Utilization of Tomato Landraces to Improve Seedling Performance under Salt Stress
Abstract
1. Introduction
2. Results
2.1. Growth Parameters
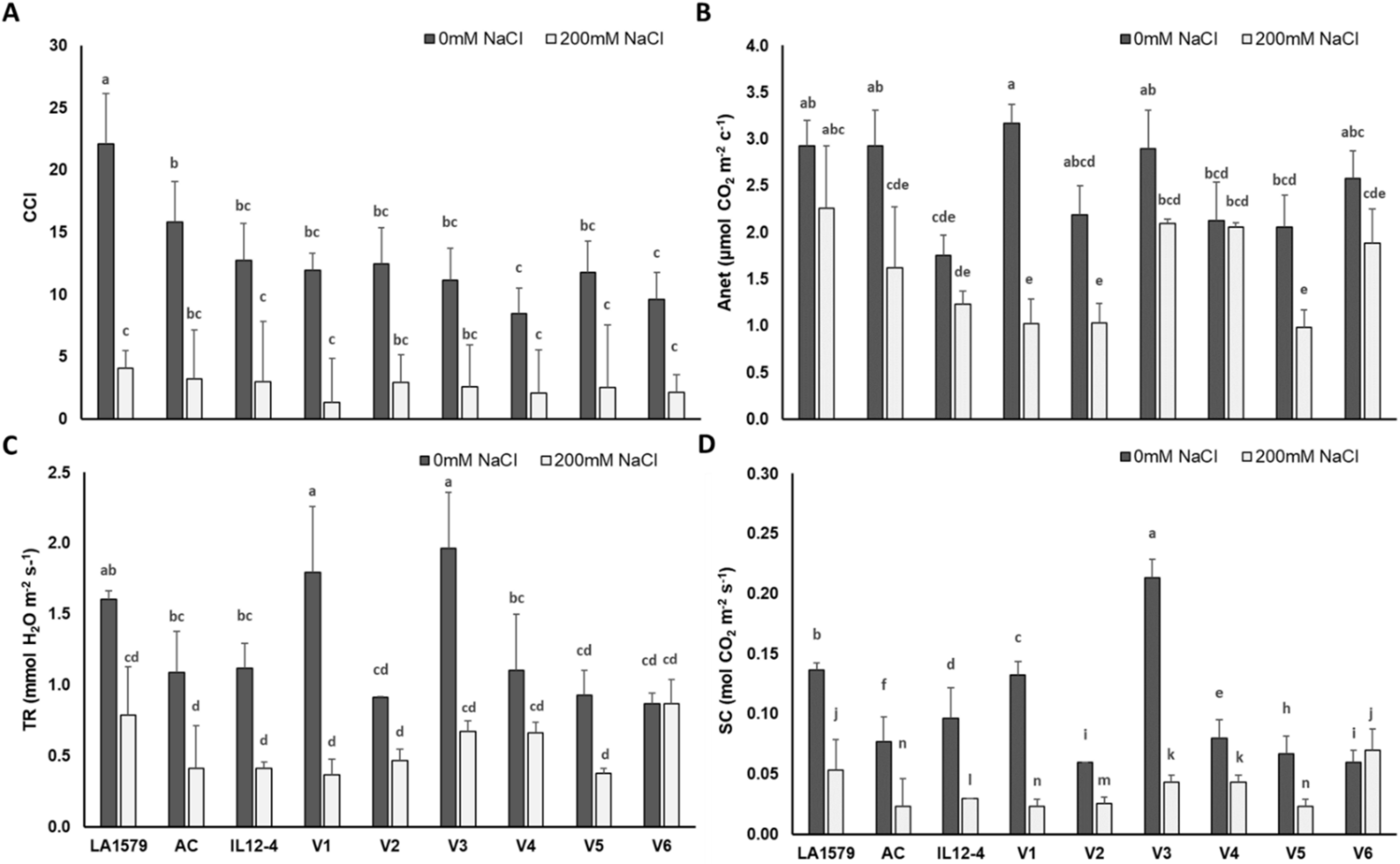
2.2. Physiological Traits
2.3. Salt Tolerance Indices
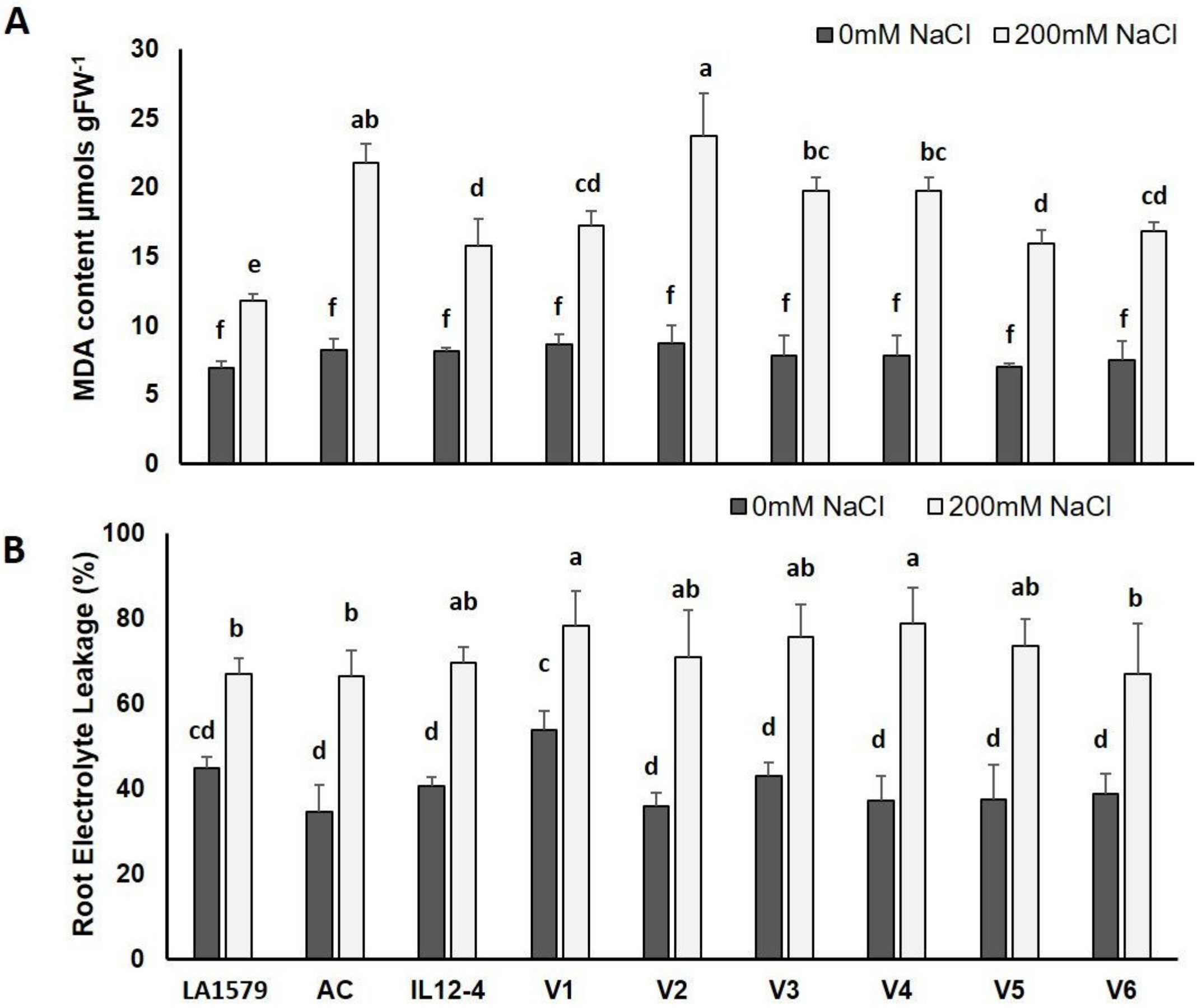
2.4. Oxidative Stress
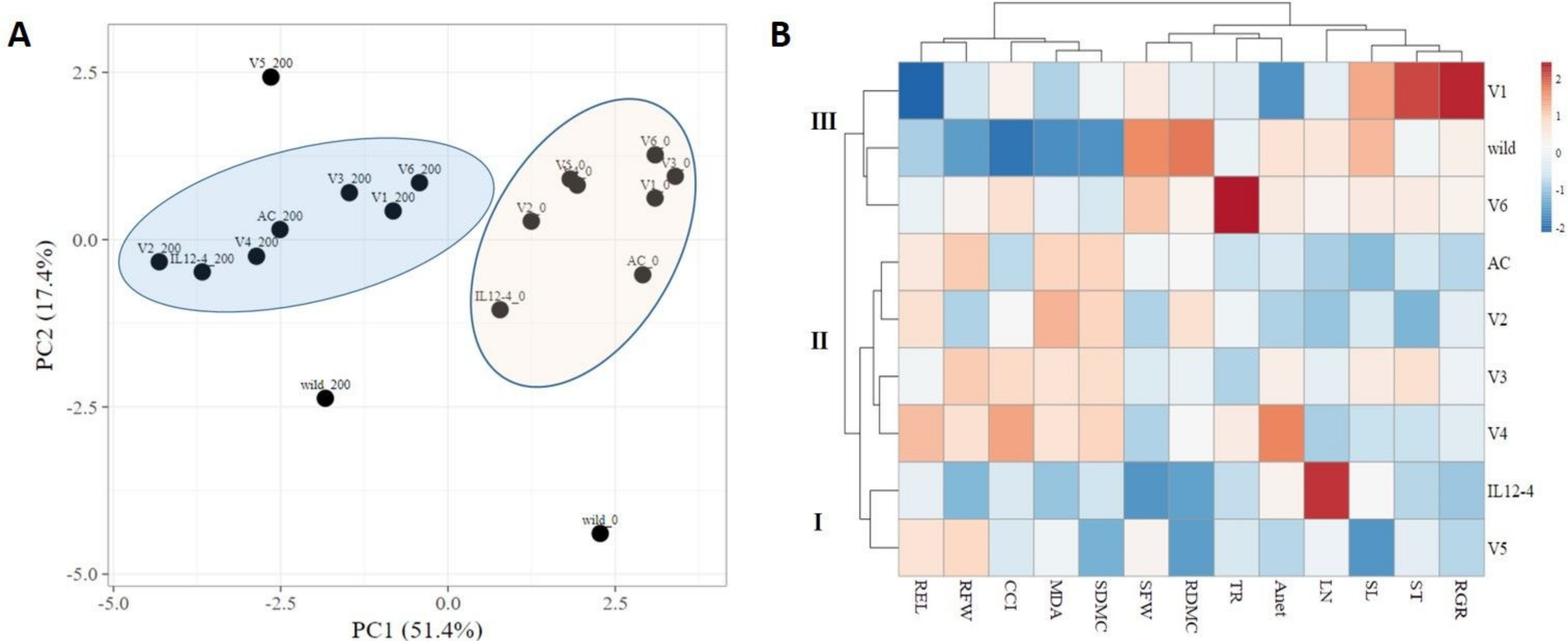
2.5. Principal Component Analysis and Hierarchical Clustering
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Plant Growth and Developmental Characteristics
4.3. Photosynthetic Parameters
4.4. Root Electrolyte Leakage
4.5. Lipid Peroxidation and Ascorbic Acid Contents
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). 2016. Available online: http://www.fao.org/soils-portal/soil-management/management-of-some-problem-soils/salt-affected-soils/more-information-on-salt-affected-soils/en/ (accessed on 20 March 2021).
- Kozminska, A.; Al Hassan, M.; Hanus-Fajerska, E.; Naranjo, M.A.; Boscaiu, M.; Vicente, O. Comparative analysis of water deficit and salt tolerance mechanisms in Silene. S. Afr. J. Bot. 2018, 117, 193–206. [Google Scholar] [CrossRef]
- Corwin, D.L. Climate change impacts on soil salinity in agricultural areas. Eur. J. Soil Sci. 2021, 72, 842–862. [Google Scholar] [CrossRef]
- Mellidou, I.; Moschou, P.N.; Ioannidis, N.E.; Pankou, C.; Gėmes, K.; Valassakis, C.; Andronis, E.A.; Beris, D.; Haralampidis, K.; Roussis, A.; et al. Silencing S-adenosyl-L-methionine decarboxylase (SAMDC) in Nicotiana tabacum points at a polyamine-dependent trade-off between growth and tolerance responses. Front. Plant Sci. 2016, 7, 379. [Google Scholar] [CrossRef] [PubMed]
- Abid, M.; Zhang, Y.J.; Li, Z.; Bai, D.F.; Zhong, Y.P.; Fang, J.B. Effect of salt stress on growth, physiological and biochemical characters of four kiwifruit genotypes. Sci. Hortic. 2020, 271, 109473. [Google Scholar] [CrossRef]
- Shabala, S. Learning from halophytes: Physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot. 2013, 112, 1209–1221. [Google Scholar] [CrossRef]
- Gémes, K.; Mellidou, Ι.; Karamanoli, K.; Beris, D.; Park, K.Y.; Matsi, T.; Haralampidis, K.; Constantinidou, H.I.; Roubelakis-Angelakis, K.A. Deregulation of apoplastic polyamine oxidase affects development and salt response of tobacco plants. J. Plant Physiol. 2017, 211, 1–12. [Google Scholar] [CrossRef]
- Devkar, V.; Thirumalaikumar, V.P.; Xue, G.P.; Vallarino, J.G.; Turečková, V.; Strnad, M.; Fernie, A.R.; Hoefgen, R.; Mueller-Roeber, B.; Balazadeh, S. Multifaceted regulatory function of tomato SlTAF1 in the response to salinity stress. New Phytol. 2020, 225, 1681–1698. [Google Scholar] [CrossRef]
- Lotfi, N.; Vahdati, K.; Kholdebarin, B.; Ashrafi, E.N. Germination, mineral composition, and ion uptake in walnut under salinity conditions. HortScience 2009, 44, 1352–1357. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Mellidou, I.; Ainalidou, A.; Papadopoulou, A.; Leontidou, K.; Genitsaris, S.; Karagiannis, E.; Van de Poel, B.; Karamanoli, K. Comparative Transcriptomics and Metabolomics Reveal an Intricate Priming Mechanism Involved in PGPR-Mediated Salt Tolerance in Tomato. Front. Plant Sci. 2021, 12, 1–22. [Google Scholar] [CrossRef]
- Radoglou, K.; Cabral, R.T.; Hasanagas, N.; Sutinen, M.-L.; Waisel, Y. Appraisal of root leakage as a method for estimation of root viability. Plant Biosyst. 2007, 141, 443–459. [Google Scholar] [CrossRef][Green Version]
- Lotfi, N.; Soleimani, A.; Vahdati, K.; Çakmakçı, R. Comprehensive biochemical insights into the seed germination of walnut under drought stress. Sci. Hortic. 2019, 250, 329–343. [Google Scholar] [CrossRef]
- Singh, J.; Sastry, E.V.D.; Singh, V. Effect of salinity on tomato (Lycopersicon esculentum Mill.) during seed germination stage. Physiol. Mol. Biol. Plants 2012, 18, 45–50. [Google Scholar] [CrossRef]
- Massaretto, I.L.; Albaladejo, I.; Purgatto, E.; Flores, F.B.; Plasencia, F.; Egea-Fernández, J.M.; Bolarin, M.C.; Egea, I. Recovering tomato landraces to simultaneously improve fruit yield and nutritional quality against salt stress. Front. Plant Sci. 2018, 9, 1778. [Google Scholar] [CrossRef]
- Maksimovic, I.; Ilin, Ž. Effects of salinity on vegetable growth and nutrients uptake. In Irrigation Systems and Practices in Challenging Environments; Lee, T.S., Ed.; InTech: Rijeka, Croatia, 2012; pp. 169–190. [Google Scholar]
- Campos, C.A.B.; Fernandes, P.D.; Gheyi, H.R.; Blanco, F.F.; Belem, G.C.; Ferreira, C.S.A. Yield and fruit quality of industrial tomato under saline irrigation. Sci. Agric. 2006, 63, 146–152. [Google Scholar] [CrossRef]
- Karlberg, L.; Ben-Gal, A.; Jansson, P.-E.; Shani, U. Modelling transpiration and growth in salinity-stressed tomato under different climatic conditions. Ecol. Modell. 2006, 190, 15–40. [Google Scholar] [CrossRef]
- Raza, M.A.; Saeed, A.; Munir, H.; Ziaf, K.; Shakeel, A.; Saeed, N.; Munawar, A.; Rehman, F. Screening of tomato genotypes for salinity tolerance based on early growth attributes and leaf inorganic osmolytes. Arch. Agron. Soil Sci. 2017, 63, 501–512. [Google Scholar] [CrossRef]
- Al-Taisan, W.A. Comparative effects of drought and salt stress on germination and seedling growth of Pennisetum divisum (Gmel.) Henr. Am. J. Appl. Sci. 2010, 7, 640–646. [Google Scholar] [CrossRef][Green Version]
- Reina-Sánchez, A.; Romero-Aranda, R.; Cuartero, J. Plant water uptake and water use efficiency of greenhouse tomato cultivars irrigated with saline water. Agric. Water Manag. 2005, 78, 54–66. [Google Scholar] [CrossRef]
- Cuartero, J.; Fernández-Muñoz, R. Tomato and salinity. Sci. Hortic. 1998, 78, 83–125. [Google Scholar] [CrossRef]
- Leontidou, K.; Genitsaris, S.; Papadopoulou, A.; Kamou, N.; Bosmali, I.; Matsi, T.; Madesis, P.; Vokou, D.; Karamanoli, K.; Mellidou, I. Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: Genomic characterisation and exploration of phyto-beneficial traits. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Ibrahim, W.; Ahmed, I.M.; Chen, X.; Cao, F.; Zhu, S.; Wu, F. Genotypic differences in photosynthetic performance, antioxidant capacity, ultrastructure and nutrients in response to combined stress of salinity and Cd in cotton. Biometals 2015, 28, 1063–1078. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Zaki, H.E.M.; Yokoi, S. A comparative in vitro study of salt tolerance in cultivated tomato and related wild species. Plant Biotechnol. 2016, 33, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Razali, R.; Bougouffa, S.; Morton, M.J.L.; Lightfoot, D.J.; Alam, I.; Essack, M.; Arold, S.T.; Kamau, A.; Schmöckel, S.M.; Pailles, Y.; et al. The genome sequence of the wild tomato Solanum pimpinellifolium provides insights into salinity tolerance. Front. Plant Sci. 2018, 9, 1402. [Google Scholar] [CrossRef]
- Ntatsi, G.; Aliferis, K.A.; Rouphael, Y.; Napolitano, F.; Makris, K.; Kalala, G.; Katopodis, G.; Savvas, D. Salinity source alters mineral composition and metabolism of Cichorium spinosum. Environ. Exp. Bot. 2017, 141, 113–123. [Google Scholar] [CrossRef]
- Sikder, R.K.; Wang, X.; Jin, D.; Zhang, H.; Gui, H.; Dong, Q.; Pang, N.; Zhang, X.; Song, M. Screening and evaluation of reliable traits of upland cotton (Gossypium hirsutum L.) genotypes for salt tolerance at the seedling growth stage. J. Cott. Res. 2020, 3, 11. [Google Scholar] [CrossRef]
- Chunthaburee, S.; Dongsansuk, A.; Sanitchon, J.; Pattanagul, W.; Theerakulpisut, P. Physiological and biochemical parameters for evaluation and clustering of rice cultivars differing in salt tolerance at seedling stage, Saudi. J. Biol. Sci. 2016, 23, 467–477. [Google Scholar]
- Chang, J.; Cheong, B.E.; Natera, S.; Roessner, U. Morphological and metabolic responses to salt stress of rice (Oryza sativa L.) cultivars which differ in salinity tolerance. Plant Physiol. Biochem. 2019, 144, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Akinci, S.; Yilmaz, K.; Akinci, I.E. Response of tomato (Lycopersicon esculentum Mill.) to salinity in the early growth stages for agricultural cultivation in saline environments. J. Environ. Biol. 2004, 25, 351–357. [Google Scholar]
- Rashid, M.H.O.; Islam, S.; Bari, M. In vitro screening for salt stress tolerance of native and exotic potato genotypes by morphological and physiological parameters. J. Bio-Sci. 2019, 28, 21–32. [Google Scholar] [CrossRef]
- Wu, H.; Guo, J.; Wang, C.; Li, K.; Zhang, X.; Yang, Z.; Li, M.; Wang, B. An effective screening method and a reliable screening trait for salt tolerance of Brassica napus at the germination stage. Front. Plant Sci. 2019, 10, 530. [Google Scholar] [CrossRef] [PubMed]
- Doğru, A.; Yılmaz Kaçar, M. A preliminary study on salt tolerance of some barley genotypes. Sak. Univ. J. Sci. 2019, 23, 755–762. [Google Scholar]
- Rajabi Dehnavi, A.; Zahedi, M.; Ludwiczak, A.; Cardenas Perez, S.; Piernik, A. Effect of salinity on seed germination and seedling development of Sorghum (Sorghum bicolor (L.) Moench) genotypes. Agronomy 2020, 10, 859. [Google Scholar] [CrossRef]
- Stevens, J.; Senaratna, T.; Sivasithamparam, K. Salicylic Acid Induces Salinity Tolerance in Tomato (Lycopersicon esculentum cv. Roma): Associated Changes in Gas Exchange, Water Relations and Membrane Stabilisation. Plant Growth Regul. 2006, 49, 77–83. [Google Scholar]
- Mellidou, I.; Karamanoli, K.; Constantinidou, H.-I.A.; Roubelakis-Angelakis, K.A. Antisense-mediated S-adenosyl-L-methionine decarboxylase silencing affects heat stress responses of tobacco plants. Funct. Plant Biol. 2020, 47, 651–658. [Google Scholar] [CrossRef]
- De la Torre-González, A.; Navarro-León, E.; Albacete, A.; Blasco, B.; Ruiz, J.M. Study of phytohormone profile and oxidative metabolism as key process to identification of salinity response in tomato commercial genotypes. J. Plant Physiol. 2017, 216, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Wungrampha, S.; Joshi, R.; Singla-Pareek, S.L.; Pareek, A. Photosynthesis and salinity: Are these mutually exclusive? Photosynthetica 2018, 56, 366–381. [Google Scholar] [CrossRef]
- Haghighi, M.; Pessarakli, M. Influence of silicon and nano-silicon on salinity tolerance of cherry tomatoes (Solanum lycopersicum L.) at early growth stage. Sci. Hortic. 2013, 161, 111–117. [Google Scholar] [CrossRef]
- Rivero, R.M.; Mestre, T.C.; Mittler, R.; Rubio, F.; Garcia-Sanchez, F.; Martinez, V. The combined effect of salinity and heat reveals a specific physiological, biochemical and molecular response in tomato plants. Plant Cell Environ. 2014, 37, 1059–1073. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Yan, F.; Hu, L.P.; Zhou, X.T.; Zou, Z.R.; Cui, L.R. Effects of exogenous 5-aminolevulinic acid on photosynthesis, stomatal conductance, transpiration rate, and PIP gene expression of tomato seedlings subject to salinity stress. Genet. Mol. Res. 2015, 14, 6401–6412. [Google Scholar] [CrossRef] [PubMed]
- Mellidou, I.; Karamanoli, K.; Beris, D.; Haralampidis, K.; Constantinidou, H.-I.A.; Roubelakis-Angelakis, K. Underexpression of apoplastic polyamine oxidase improves thermotolerance in Nicotiana tabacum. J. Plant Physiol. 2017, 218, 171–174. [Google Scholar] [CrossRef]
- Taiz, L.; Zeiger, E.; Møller, I.M.; Murphy, A. Plant Physiology and Development, 6th ed.; Sinauer Associates, Inc., Publishers: Sunderland, MA, USA, 2015; pp. 269–279. [Google Scholar]
- Misra, A.N.; Srivastava, A.; Strasser, R.J. Utilization of fast chlorophyll a fluorescence technique in assessing the salt/ion sensitivity of mung bean and Brassica seedlings. J. Plant Physiol. 2001, 158, 1173–1181. [Google Scholar] [CrossRef]
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.J.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Mahlooji, M.; Seyed Sharifi, R.; Razmjoo, J.; Sabzalian, M.R.; Sedghi, M. Effect of salt stress on photosynthesis and physiological parameters of three contrasting barley genotypes. Photosynthetica 2018, 56, 549–556. [Google Scholar] [CrossRef]
- Shao, H.; Chu, L.; Jaleel, C.A.; Zhao, C. Water-deficit stress-induced anatomical changes in higher plants. C. R. Biol. 2008, 331, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Aghaie, P.; Hosseini Tafreshi, S.A.; Ebrahimi, M.A.; Haerinasab, M. Tolerance evaluation and clustering of fourteen tomato cultivars grown under mild and severe drought conditions. Sci. Hortic. 2018, 232, 1–12. [Google Scholar] [CrossRef]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M. Abiotic Stress and Reactive Oxygen Species: Generation. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Davey, M.W.; Stals, E.; Panis, B.; Keulemans, J.; Swennenet, R.L. High-throughput determination of malondialdehyde in plant tissues. Anal. Biochem. 2005, 347, 201–207. [Google Scholar] [CrossRef]
- Zeeshan, M.; Lu, M.; Sehar, S.; Holford, P.; Wu, F. Comparison of biochemical, anatomical, morphological, and physiological responses to salinity stress in wheat and barley genotypes deferring in salinity tolerance. Agronomy 2020, 10, 127. [Google Scholar] [CrossRef]
- Katsuhara, M.; Otsuka, T.; Ezaki, B. Salt stress-induced lipid peroxidation is reduced by glutathione S-transferase, but this reduction of lipid peroxides is not enough for a recovery of root growth in Arabidopsis. Plant Sci. 2005, 169, 369–373. [Google Scholar] [CrossRef]
- Ashraf, M.; Ali, Q. Relative membrane permeability and activities of some antioxidant enzymes as the key determinants of salt tolerance in canola (Brassica napus L.). Environ. Exp. Bot. 2008, 63, 266–273. [Google Scholar] [CrossRef]
- Farooq, M.; Hussain, M.; Wakeel, A.; Siddique, K.H.M. Salt stress in maize: Effects, resistance mechanisms, and management. A review. Agron. Sustain. Dev. 2015, 35, 461–481. [Google Scholar] [CrossRef]
- Alves, R.d.C.; Rossatto, D.R.; da Silva, J.S.; Checchio, M.V.; de Oliveira, K.R.; Oliveira, F.A.; de Queiroz, S.F.; da Cruz, M.C.P.; Gratão, P.L. Seed priming with ascorbic acid enhances salt tolerance in micro-tom tomato plants by modifying the antioxidant defense system components. Biocatal. Agric. Biotechnol. 2021, 31, 101927. [Google Scholar] [CrossRef]
- Mellidou, I.; Krommydas, K.; Nianiou-Obeidat, I.; Ouzounidou, G.; Kalivas, A.; Ganopoulos, I. Exploring morpho-physiological profiles of a collection of tomato (Solanum lycopersicum) germplasm using multivariate statistics. Plant Genet. Resour. Characterisation Util. 2020, 18, 88–97. [Google Scholar] [CrossRef]
- McKay, H. Root electrolyte leakage and root growth potential as indicators of spruce and larch establishment. Silva Fenn. 1998, 32, 241–252. [Google Scholar] [CrossRef][Green Version]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Pateraki, I.; Sanmartin, M.; Kalamaki, M.S.; Gerasopoulos, D.; Kanellis, A. Molecular characterization and expression studies during melon fruit development and ripening of L-galactono-1,4-lactone dehydrogenase. J. Exp. Bot. 2004, 55, 1623–1633. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]

| Genotypes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trait | NaCl Level | LA1579 | AC | IL12-4 | V1 | V2 | V3 | V4 | V5 | V6 |
| SL (cm) | 0 mM | 28.3 ab | 28.4 ab | 23.7 cdefg | 21.9 fghi | 23.7 cdefg | 25.3 bcdef | 23.1 defgh | 26.8 abc | 29.2 a |
| 200 mM | 26.5 abcd | 22.3 fghi | 20.7 ghi | 20.9 ghi | 19.6 hi | 22.6 efgh | 18.9 i | 20.3 ghi | 26.0 abcde | |
| % Change | −6.5 | −21.6 * | −10.9 | −4.6 | −17.5 * | −10.7 | −18.2 * | −24.3 * | −11 | |
| LN | 0 mM | 6.6 abc | 7.4 a | 5.1 efg | 7.0 ab | 5.1 efg | 5.3 defg | 5.9 cdef | 4.9 fg | 5.2 efg |
| 200 mM | 6.2 bcde | 6.3 abcd | 5.3 defg | 6.2 bcde | 4.3 g | 4.7 g | 5.0 fg | 4.4 g | 4.8 fg | |
| % Change | −5.3 | −14.4 | 2.8 | −11.4 | −15.7 | −11.3 | −14.6 | −8.5 | −7.7 | |
| ST (mm) | 0 mM | 3.89 fg | 4.79 b | 4.67 bc | 4.70 bc | 4.78 b | 4.77 b | 4.85 ab | 4.91 ab | 5.07 a |
| 200 mM | 3.15 i | 3.70 gh | 3.49 h | 4.49 cd | 3.46 h | 4.12 ef | 3.71 gh | 3.87 fg | 4.25 de | |
| % Change | −18.9 * | −22.7 * | −25.3 * | −4.5 | −27.7 * | −13.7 * | −23.5 * | −21.2 * | −16.2 * | |
| RGR | 0 mM | 0.119 a | 0.042 h | 0.087 c | 0.050 g | 0.076 e | 0.082 d | 0.081 d | 0.067 f | 0.091 b |
| 200 mM | 0.089 bc | 0.015 k | 0.026 i | 0.023 j | 0.042 h | 0.050 g | 0.043 h | 0.024 ij | 0.066 f | |
| % Change | −25.2 * | −63.7 * | −70.5 * | −52.8 * | −45.1 * | −39 * | −46.1 * | −63.7 * | −27 * | |
| Genotypes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trait | NaCl Level | LA1579 | AC | IL12-4 | V1 | V2 | V3 | V4 | V5 | V6 |
| SFW (g) | 0 mM | 4.29 bcde | 5.58 ab | 4.77 abcd | 6.26 a | 5.16 abc | 6.11 a | 5.63 ab | 5.35 abc | 6.12 a |
| 200 mM | 3.82 cde | 4.14 bcde | 2.81 e | 4.89 abcd | 3.43 de | 4.26 bcde | 3.69 cde | 4.07 bcde | 5.13 abcd | |
| % Change | −10.9 | −25.8 | −41.2 * | −21.8 | −33.5 * | −30.3 * | −34.5 * | −24 | −16.3 | |
| % SDMC | 0 mM | 14.18 ab | 11.93 cde | 14.70 a | 12.52 bcde | 12.48 bcde | 11.68 de | 13.01 abcd | 12.22 cde | 11.96 cde |
| 200 mM | 12.44 bcde | 12.88 bcde | 14.07 ab | 12.65 bcde | 13.48 abc | 12.49 bcde | 14.07 ab | 11.13 e | 11.66 de | |
| % Change | −12.3 | 8 | −4.3 | 1 | 8 | 7 | 8.1 | −9 | −2.6 | |
| RFW (g) | 0 mM | 1.18 cdef | 1.38 bcd | 1.36 bcd | 1.96 a | 1.21 cde | 1.59 abc | 1.63 abc | 1.33 bcd | 1.72 ab |
| 200 mM | 0.61 g | 1.04 defg | 0.75 efg | 1.18 cdef | 0.70 fg | 1.19 cdef | 1.18 cdef | 0.99 defg | 1.17 cdef | |
| % Change | −48.7 * | −24.5 | −44.9 * | −39.6 * | −42.2 * | −25.1 | −27.6 | −26 | −31.9 * | |
| % RDMC | 0 mM | 10.38 bcdef | 8.92 defg | 10.73 bcdef | 8.53 efg | 9.70 cdefg | 7.89 g | 9.46 cdefg | 8.55 efg | 8.43 fg |
| 200 mM | 14.75 a | 10.88 bcde | 11.01 bcd | 10.09 cdefg | 12.50 ab | 9.35 cdefg | 11.53 bc | 8.75 defg | 10.43 bcdef | |
| % Change | 42 * | 22 | 2.6 | 18.2 | 28.8 * | 18.5 | 21.8 | 2.3 | 23.8 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadoglidou, K.; Xanthopoulou, A.; Kalyvas, A.; Mellidou, I. Utilization of Tomato Landraces to Improve Seedling Performance under Salt Stress. Stresses 2021, 1, 238-252. https://doi.org/10.3390/stresses1040017
Kadoglidou K, Xanthopoulou A, Kalyvas A, Mellidou I. Utilization of Tomato Landraces to Improve Seedling Performance under Salt Stress. Stresses. 2021; 1(4):238-252. https://doi.org/10.3390/stresses1040017
Chicago/Turabian StyleKadoglidou, Kalliopi, Aliki Xanthopoulou, Apostolos Kalyvas, and Ifigeneia Mellidou. 2021. "Utilization of Tomato Landraces to Improve Seedling Performance under Salt Stress" Stresses 1, no. 4: 238-252. https://doi.org/10.3390/stresses1040017
APA StyleKadoglidou, K., Xanthopoulou, A., Kalyvas, A., & Mellidou, I. (2021). Utilization of Tomato Landraces to Improve Seedling Performance under Salt Stress. Stresses, 1(4), 238-252. https://doi.org/10.3390/stresses1040017







