromatin immunoprecipitation sequencing |
| GWA | genome-wide association |
| GS | genomic selection |
References
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J. Abiotic stress responses in plants. Nat. Rev. Genet. 2021, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Uddling, J.; Broberg, M.C.; Feng, Z.; Pleijel, H. Crop Quality under Rising Atmospheric CO2. Curr. Opin. Plant Biol. 2018, 45, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Poorter, H.; Knopf, O.; Wright, I.J.; Temme, A.A.; Hogewoning, S.W.; Graf, A.; Cernusak, L.A.; Pons, T.L. A meta-analysis of responses of C3 plants to atmospheric CO2: Dose–response curves for 85 traits ranging from the molecular to the whole-plant level. New Phytol. 2021, 233, 1560–1596. [Google Scholar] [CrossRef] [PubMed]
- Menezes-Silva, P.E.; Sanglard, L.M.; Ávila, R.T.; Morais, L.E.; Martins, S.C.; Nobres, P.; Patreze, C.M.; Ferreira, M.A.; Araújo, W.L.; Fernie, A.R. Photosynthetic and metabolic acclimation to repeated drought events play key roles in drought tolerance in coffee. J. Exp. Bot. 2017, 68, 4309–4322. [Google Scholar] [CrossRef] [PubMed]
- Bryant, C.; Fuenzalida, T.I.; Brothers, N.; Mencuccini, M.; Sack, L.; Binks, O.; Ball, M.C. Shifting access to pools of shoot water sustains gas exchange and increases stem hydraulic safety during seasonal atmospheric drought. Plant Cell Environ. 2021, 44, 2898–2911. [Google Scholar] [CrossRef]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2014, 5, 771. [Google Scholar] [CrossRef]
- Demidchik, V. Mechanisms of oxidative stress in plants: From classical chemistry to cell biology. Environ. Exp. Bot. 2015, 109, 212–228. [Google Scholar] [CrossRef]
- Makarevitch, I.; Waters, A.J.; West, P.T.; Stitzer, M.; Hirsch, C.N.; Ross-Ibarra, J.; Springer, N.M. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 2015, 11, e1004915. [Google Scholar]
- Hou, Q.; Ufer, G.; Bartels, D. Lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 2016, 39, 1029–1048. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Saito, K. Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 2015, 24, 10–16. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive Oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, S.; Sawant, K. Drought stress adaptation: Metabolic adjustment and regulation of gene expression. Plant Breed. 2013, 132, 21–32. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. Overcoming water challenges in agriculture. In The State of Food and Agriculture 2020; FAO: Rome, Italy, 2020. [Google Scholar]
- Gimenez, C.; Gallardo, M.; Thompson, R.B. Plant water relations. In Encyclopedia of Soils in the Environment; Hillel, D., Ed.; Elsevier: Oxford, UK, 2005; pp. 231–238. [Google Scholar]
- Kabbadj, A.; Makoudi, B.; Mouradi, M.; Pauly, N.; Frendo, P.; Ghoulam, C. Physiological and biochemical responses involved in water deficit tolerance of nitrogen-fixing Vicia faba. PLoS ONE 2017, 12, e0190284. [Google Scholar] [CrossRef] [PubMed]
- Salehi-Lisar, S.Y.; Motafakkerazad, R.; Hossain, M.M.; Rahman, I.M.M. Water stress in plants: Causes, effects and responses. In Water Stress; Rahman, M., Hasegawa, H., Eds.; InTech: Rijeka, Croatia, 2012; pp. 1–14. [Google Scholar]
- Salehi-Lisar, S.Y.; Bakhshayeshan-Agdam, H. Drought stress in plants: Causes, consequences, and tolerance. In Drought Stress Tolerance in Plants; Springer: Berlin, Germany, 2016; Volume 1, pp. 1–16. [Google Scholar]
- Bray, E. Plant response to water deficit stress. In Encyclopedia of Life Sciences (ELS); John Wiley & Sons: Hoboken, NJ, USA, 2007; Volume 1, pp. 1–7. [Google Scholar]
- O’Connell, E. Towards adaptation of water resource Systems to climatic and socio-economic Chang. Water Resour. Manag. 2017, 31, 2965–2984. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Dold, C. Water-Use Efficiency: Advances and Challenges in a Changing Climate. Front. Plant Sci. 2019, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Schulz, P.; Piepenburg, K.; Lintermann, R.; Herde, M.; Schottler, M.A.; Schmidt, L.K.; Ruf, S.; Kudla, J.; Romeis, T.; Bock, R. Improving plant drought tolerance and growth under water limitation through combinatorial engineering of signaling networks. Plant Biotechnol. J. 2021, 19, 74–86. [Google Scholar] [CrossRef]
- Nuccio, M.L.; Wu, J.; Mowers, R.; Zhou, H.-P.; Meghji, M.; Primavesi, L.F.; Paul, M.J.; Chen, X.; Gao, Y.; Haque, E.; et al. Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat. Biotechnol. 2015, 33, 862–869. [Google Scholar] [CrossRef]
- Damerum, A.; Smith, H.K.; Clarkson, G.; Truco, M.J.; Michelmore, R.W.; Taylor, G. The genetic basis of water-use efficiency and yield in lettuce. BMC Plant Biol. 2021, 21, 237. [Google Scholar] [CrossRef]
- Jia, X.; Mao, K.; Wang, P.; Wang, Y.; Jia, X.; Huo, L.; Sun, X.; Che, R.; Gong, X.; Ma, F. Overexpression of MdATG8i improves water use efficiency in transgenic apple by modulating photosynthesis, osmotic balance, and autophagic activity under moderate water deficit. Hortic. Res. 2021, 8, 81. [Google Scholar] [CrossRef]
- Baidyussen, A.; Jatayev, S.; Khassanova, G.; Amantayev, B.; Sereda, G.; Sereda, S.; Gupta, N.K.; Gupta, S.; Schramm, C.; Anderson, P.; et al. Expression of Specific Alleles of Zinc-Finger Transcription Factors, HvSAP8 and HvSAP16, and Corresponding SNP Markers, Are Associated with Drought Tolerance in Barley Populations. Int. J. Mol. Sci. 2021, 22, 12156. [Google Scholar] [CrossRef]
- Cai, K.; Chen, X.; Han, Z.; Wu, X.; Zhang, S.; Li, Q.; Nazir, M.M.; Zhang, G.; Zeng, F. Screening of Worldwide Barley Collection for Drought Tolerance: The Assessment of Various Physiological Measures as the Selection Criteria. Front. Plant Sci. 2020, 29, 1159. [Google Scholar] [CrossRef]
- Faillace, G.R.; Caruso, P.B.; Timmers, L.F.S.M.; Favero, D.; Guzman, F.L.; Rechenmacher, C.; de Oliveira-Busatto, L.A.; de Souza, O.N.; Bredemeier, C.; Bodanese-Zanettini, M.H. Molecular Characterisation of Soybean Osmotins and Their Involvement in Drought Stress Response. Front. Genet. 2021, 25, 632685. [Google Scholar] [CrossRef] [PubMed]
- Kamphorst, S.H.; Gonçalves, G.M.B.; Amaral Júnior, A.T.D.; Lima, V.J.D.; Schmitt, K.F.M.; Leite, J.T.; Azeredo, V.C.; Gomes, L.P.; Silva, J.G.d.S.; Carvalho, C.M.; et al. Supporting Physiological Trait for Indirect Selection for Grain Yield in Drought-Stressed Popcorn. Plants 2021, 10, 1510. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.D.O.; Amaral Junior, A.T.D.; Bispo, R.B.; Lima, V.J.; Kamphorst, S.H.; Leite, J.T.; Júnior, D.R.d.S.; Santos, P.H.A.D.; de Oliveira, U.A.; Schmitt, K.F.M.; et al. Phenotyping Latin American Open-Pollinated Varieties of Popcorn for Environments with Low Water Availability. Plants 2021, 10, 1211. [Google Scholar] [CrossRef] [PubMed]
- Barik, S.R.; Pandit, E.; Pradhan, S.K.; Mohanty, S.P.; Mohapatra, T. Genetic mapping of morpho-physiological traits involved during reproductive stage drought tolerance in rice. PLoS ONE. 2019, 14, e0214979. [Google Scholar] [CrossRef]
- Ghazy, M.I.; Salem, K.F.M.; Sallam, A. Utilization of genetic diversity and marker-trait to improve drought tolerance in rice (Oryza sativa L.). Mol. Biol. Rep. 2021, 48, 157–170. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Mohammadi, R.; Etminan, A.; Shooshtari, L.; Maleki-Tabrizi, N.; Poczai, P. Effects of Drought Stress on Some Agronomic and Morpho-Physiological Traits in Durum Wheat Genotypes. Sustainability 2020, 12, 5610. [Google Scholar] [CrossRef]
- Li, P.; Ma, B.; Palta, J.A.; Ding, T.; Cheng, Z.; Lv, G.; Xiong, Y. Wheat breeding highlights drought tolerance while ignores the advantages of drought avoidance: A meta-analysis. Eur. J. Agron. 2021, 122, 126196. [Google Scholar] [CrossRef]
- Kaur, H.; Kohli, S.K.; Khanna, K.; Bhardwaj, R. Scrutinizing the impact of water deficit in plants: Transcriptional regulation, signaling, photosynthetic efficacy, and management. Physiol. Plant 2021, 172, 935–962. [Google Scholar] [CrossRef]
- Chevilly, S.; Dolz-Edo, L.; López-Nicolás, J.M.; Morcillo, L.; Vilagrosa, A.; Yenush, L.; Mulet, J.M. Physiological and Molecular Characterization of the Differential Response of Broccoli (Brassica oleracea var. Italica) Cultivars Reveals Limiting Factors for Broccoli Tolerance to Drought Stress. J. Agric. Food Chem. 2021, 69, 10394–10404. [Google Scholar] [CrossRef]
- Mehari, T.G.; Xu, Y.; Umer, M.J.; Shiraku, M.L.; Hou, Y.; Wang, Y.; Yu, S.; Zhang, X.; Wang, K.; Cai, X.; et al. Multi-Omics-Based Identification and Functional Characterization of Gh_A06G1257 Proves Its Potential Role in Drought Stress Tolerance in Gossypium hirsutum. Front. Plant Sci. 2021, 21, 746771. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, D.; Bhardwaj, S.; Landi, M.; Sharma, A.; Ramakrishnan, M.; Sharma, A. The impact of drought in plant metabolism: How to exploit tolerance mechanisms to increase crop production. Appl. Sci. 2020, 10, 5692. [Google Scholar] [CrossRef]
- Larcher, W. Climatic constraints drive the evolution of low temperature resistance in woody plants. J. Agric. Meteorol. 2005, 61, 189–202. [Google Scholar] [CrossRef]
- Zingaretti, S.M.; Rodrigues, F.A.; Graca, J.P.; Pereira, L.M.; Lourenco, M.V. Sugarcane responses at water deficit conditions. In Water Stress, 1st ed.; Rahman, I.M.M., Ed.; IntechOpen: Shanghai, China, 2012; pp. 255–276. [Google Scholar]
- Fang, Y.; Xiong, L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Cameron, K.D.; Teece, M.A.; Smart, L.B. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 2006, 140, 176–183. [Google Scholar] [CrossRef]
- Perlikowski, D.; Kosmala, A. Mechanisms of drought resistance in introgression forms of Lolium multiflorum/Festuca arundinacea. Biol. Plant. 2020, 64, 497–503. [Google Scholar] [CrossRef]
- Luo, L.J. Breeding for water-saving and drought-resistance rice (WDR) in China. J. Exp. Bot. 2010, 61, 3509–3517. [Google Scholar] [CrossRef]
- Wittenmayer, L.; Merbach, W. Plant responses to drought and phosphorus deficiency: Contribution of phytohormones in root-related processes. J. Plant Nutr. Soil Sci. 2010, 168, 531–540. [Google Scholar] [CrossRef]
- Takahashi, F.; Kuromori, T.; Urano, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Drought Stress Responses and Resistance in Plants: From Cellular Responses to Long-Distance Intercellular Communication. Front. Plant Sci. 2020, 11, 556972. [Google Scholar] [CrossRef]
- Yang, X.; Lu, M.; Wang, Y.; Wang, Y.; Liu, Z.; Chen, S. Response mechanism of plants to drought stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Chan, Z. Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics 2012, 100, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; He, Y.; Zhu, M.; Ahmad, A.; Xu, S.; He, Z.; Jiang, S.; Huang, J.; Li, Z.; Liu, S.; et al. Ipa1 improves rice drought tolerance at seedling stage mainly through activating abscisic acid pathway. Plant Cell Rep. 2022, 41, 221–232. [Google Scholar] [CrossRef]
- Razi, K.; Muneer, S. Drought stress-induced physiological mechanisms, signaling pathways and molecular response of chloroplasts in common vegetable crops. Crit. Rev. Biotechnol. 2021, 41, 669–691. [Google Scholar] [CrossRef]
- Pinheiro, C.; Chaves, M.M. Photosynthesis and drought: Can we make metabolic connections from available data? J. Exp. Bot. 2011, 62, 869–882. [Google Scholar] [CrossRef] [PubMed]
- Galmés, J.; Medrano, H.; Flexas, J. Photosynthetic limitations in response to water stress and recovery in Mediterranean plants with different growth forms. New Phytol. 2007, 175, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Flexas, J.; Galmes, J.; Ribas-Carbo, M.; Medrano, H. The effects of water stress on plant respiration. In Plant Respiration: From Cell to Ecosystem; Lambers, H., Ribas-Carbo, M., Eds.; Springer: Berlin, Germany, 2005; pp. 85–94. [Google Scholar]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Vanlerberghe, G.C.; Martyn, G.D.; Dahal, K. Alternative oxidase: A respiratory electron transport chain pathway essential for maintaining photosynthetic performance during drought stress. Physiol. Plant 2016, 157, 322–337. [Google Scholar] [CrossRef]
- Fait, A.; Fromm, H.; Walter, D.; Galili, G.; Fernie, A.R. Highway or byway: The metabolic role of the GABA shunt in plants. Trends Plant Sci. 2008, 13, 14–19. [Google Scholar] [CrossRef]
- Khan, N.; Ali, S.; Shahid, M.A.; Mustafa, A.; Sayyed, R.Z.; Curá, J.A. Insights into the interactions among roots, rhizosphere, and rhizobacteria for improving plant growth and tolerance to abiotic stresses: A review. Cells 2021, 10, 1551. [Google Scholar] [CrossRef]
- Xu, B.; Long, Y.; Feng, X.; Zhu, X.; Sai, N.; Chirkova, L.; Betts, A.; Herrmann, J.; Edwards, E.J.; Okamoto, M.; et al. GABA signalling modulates stomatal opening to enhance plant water use efficiency and drought resilience. Nat. Commun. 2021, 29, 1952. [Google Scholar] [CrossRef]
- El Sabagh, A.; Hossain, A.; Barutcular, C.; Gormus, O.; Ahmad, Z.; Hussain, S.; Islam, M.; Alharby, H.; Bamagoos, A.; Kumar, N.; et al. Effects of drought stress on the quality of major oilseed crops: Implications and possible mitigation strategies—A review. Appl. Ecol. Environ. Res. 2019, 17, 4019–4043. [Google Scholar] [CrossRef]
- Ghaffari, H.; Tadayon, M.R.; Nadeem, M.; Cheema, M.; Razmjoo, J. Proline-mediated changes in antioxidant enzymatic activities and the physiology of sugar beet under drought stress. Acta Physiol. Plant 2019, 41, 23. [Google Scholar] [CrossRef]
- Huan, L.; Jin-Qiang, W.; Qing, L. Photosynthesis product allocation and yield in sweet potato with spraying exogenous hormones under drought stress. J. Plant Physiol. 2020, 253, 153265. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.; Ebeed, H.; Aljaarany, A. Exogenous application of spermine and putrescine mitigate adversities of drought stress in wheat by protecting membranes and chloroplast ultra-structure. Physiol. Mol. Biol Plants. 2020, 26, 233–245. [Google Scholar] [CrossRef]
- Deshpande, S.; Manoharan, R.; Mitra, S. Exogenous β-cyclocitral treatment primes tomato plants against drought by inducing tolerance traits, independent of abscisic acid. Plant Biol. 2021, 23, 170–180. [Google Scholar] [CrossRef]
- Xie, H.; Bai, G.; Lu, P.; Li, H.; Fei, M.; Xiao, B.G.; Chen, X.J.; Tong, Z.J.; Wang, Z.Y.; Yang, D.H. Exogenous citric acid enhances drought tolerance in tobacco (Nicotiana tabacum). Plant Biol. 2021, 24, 333–343. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef]
- Cattivelli, L.; Rizza, F.; Badeck, F.-W.; Mazzucotelli, E.; Mastrangelo, A.M.; Francia, E.; Marè, C.; Tondelli, A.; Stanca, A.M. Drought tolerance improvement in crop plants: An integrated view from breeding to genomics. Field Crops Res. 2008, 105, 1–14. [Google Scholar] [CrossRef]
- Rosero, A.; Granda, L.; Berdugo-Cely, J.A.; Šamajová, O.; Šamaj, J.; Cerkal, R. A dual strategy of breeding for drought tolerance and introducing drought-tolerant, underutilized crops into production systems to enhance their resilience to water deficiency. Plants 2020, 9, 1263. [Google Scholar] [CrossRef]
- Ashraf, M. Inducing drought tolerance in plants: Some recent advances. Biotechnol. Adv. 2010, 28, 169–183. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Samuel, C.; Fatai, A.; Magaji, U.; Kareem, I.; Kamarudin, Z.S.; Muhammad, I.I.; Kolapo, K. Drought Resistance in Rice from Conventional to Molecular Breeding: A Review. Int. J. Mol. Sci. 2019, 20, 3519. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Qin, F. Genetic dissection of maize drought tolerance for trait improvement. Mol. Breed. 2021, 41, 8. [Google Scholar] [CrossRef]
- Kopittke, P.N.; Menzies, P.; Wang, B.A.; McKenna, E. Lombi Soil and the intensification of agriculture for global food security. Environ. Int. 2019, 132, 105078. [Google Scholar] [PubMed]
- Munns, R.; Gilliham, M. Salinity tolerance of crops–what is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, C.H.; Song, J.K.; Zhu, S. Shabala. Mechanisms of plant responses and adaptation to soil salinity. Innovation 2020, 1, 100017. [Google Scholar]
- Parihar, P.; Singh, S.S.; Singh, R.; Singh, V.P.; Prasad, S.M. Effect of salinity stress on plants and its tolerance strategies: A review. Environ. Sci. Pollut. Res. 2015, 22, 4056–4075. [Google Scholar] [CrossRef]
- İbrahimova, U.; Kumari, P.; Yadav, S.; Rastogi, A.; Antala, M.; Suleymanova, Z.; Zivcak, M.; Arif, T.-U.; Hussain, S.; Abdelhamid, M.; et al. Progress in understanding salt stress response in plants using biotechnological tools. J. Biotechnol. 2021, 329, 180–191. [Google Scholar] [CrossRef]
- Liang, W.; Ma, X.; Wan, P.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Ashraf, M.H.P.J.C.; Harris, P.J. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef]
- Duarte, B.; Sleimi, N.; Caçador, I. Biophysical and biochemical constraints imposed by salt stress: Learning from halophytes. Front. Plant Sci. 2014, 5, 746. [Google Scholar] [CrossRef] [PubMed]
- Ventura, Y.; Eshel, A.; Pasternak, D.; Sagi, M. The development of halophyte based agriculture: Past and present. Ann. Bot. 2015, 115, 529–540. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Li, P.; Yang, X.; Wang, H.; Pan, T.; Wang, Y.; Xu, Y.; Xu, C.; Yang, Z. Genetic control of root plasticity in response to salt stress in maize. Theor. Appl. Genet. 2021, 134, 1475–1492. [Google Scholar] [CrossRef] [PubMed]
- Pierik, R.; Testerink, C. The art of being flexible: How to escape from shade; salt; and drought. Plant Physiol. 2014, 166, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Chun, H.J.; Baek, D.; Cho, H.M.; Jung, H.S.; Jeong, M.S.; Jung, W.-H.; Choi, C.W.; Lee, S.H.; Jin, B.J.; Park, M.S.; et al. Metabolic adjustment of Arabidopsis root suspension cells during adaptation to salt stress and mitotic stress memory. Plant Cell Physiol. 2019, 60, 612–625. [Google Scholar] [CrossRef] [PubMed]
- Cassaniti, C.; Leonardi, C.; Flowers, T.J. The effects of sodium chloride ornamental shrubs. Sci. Hortic. 2009, 122, 586–593. [Google Scholar] [CrossRef]
- Kumar, S.; Li, G.; Yang, J.; Huang, X.; Ji, Q.; Liu, Z.; Ke, W.; Hou, H. Effect of Salt Stress on Growth; Physiological Parameters; and Ionic Concentration of Water Dropwort (Oenanthe javanica) Cultivars. Front. Plant Sci. 2021, 12, 660409. [Google Scholar] [CrossRef]
- Rahneshan, Z.; Nasibi, F.; Moghadam, A.A. Effects of salinity stress on some growth; physiological; biochemical parameters and nutrients in two pistachio (Pistacia vera L.) rootstocks. J. Plant Interact. 2018, 13, 73–82. [Google Scholar] [CrossRef]
- dos Santos, T.B.; Budzinski, I.G.; Marur, C.J.; Petkowicz, C.L.; Pereira, L.F.; Vieira, L.G. Expression of three galactinol synthase isoforms in Coffea arabica L. and accumulation of raffinose and stachyose in response to abiotic stresses. Plant Physiol. Biochem. 2011, 49, 441–448. [Google Scholar] [CrossRef]
- Abid, M.; Zhang, Y.J.; Li, Z.; Bai, D.F.; Zhong, Y.P.; Fang, J.B. Effect of salt stress on growth; physiological and biochemical characters of four kiwifruit genotypes. Sci. Hortic. 2020, 271, 109473. [Google Scholar] [CrossRef]
- Molazem, D.; Azimi, J. Morpho-Physiological Characterization in Eight Varieties of Maize (Zea mays L.) under Soil Salinity. Pol. J. Environ. Stud. 2015, 24, 2537–2542. [Google Scholar] [CrossRef][Green Version]
- Phang, T.H.; Shao, G.; Lam, H.M. Salt tolerance in soybean. J. Integr. Plant Biol. 2008, 50, 1196–1212. [Google Scholar] [CrossRef] [PubMed]
- Płażek, A.; Tatrzańska, M.; Maciejewski, M.; Kościelniak, J.; Gondek, K.; Bojarczuk, J.; Dubert, F. Investigation of the salt tolerance of new Polish bread and durum wheat cultivars. Acta Physiologiae Plantarum 2013, 35, 2513–2523. [Google Scholar] [CrossRef]
- Zouari, M.; Ahmed, C.B.; Elloumi, N.; Bellassoued, K.; Delmail, D.; Labrousse, P.; Abdallah, F.B.; Rouina, B.B. Impact of proline application on cadmium accumulation; mineral nutrition and enzymatic antioxidant defense system of Olea europaea L. cv Chemlali exposed to cadmium stress. Ecotoxicol. Environ. Saf. 2016, 128, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Lima, R.B.; dos Santos, T.B.; Vieira, L.G.E.; Ferrarese, M.D.L.L.; Ferrarese-Filho, O.; Donatti, L.; de Oliveira Petkowicz, C.L. Salt stress alters the cell wall polysaccharides and anatomy of coffee (Coffea arabica L.) leaf cells. Carbohydr. Polym. 2014, 112, 686–694. [Google Scholar] [CrossRef]
- Mbarki, S.; Sytar, O.; Cerda, A.; Zivcak, M.; Rastogi, A.; He, X.; Zoghlami, A.; Abdelly, C.; Brestic, M. Strategies to mitigate the salt stress effects on photosynthetic apparatus and productivity of crop plants. In Salinity Responses and Tolerance in Plants; Springer: Cham, Switzerland, 2018; Volume 1, pp. 85–136. [Google Scholar]
- Hnilickova, H.; Hnilicka, F.; Martinkova, J.; Kraus, K. Effects of Salt Stress on Water Status; Photosynthesis and Chlorophyll Fluorescence of Rocket. Plant Soil Environ. 2017, 63, 362–367. [Google Scholar]
- Mehta, P.; Jajoo, A.; Mathur, S.; Bharti, S. Chlorophyll a Fluorescence Study Revealing Effects of High Salt Stress on Photosystem II in Wheat Leaves. Plant Physiol. Biochem. 2010, 48, 16–20. [Google Scholar] [CrossRef]
- Kalaji, H.M.; Rastogi, A.; Zivcak, M.; Brestic, M.; Daszkowska-Golec, A.; Sitko, K.; Alsharafa, K.Y.; Lotfi, R.; Stypiński, P.; Samborska, I.A.; et al. Prompt Chlorophyll Fluorescence as a Tool for Crop Phenotyping: An Example of Barley Landraces Exposed to Various Abiotic Stress Factors. Photosynthetica 2018, 56, 953–961. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Huang, J.; Peng, S.; Xiong, D. Diffusional conductance to CO2 is the key limitation to photosynthesis in salt-stressed leaves of rice (Oryza sativa). Physiol. Plant 2018, 163, 45–58. [Google Scholar] [CrossRef]
- Hussain, N.; Ghaffar, A.; Zafar, Z.U.; Javed, M.; Shah, K.H.; Noreen, S.; Manzoor, H.; Iqbal, M.; Hassan, I.F.Z.; Bano, H.; et al. Identification of novel source of salt tolerance in local bread wheat germplasm using morpho-physiological and biochemical attributes. Sci. Rep. 2021, 11, 10854. [Google Scholar] [CrossRef] [PubMed]
- Hnilickova, H.; Kraus, K.; Vachova, P.; Hnilicka, F. Salinity Stress Affects Photosynthesis; Malondialdehyde Formation; and Proline Content in Portulaca oleracea L. Plants 2021, 10, 845. [Google Scholar] [CrossRef] [PubMed]
- Chutipaijit, S.; Cha-um, S.; Sompornpailin, K. High contents of proline and anthocyanin increase protective response to salinity in Oryza sativa L. spp. ‘indica’. Aust. J. Crop. Sci. 2011, 5, 1191–1198. [Google Scholar]
- Amirjani, M.R. Effect of salinity stress on growth; sugar content; pigments and enzyme activity of rice. Int. J. Bot. Stud. 2011, 7, 73–81. [Google Scholar] [CrossRef]
- Hernández, J.A.; Jiménez, A.; Mullineaux, P.M.; Sevilla, F. Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defenses. Plant Cell Environ. 2000, 23, 853–862. [Google Scholar] [CrossRef]
- Sgherri, C.; Pinzino, C.; Quartacci, M.F. Reactive oxygen species and photosynthetic functioning: Past and present. In Reactive Oxygen Species in Plants: Boon or Bane—Revisiting the Role of ROS; Wiley: Chichester, UK, 2018; pp. 137–155. [Google Scholar]
- AbdElgawad, H.; Zinta, G.; Hegab, M.M.; Pandey, R.; Asard, H.; Abuelsoud, W. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front. Plant Sci. 2016, 7, 276. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Rakhmankulova, Z.F.; Shuyskaya, E.V.; Shcherbakov, A.V.; Fedyaev, V.V.; Biktimerova, G.Y.; Khafisova, R.R.; Usmanov, I.Y. Content of proline and flavonoids in the shoots of halophytes inhabiting the South Urals. Russ. J. Plant Physiol. 2015, 62, 71–79. [Google Scholar] [CrossRef]
- Giordano, M.; Petropoulos, S.A.; Rouphael, Y. Response and defence mechanisms of vegetable crops against drought, heat and salinity stress. Agriculture 2021, 11, 463. [Google Scholar] [CrossRef]
- Chung, Y.S.; Kim, K.S.; Hamayun, M.; Kim, Y. Silicon confers soybean resistance to salinity stress through regulation of reactive oxygen and reactive nitrogen species. Front. Plant Sci. 2020, 10, 1725. [Google Scholar] [CrossRef]
- Abdelaziz, M.E.; Abdelsattar, M.; Abdeldaym, E.A.; Atia, M.A.; Mahmoud, A.W.M.; Saad, M.M.; Hirt, H. Piriformospora indica alters Na+/K+ homeostasis, antioxidant enzymes and LeNHX1 expression of greenhouse tomato grown under salt stress. Sci. Hortic. 2019, 256, 108532. [Google Scholar] [CrossRef]
- Gengmao, Z.; Shihui, L.; Xing, S.; Yizhou, W.; Zipan, C. The role of silicon in physiology of the medicinal plant (Lonicera japonica L.) under salt stress. Sci. Rep. 2015, 5, 12696. [Google Scholar] [CrossRef] [PubMed]
- Jahantigh, O.; Najafi, F.; Badi, H.N.; Khavari-Nejad, R.A.; Sanjarian, F. Changes in antioxidant enzymes activities and proline, total phenol and anthocyanine contents in Hyssopus officinalis L. plants under salt stress. Acta Biol. Hung. 2016, 67, 195–204. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bonacina, C.; Trevizan, C.B.; Stracieri, J.; dos Santos, T.B.; Goncalves, J.E.; Gazim, Z.C.; Souza, S.G.H. Changes in growth, oxidative metabolism and essential oil composition of lemon balm (‘Melissa officinalis’ L.) subjected to salt stress. Aust. J. Crop. Sci. 2017, 11, 1665–1674. [Google Scholar] [CrossRef]
- Bonacina, C.; Cruz, R.M.S.; Nascimento, A.B.; Barbosa, L.N.; Gonçalves, J.E.; Gazim, Z.C.; Magalhães, H.M.; Souza, S.G.H. Salinity modulates growth, oxidative metabolism, and essential oil profile in Curcuma longa L. (Zingiberaceae) rhizomes. S. Afr. J. Bot. 2022, 146, 1–11. [Google Scholar] [CrossRef]
- Qu, C.; Liu, C.; Gong, X.; Li, C.; Hong, M.; Wang, L.; Hong, F. Impairment of maize seedling photosynthesis caused by a combination of potassium deficiency and salt stress. Environ. Exp. Bot. 2012, 75, 134–141. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; He, X.; Li, Q.; Liu, B.; Ai, X.; Zhang, D. Response of water balance and nitrogen assimilation in cucumber seedlings to CO2 enrichment and salt stress. Plant Physiol. Biochem. 2019, 139, 256–263. [Google Scholar] [CrossRef]
- Singh, R.K.; Prasad, A.; Muthamilarasan, M.; Parida, S.K.; Prasad, M. Breeding and biotechnological interventions for trait improvement: Status and prospects. Planta 2020, 252, 54. [Google Scholar] [CrossRef]
- Nakhla, W.R.; Sun, W.; Fan, K.; Yang, K.; Zhang, C.; Yu, S. Identification of QTLs for Salt Tolerance at the Germination and Seedling Stages in Rice. Plants 2021, 10, 428. [Google Scholar] [CrossRef]
- Cho, K.H.; Kim, M.Y.; Kwon, H.; Yang, X.; Lee, S.H. Novel QTL identification and candidate gene analysis for enhancing salt tolerance in soybean (Glycine max (L.) Merr.). Plant Sci. 2021, 313, 111085. [Google Scholar] [CrossRef]
- Lopez, C.; Orazaly, M.; Mozzoni, L.; Korth, K.L.; Chen, P. Quantitative trait loci for salt tolerance in soybean. J. Crop. Improv. 2018, 32, 766–780. [Google Scholar] [CrossRef]
- Luo, M.; Zhang, Y.; Chen, K.; Kong, M.; Song, W.; Lu, B.; Shi, Y.; Zhao, Y.; Zhao, J. Mapping of quantitative trait loci for seedling salt tolerance in maize. Mol. Breed. 2019, 39, 64. [Google Scholar] [CrossRef]
- Singh, R.K.; Kota, S.; Flowers, T.J. Salt tolerance in rice: Seedling and reproductive stage QTL mapping come of age. Theor. Appl. Genet. 2021, 134, 3495–3533. [Google Scholar] [CrossRef] [PubMed]
- Ahmadizadeh, M.; Babaeian-Jelodar, N.; Mohammadi-Nejad, G.; Singh, R.K. High-density linkage mapping for agronomic and physiological traits of rice (Oryza sativa L.) under reproductive-stage salt stress. J. Genet. 2021, 100, 51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced Rice Salinity Tolerance via CRISPR/Cas9-Targeted Mutagenesis of the OsRR22 Gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef]
- Kumar, V.V.S.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR-Cas9 Mediated Genome Editing of Drought and Salt Tolerance (OsDST) Gene in Indica Mega Rice Cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Tran, M.T.; Doan, D.T.H.; Kim, J.; Song, Y.J.; Sung, Y.W.; Das, S.; Kim, E.-J.; Son, G.H.; Kim, S.H.; Van Vu, T.; et al. CRISPR/Cas9-Based Precise Excision of SlHyPRP1 Domain(s) to Obtain Salt Stress-Tolerant Tomato. Plant Cell Rep. 2021, 40, 999–1011. [Google Scholar] [CrossRef]
- Roy, S.J.; Negrao, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef]
- Huang, Y.; Guan, C.; Liu, Y.; Chen, B.; Yuan, S.; Cui, X.; Zhang, Y.; Yang, F. Enhanced growth performance and salinity tolerance in transgenic switchgrass via overexpressing vacuolar Na+ (K+)/H+ antiporter gene (PvNHX1). Front. Plant Sci. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Xu, C.; Luo, M.; Sun, X.; Yan, J.; Shi, H.; Yan, H.; Yan, R.; Wang, S.; Tang, W.; Zhou, Y.; et al. SiMYB19 from Foxtail Millet (Setaria italica) Confers Transgenic Rice Tolerance to High Salt Stress in the Field. Int. J. Mol. Sci. 2022, 23, 756. [Google Scholar] [CrossRef]
- Li, M.; Chen, R.; Jiang, Q.; Sun, X.; Zhang, H.; Hu, Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 2021, 105, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, N.; Bhandari, K.; Siddique, K.H.; Nayyar, H. Food crops face rising temperatures: An overview of responses, adaptive mechanisms, and approaches to improve heat tolerance. Cogent Food Agric. 2016, 2, 1134380. [Google Scholar] [CrossRef]
- Qu, A.-L.; Ding, Y.-F.; Jiang, Q.; Zhu, C. Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 2013, 432, 203–207. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, A.; Yadav, S.; Singh, I.K. Reactive oxygen species-mediated signaling during abiotic stress. Plant Gene 2019, 18, 100–173. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84. [Google Scholar] [CrossRef]
- Singsaas, E.L.; Laporte, M.M.; Shi, J.-Z.; Monson, R.K.; Bowling, D.R.; Johnson, K.; Lerdau, M.; Jasentuliytana, A.; Sharkey, T.D. Kinetics of leaf temperature fluctuation affect isoprene emission from red oak (Quercus rubra) leaves. Tree Physiol. 1999, 19, 917–924. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Akram, N.A.; Ashraf, M.; Alyemeni, M.N.; Wijaya, L.; Ahmad, P. Plant responses to environmental stresses—From gene to biotechnology. AoB Plants 2017, 9, 1–17. [Google Scholar] [CrossRef]
- Li, N.; Euring, D.; Cha, J.Y.; Lin, Z.; Lu, M.; Huang, L.J.; Kim, W.Y. Plant hormone-mediated regulation of heat tolerance in response to global climate change. Front. Plant Sci. 2021, 11, 2318. [Google Scholar] [CrossRef]
- Sicher, R.C.; Timlin, D.; Bailey, B. Responses of growth and primary metabolism of water-stressed barley roots to rehydration. J. Plant Physiol. 2012, 169, 686–695. [Google Scholar] [CrossRef]
- Goufo, P.; Moutinho-Pereira, J.M.; Jorge, T.F.; Correia, C.M.; Oliveira, M.R.; Rosa, E.A.S.; António, C.; Trindade, H. Cowpea (Vigna unguiculata L. Walp.) Metabolomics: Osmoprotection as a Physiological Strategy for Drought Stress Resistance and Improved Yield. Front. Plant Sci. 2017, 8, 586. [Google Scholar] [CrossRef]
- Lima, R.B.; dos Santos, T.B.; Vieira, L.G.E.; de Lourdes Lúcio Ferrarese, M.; Ferrarese-Filho, O.; Donatti, L.; Boeger, M.R.T.; de Oliveira Petkowicz, C.L. Heat stress causes alterations in the cell-wall polymers and anatomy of coffee leaves (Coffea arabica L.). Carbohydr. Polym. 2013, 93, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Wahid, A. Physiological Implications of Metabolite Biosynthesis for Net Assimilation and Heat-Stress Tolerance of Sugarcane (Saccharum officinarum) Sprouts. J. Plant Res. 2007, 120, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Agric. 2009, 29, 153–188. [Google Scholar]
- Zandalinas, S.I.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A.; Inupakutika, M.A.; Mittler, R. ABA is required for the accumulation of APX1 and MBF1c during a combination of water deficit and heat stress. J. Exp. Bot. 2016, 67, 5381–5390. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hou, L.; Lu, Y.; Wu, B.; Gong, X.; Liu, M.; Wang, J.; Sun, Q.; Vierling, E.; Xu, S. Metabolic adaptation of wheat grain contributes to a stable filling rate under heat stress. J. Exp. Bot. 2018, 69, 5531–5545. [Google Scholar] [CrossRef] [PubMed]
- Raza, A. Metabolomics: A systems biology approach for enhancing heat stress tolerance in plants. Plant Cell Rep. 2020, 1–23. [Google Scholar] [CrossRef]
- Yamori, W.; Hikosaka, K.; Way, D.A. Temperature response of photosynthesis in C3, C4, and CAM plants: Temperature acclimation and temperature adaptation. Photosynth. Res. 2014, 119, 101–117. [Google Scholar] [CrossRef]
- Betti, M.; Bauwe, H.; Busch, F.; Fernie, A.R.; Keech, O.; Levey, M.; Ort, D.R.; Parry, M.A.J.; Sage, R.; Timm, S.; et al. Manipulating photorespiration to increase plant productivity: Recent advances and perspectives for crop improvement. J. Exp. Bot. 2016, 67, 2977–2988. [Google Scholar] [CrossRef]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.-R.; Bäurle, I. Arabidopsis miR156 Regulates Tolerance to Recurring Environmental Stress through SPL Transcription Factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef]
- Liu, J.; Feng, L.; Li, J.; He, Z. Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 2015, 6, 267. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Serban, A.J.; Wachter, R.M.; Moerner, W.E. Single-Molecule Diffusometry Reveals the Nucleotide-Dependent Oligomerization Pathways of Nicotiana Tabacum Rubisco Activase. J. Chem. Phys. 2018, 148, 123319. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.-Z.; Guo, F.-Q. Chloroplast Retrograde Regulation of Heat Stress Responses in Plants. Front. Plant Sci. 2016, 7, 398. [Google Scholar] [CrossRef] [PubMed]
- Todaka, D.; Nakashima, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Toward understanding transcriptional regulatory networks in abiotic stress responses and tolerance in rice. Rice J. 2012, 5, 6. [Google Scholar] [CrossRef]
- Moustafa, K.; Abuqamar, S.; Jarrar, M.; Al-Rajab, A.J.; Trémouillaux-Guiller, J. MAPK cascades and major abiotic stresses. Plant Cell Rep. 2014, 33, 1217–1225. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, J.; Zhang, N.; Xin, M.; Peng, H.; Hu, Z.; Ni, Z.; Du, J. The Wheat NAC Transcription Factor TaNAC2L Is Regulated at the Transcriptional and Post-Translational Levels and Promotes Heat Stress Tolerance in Transgenic Arabidopsis. PLoS ONE 2015, 10, e0135667. [Google Scholar] [CrossRef]
- Liu, Z.-M.; Yue, M.-M.; Yang, D.-Y.; Zhu, S.-B.; Ma, N.-N.; Meng, Q.-W. Over-Expression of SlJA2 Decreased Heat Tolerance of Transgenic Tobacco Plants via Salicylic Acid Pathway. Plant Cell Rep. 2017, 36, 529–542. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, Y.; Wang, C.; Wang, Y.C.; Wang, L.Q. ThNAC12 from Tamarix hispida directly regulates ThPIP2; 5 to enhance salt tolerance by modulating reactive oxygen species. Plant Physiol. Biochem. 2021, 163, 27–35. [Google Scholar] [CrossRef]
- Talukder, S.K.; Babar, M.A.; Vijayalakshmi, K.; Poland, J.; Prasad, P.V.V.; Bowden, R.; Fritz, A. Mapping QTL for the traits associated with heat tolerance in wheat (Triticum aestivum L.). BMC Genet. 2014, 15, 97. [Google Scholar] [CrossRef]
- Acuña-Galindo, M.A.; Mason, R.E.; Subramanian, N.K.; Hays, D.B. Meta-Analysis of Wheat QTL Regions Associated with Adaptation to Drought and Heat Stress. Crop Sci. 2015, 55, 477–492. [Google Scholar] [CrossRef]
- Sharma, D.K.; Torp, A.M.; Rosenqvist, E.; Ottosen, C.O.; Andersen, S.B. QTLs and potential candidate genes for heat stress tolerance identified from the mapping populations specifically segregating for F v/F m in wheat. Front. Plant Sci. 2017, 8, 1668. [Google Scholar] [CrossRef] [PubMed]
- Bhusal, N.; Sarial, A.K.; Sharma, P.; Sareen, S. Mapping QTLs for grain yield components in wheat under heat stress. PLoS ONE 2017, 12, e0189594. [Google Scholar] [CrossRef]
- Hassan, F.S.C.; Solouki, M.; Fakheri, B.A.; Nezhad, N.M.; Masoudi, B. Mapping QTLs for physiological and biochemical traits related to grain yield under control and terminal heat stress conditions in bread wheat (Triticum aestivum L.). Physiol. Mol. Biol. Plants 2018, 24, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Gous, P.W.; Hickey, L.; Christopher, J.T.; Franckowiak, J.; Fox, G.P. Discovery of QTL for stay-green and heat-stress in barley (Hordeum vulgare) grown under simulated abiotic stress conditions. Euphytica 2016, 207, 305–317. [Google Scholar] [CrossRef]
- Kushwah, A.; Bhatia, D.; Singh, I.; Thudi, M.; Singh, G.; Bindra, S.; Vij, S.; Gill, B.; Bharadwaj, C.; Singh, S. Identification of stable heat tolerance QTLs using inter-specific recombinant inbred line population derived from GPF 2 and ILWC 292. PLoS ONE 2021, 16, e0254957. [Google Scholar] [CrossRef]
- Du, L.; Cai, C.; Wu, S.; Zhang, F.; Hou, S.; Guo, W. Evaluation and exploration of favorable QTL alleles for salt stress related traits in cotton cultivars (G. hirsutum L.). PLoS ONE 2016, 11, e0151076. [Google Scholar] [CrossRef]
- Majeed, S.; Rana, I.A.; Atif, R.M.; Zulfiqar, A.; Hinze, L.; Azhar, M.T. Role of SNPs in determining QTLs for major traits in cotton. J. Cotton Res. 2019, 2, 5. [Google Scholar] [CrossRef]
- Driedonks, N.; Wolters-Arts, M.; Huber, H.; de Boer, G.J.; Vriezen, W.; Mariani, C.; Rieu, I. Exploring the natural variation for reproductive thermotolerance in wild tomato species. Euphytica 2018, 214, 67. [Google Scholar] [CrossRef]
- Bineau, E.; Diouf, I.; Carretero, Y.; Duboscq, R.; Bitton, F.; Djari, A.; Zouine, M.; Causse, M. Genetic diversity of tomato response to heat stress at the QTL and transcriptome levels. Plant J. 2021, 107, 1213–1227. [Google Scholar] [CrossRef]
- Ravikiran, K.; Krishnan, S.G.; Vinod, K.; Dhawan, G.; Dwivedi, P.; Kumar, P.; Bansal, V.P.; Nagarajan, M.; Bhowmick, P.K.; Ellur, R. A trait specific QTL survey identifies NL44, a NERICA cultivar as a novel source for reproductive stage heat stress tolerance in rice. Plant Physiol. Rep. 2020, 25, 664–676. [Google Scholar] [CrossRef]
- Majeed, S.; Rana, I.A.; Mubarik, M.S.; Atif, R.M.; Yang, S.-H.; Chung, G.; Jia, Y.; Du, X.; Hinze, L.; Azhar, M.T. Heat Stress in Cotton: A Review on Predicted and Unpredicted Growth-Yield Anomalies and Mitigating Breeding Strategies. Agronomy 2021, 11, 1825. [Google Scholar] [CrossRef]
- Ashkani, S.; Rafii, M.Y.; Shabanimofrad, M.; Miah, G.; Sahebi, M.; Azizi, P.; Tanweer, F.A.; Akhtar, M.S.; Nasehi, A. Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Front. Plant Sci. 2015, 6, 886. [Google Scholar] [CrossRef] [PubMed]
- Templer, S.E.; Ammon, A.; Pscheidt, D.; Ciobotea, O.; Schuy, C.; McCollum, C.; Sonnewald, U.; Hanemann, A.; Förster, J.; Ordon, F.; et al. Metabolite profiling of barley flag leaves under drought and combined heat and drought stress reveals metabolic QTLs for metabolites associated with antioxidant defense. J. Exp. Bot. 2017, 68, 1697–1713. [Google Scholar] [CrossRef] [PubMed]
- Thomason, K.; Babar, M.A.; Erickson, J.E.; Mulvaney, M.; Beecher, C.; MacDonald, G. Comparative physiological and metabolomics analysis of wheat (Triticum aestivum L.) following post-anthesis heat stress. PLoS ONE 2018, 13, e0197919. [Google Scholar] [CrossRef] [PubMed]
- Lawas, L.M.F.; Li, X.; Erban, A.; Kopka, J.; Jagadish, S.K.; Zuther, E.; Hincha, D.K. Metabolic responses of rice cultivars with different tolerance to combined drought and heat stress under field conditions. Gigascience 2019, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Dhatt, B.K.; Abshire, N.; Paul, P.; Hasanthika, K.; Sandhu, J.; Zhang, Q.; Obata, T.; Walia, H. Metabolic dynamics of developing rice seeds under high night-time temperature stress. Front. Plant Sci. 2019, 10, 1443. [Google Scholar] [CrossRef]
- Wang, L.; Ma, K.-B.; Lu, Z.-G.; Ren, S.-X.; Jiang, H.-R.; Cui, J.-W.; Chen, G.; Teng, N.-J.; Lam, H.-M.; Jin, B. Differential physiological, transcriptomic and metabolomic responses of Arabidopsis leaves under prolonged warming and heat shock. BMC Plant Biol. 2020, 20, 86. [Google Scholar] [CrossRef]
- Wei, S.; Yang, X.; Huo, G.; Ge, G.; Liu, H.; Luo, L.; Hu, J.; Huang, D.; Long, P. Distinct metabolome changes during seed germination of lettuce (Lactuca sativa L.) in response to thermal stress as revealed by untargeted metabolomics analysis. Int. J. Mol. Sci. 2020, 21, 1481. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Burritt, D.J.; Gupta, A.; Tsujimoto, H.; Tran, L.-S.P. Heat stress effects on source–sink relationships and metabolome dynamics in wheat. J. Exp. Bot. 2020, 71, 543–554. [Google Scholar] [CrossRef]
- Bhatnagar-Mathur, P.; Vadez, V.; Sharma, K.K. Transgenic approaches for abiotic stress tolerance in plants: Retrospect and prospects. Plant Cell Rep. 2008, 27, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Sinha, K.; Bhunia, R.K. Can Wheat Survive in Heat? Assembling Tools towards Successful Development of Heat Stress Tolerance in Triticum Aestivum, L. Mol. Biol. Rep. 2019, 46, 2577–2593. [Google Scholar] [CrossRef] [PubMed]
- Dangol, S.D.; Naeem, M.; Azimi, M.H.; Yasmeen, A.; Caliskan, M.E.; Bakhsh, A. Genetic Engineering of Solanum tuberosum L. to Enhance Resistance Against Abiotic Stresses: A Review. J. Sci. 2018, 1, 1–10. [Google Scholar]
- Batcho, A.A.; Sarwar, M.B.; Rashid, B.; Hassan, S.; Husnain, T. Heat shock protein gene identified from Agave sisalana (As HSP70) confers heat stress tolerance in transgenic cotton (Gossypium hirsutum). Theor. Exp. Plant Physiol. 2021, 33, 141–156. [Google Scholar] [CrossRef]
- Fragkostefanakis, S.; Roeth, S.; Schleiff, E.; Scharf, K. Prospects of Engineering Thermotolerance in Crops Through Modulation of Heat Stress Transcription Factor and Heat Shock Protein Networks. Plant Cell Environ. 2015, 38, 1881–1895. [Google Scholar] [CrossRef]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef]
- Xu, Q.; Xu, X.; Shi, Y.; Xu, J.; Huang, B. Transgenic Tobacco Plants Overexpressing a Grass PpEXP1 Gene Exhibit Enhanced Tolerance to Heat Stress. PLoS ONE 2014, 9, e100792. [Google Scholar] [CrossRef]
- dos Santos, T.B.; Vieira, L.G.E. Involvement of the galactinol synthase gene in abiotic and biotic stress responses: A review on current knowledge. Plant Gene 2020, 24, 100258. [Google Scholar] [CrossRef]
- Mubarik, M.S.; Khan, S.H.; Ahmad, A.; Khan, Z.; Sajjad, M.; Khan, I.A. Disruption of Phytoene Desaturase Gene using Transient Expression of Cas9: gRNA Complex. Int. J. Agric. Biol. 2016, 18. [Google Scholar]
- Yin, K.; Gao, C.; Qiu, J.L. Progress and prospects in plant genome editing. Nat. Plants 2017, 3, 17107. [Google Scholar] [CrossRef]
- Li, H.; Hu, T.; Amombo, E.; Fu, J. Genome-wide identification of heat stress-responsive small RNAs in tall fescue (Festuca arundinacea) by high-throughput sequencing. J. Plant Physiol. 2017, 213, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, S.-L.; Li, J.-L.; Hu, X.-H.; Wang, H.; Cao, X.-L.; Xu, Y.-J.; Zhao, Z.-X.; Xiao, Z.-Y.; Yang, N. Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front. Plant Sci. 2017, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Bhogireddy, S.; Babu, M.S.; Swamy, K.N.; Vishnukiran, T.; Subrahmanyam, D.; Sarla, N.; Rao, P.R.; Mangrauthia, S.K. Expression Dynamics of Genes and microRNAs at Different Growth Stages and Heat Treatments in Contrasting High Temperature Responsive Rice Genotypes. J. Plant Growth Regul. 2022, 41, 74–91. [Google Scholar] [CrossRef]
- Xu, J.; Hou, Q.M.; Khare, T.; Verma, S.K.; Kumar, V. Exploring miRNAs for developing climate-resilient crops: A perspective review. Sci. Total Environ. 2019, 653, 91–104. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 1–2. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
dos Santos, T.B.; Ribas, A.F.; de Souza, S.G.H.; Budzinski, I.G.F.; Domingues, D.S. Physiological Responses to Drought, Salinity, and Heat Stress in Plants: A Review. Stresses 2022, 2, 113-135. https://doi.org/10.3390/stresses2010009
dos Santos TB, Ribas AF, de Souza SGH, Budzinski IGF, Domingues DS. Physiological Responses to Drought, Salinity, and Heat Stress in Plants: A Review. Stresses. 2022; 2(1):113-135. https://doi.org/10.3390/stresses2010009
Chicago/Turabian Styledos Santos, Tiago Benedito, Alessandra Ferreira Ribas, Silvia Graciele Hülse de Souza, Ilara Gabriela Frasson Budzinski, and Douglas Silva Domingues. 2022. "Physiological Responses to Drought, Salinity, and Heat Stress in Plants: A Review" Stresses 2, no. 1: 113-135. https://doi.org/10.3390/stresses2010009
APA Styledos Santos, T. B., Ribas, A. F., de Souza, S. G. H., Budzinski, I. G. F., & Domingues, D. S. (2022). Physiological Responses to Drought, Salinity, and Heat Stress in Plants: A Review. Stresses, 2(1), 113-135. https://doi.org/10.3390/stresses2010009





