Comparison between the Nature and Activity of Silver Nanoparticles Produced by Active and Inactive Fungal Biomass Forms on Cervical Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Cultivation
2.2. Biomass Pretreatments
2.3. SNPs Production
2.4. Characterization of the Produced SNPs
2.5. Spectrophotometry
2.6. Transmission Electron Microscopy (TEM)
2.7. X-ray Diffraction Analysis (XRD)
2.8. Zetasizer Analysis
2.9. SNPs Purification
2.10. Determination of the SNPs Concentration
2.11. Antibacterial Activity Test
2.12. MTT Assay
2.13. Apoptosis Induction Assay
3. Results
3.1. Fungal Cultivation
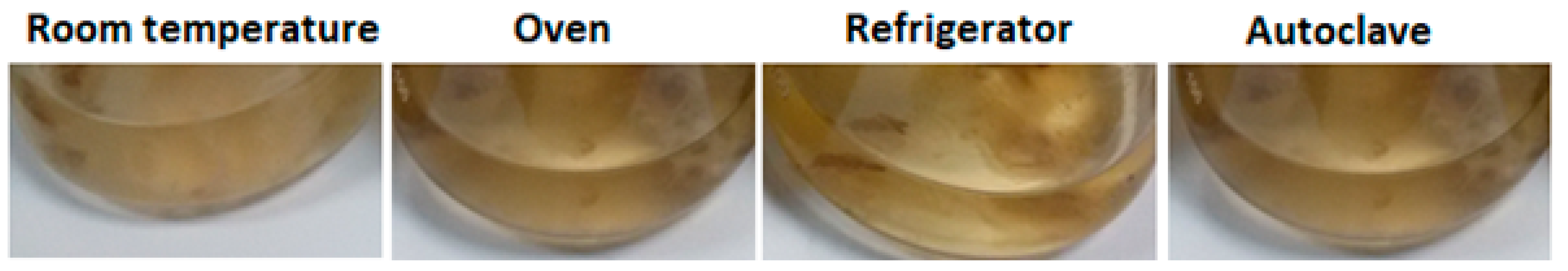
3.2. Pretreatments of the Biomass
3.3. SNPs Production
4. Characterization of the Produced SNPs
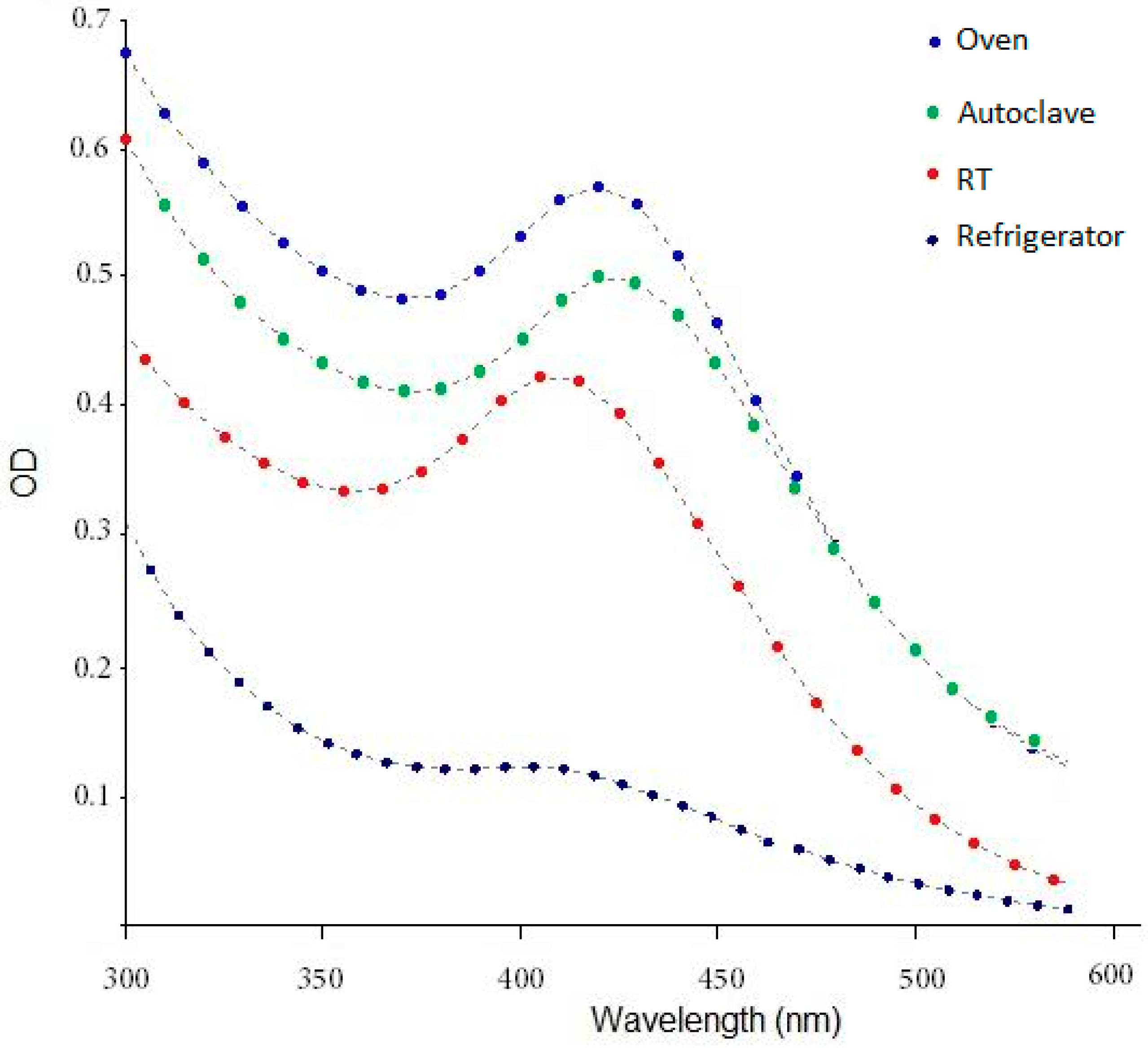
4.1. Spectrophotometry
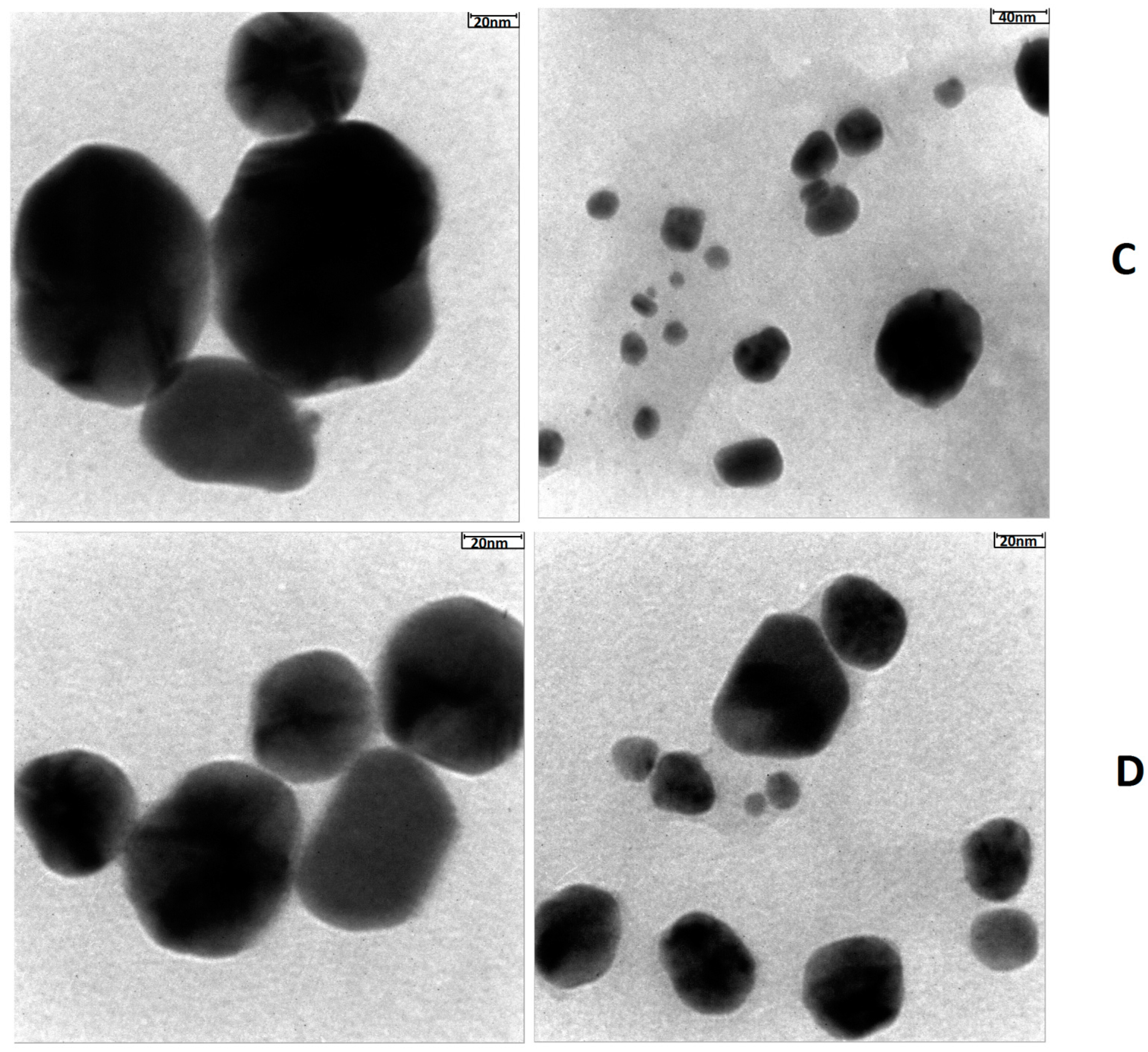
4.2. TEM Analysis Results
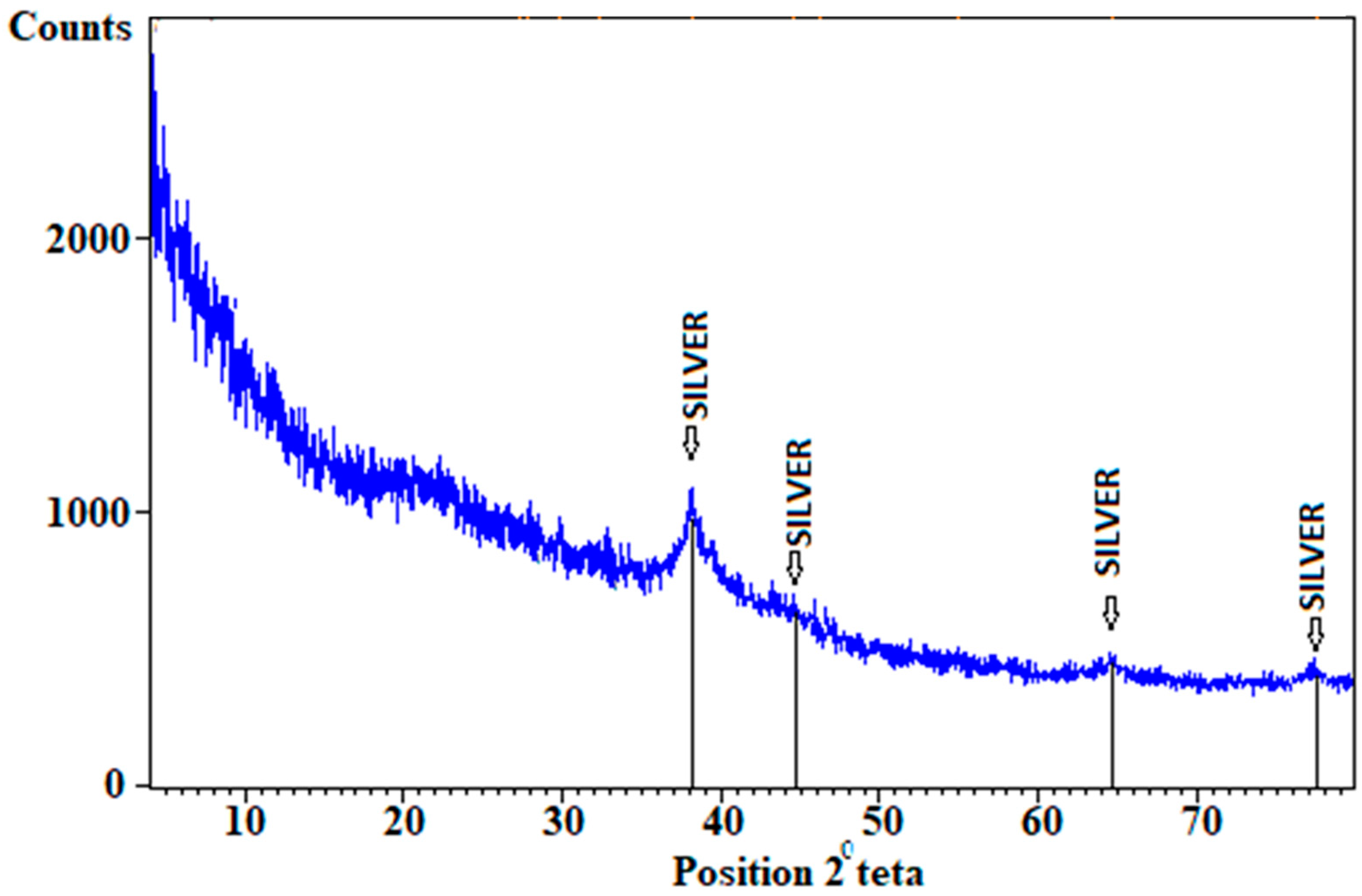
4.3. XRD Result Data
4.4. Zetasizer Analysis
4.5. Determination of the SNPs’ Concentration
4.6. Antibacterial Activity Test
4.7. MTT Assay
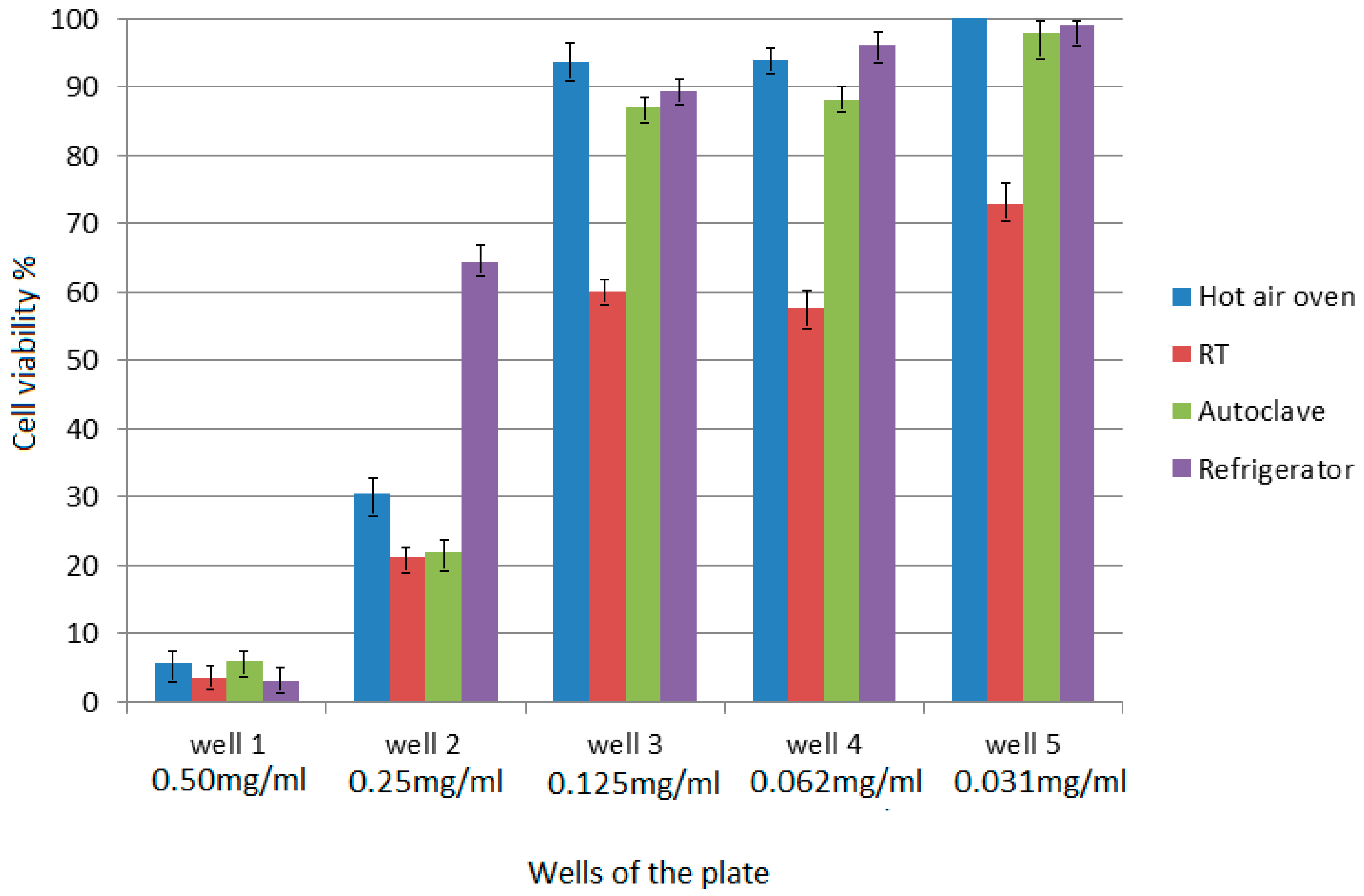
4.8. Apoptosis Induction Assay
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Alkilany, A.M.; Murphy, C.J. Toxicity and cellular uptake of gold nanoparticles: What we have learned so far? J. Nanopart. Res. 2010, 12, 2313–2333. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-S.; Hung, Y.-C.; Liau, I.; Huang, G.S. Assessment of the in vivo toxicity of gold nanoparticles. Nanoscale Res. Lett. 2009, 4, 858. [Google Scholar] [CrossRef] [PubMed]
- Sneha, K.; Sathishkumar, M.; Mao, J.; Kwak, I.; Yun, Y.-S. Corynebacterium glutamicum-mediated crystallization of silver ions through sorption and reduction processes. Chem. Eng. J. 2010, 162, 989–996. [Google Scholar] [CrossRef]
- Binupriya, A.; Sathishkumar, M.; Yun, S.-I. Biocrystallization of silver and gold ions by inactive cell filtrate of Rhizopus stolonifer. Colloids Surf. B Biointerfaces 2010, 79, 531–534. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.B.; Sakthivel, N. Biological synthesis of metal nanoparticles by microbes. Adv. Colloid Interface Sci. 2010, 156, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mata, Y.; Torres, E.; Blazquez, M.; Ballester, A.; González, F.; Munoz, J. Gold (III) biosorption and bioreduction with the brown alga Fucus vesiculosus. J. Hazard. Mater. 2009, 166, 612–618. [Google Scholar] [CrossRef]
- Fanti, J.R.; Tomiotto-Pellissier, F.; Miranda-Sapla, M.M.; Cataneo, A.H.D.; de Jesus Andrade, C.G.T.; Panis, C.; da Silva Rodrigues, J.H.; Wowk, P.F.; Kuczera, D.; Costa, I.N. Biogenic silver nanoparticles inducing Leishmania amazonensis promastigote and amastigote death in vitro. Acta Trop. 2018, 178, 46–54. [Google Scholar] [CrossRef]
- Almeida, É.S.; de Oliveira, D.; Hotza, D. Characterization of silver nanoparticles produced by biosynthesis mediated by Fusarium oxysporum under different processing conditions. Bioprocess Biosyst. Eng. 2017, 40, 1291–1303. [Google Scholar] [CrossRef]
- Ahmed, A.-A.; Hamzah, H.; Maaroof, M. Analyzing formation of silver nanoparticles from the filamentous fungus Fusarium oxysporum and their antimicrobial activity. Turk. J. Biol. 2018, 42, 54–62. [Google Scholar] [CrossRef]
- Asghari-Paskiabi, F.; Imani, M.; Razzaghi-Abyaneh, M.; Rafii-Tabar, H. Fusarium oxysporum, a bio-Factory for Nano Selenium Compounds: Synthesis and Characterization. Sci. Iran. 2018, 25, 1857–1863. [Google Scholar] [CrossRef]
- Pourali, P.; Badiee, S.H.; Manafi, S.; Noorani, T.; Rezaei, A.; Yahyaei, B. Biosynthesis of gold nanoparticles by two bacterial and fungal strains, Bacillus cereus and Fusarium oxysporum, and assessment and comparison of their nanotoxicity in vitro by direct and indirect assays. Electron. J. Biotechnol. 2017, 29, 86–93. [Google Scholar] [CrossRef]
- Pourali, P.; Yahyaei, B.; Afsharnezhad, S. Bio-Synthesis of Gold Nanoparticles by Fusarium oxysporum and Assessment of Their Conjugation Possibility with Two Types of β-Lactam Antibiotics without Any Additional Linkers. Microbiology 2018, 87, 229–237. [Google Scholar] [CrossRef]
- Yahyaei, B.; Peyvandi, N.; Akbari, H.; Arabzadeh, S.; Afsharnezhad, S.; Ajoudanifar, H.; Pourali, P. Production, assessment, and impregnation of hyaluronic acid with silver nanoparticles that were produced by Streptococcus pyogenes for tissue engineering applications. Appl. Biol. Chem. 2016, 59, 227–237. [Google Scholar] [CrossRef]
- Yahyaei, B.; Manafi, S.; Fahimi, B.; Arabzadeh, S.; Pourali, P. Production of electrospun polyvinyl alcohol/microbial synthesized silver nanoparticles scaffold for the treatment of fungating wounds. Appl. Nanosci. 2018, 8, 417–426. [Google Scholar] [CrossRef]
- Pourali, P.; Yahyaei, B.; Ajoudanifar, H.; Taheri, R.; Alavi, H.; Hoseini, A. Impregnation of the bacterial cellulose membrane with biologically produced silver nanoparticles. Curr. Microbiol. 2014, 69, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Braydich-Stolle, L.K.; Lucas, B.; Schrand, A.; Murdock, R.C.; Lee, T.; Schlager, J.J.; Hussain, S.M.; Hofmann, M.-C. Silver nanoparticles disrupt GDNF/Fyn kinase signaling in spermatogonial stem cells. Toxicol. Sci. 2010, 116, 577–589. [Google Scholar] [CrossRef]
- Pourali, P.; Razavian Zadeh, N.; Yahyaei, B. Silver nanoparticles production by two soil isolated bacteria, Bacillus thuringiensis and Enterobacter cloacae, and assessment of their cytotoxicity and wound healing effect in rats. Wound Repair Regen. 2016, 24, 860–869. [Google Scholar] [CrossRef]
- Song, H.-P.; Li, X.-G.; Sun, J.-S.; Xu, S.-M.; Han, X. Application of a magnetotactic bacterium, Stenotrophomonas sp. to the removal of Au (III) from contaminated wastewater with a magnetic separator. Chemosphere 2008, 72, 616–621. [Google Scholar] [CrossRef]
- Park, S.; Lee, W.J.; Park, S.; Choi, D.; Kim, S.; Park, N. Reversibly pH-responsive gold nanoparticles and their applications for photothermal cancer therapy. Sci. Rep. 2019, 9, 20180. [Google Scholar] [CrossRef]
- Yahyaei, B.; Pourali, P. One step conjugation of some chemotherapeutic drugs to the biologically produced gold nanoparticles and assessment of their anticancer effects. Sci. Rep. 2019, 9, 10242. [Google Scholar] [CrossRef]
- Lomelí-Marroquín, D.; Cruz, D.M.; Nieto-Argüello, A.; Crua, A.V.; Chen, J.; Torres-Castro, A.; Webster, T.J.; Cholula-Díaz, J.L. Starch-mediated synthesis of mono-and bimetallic silver/gold nanoparticles as antimicrobial and anticancer agents. Int. J. Nanomed. 2019, 14, 2171. [Google Scholar] [CrossRef]
- Pourali, P.; Baserisalehi, M.; Afsharnezhad, S.; Behravan, J.; Ganjali, R.; Bahador, N.; Arabzadeh, S. The effect of temperature on antibacterial activity of biosynthesized silver nanoparticles. Biometals 2013, 26, 189–196. [Google Scholar] [CrossRef]
- Zhou, Y.; Kong, Y.; Kundu, S.; Cirillo, J.D.; Liang, H. Antibacterial activities of gold and silver nanoparticles against Escherichia coli and bacillus Calmette-Guérin. J. Nanobiotechnol. 2012, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Qian, J.; Zhou, H.; Gan, Q.; Tang, W.; Lu, J.; Yuan, Y.; Liu, C. In vitro cytotoxicity and induction of apoptosis by silica nanoparticles in human HepG2 hepatoma cells. Int. J. Nanomed. 2011, 6, 1889. [Google Scholar]
- Altunbek, M.; Kuku, G.; Culha, M. Gold nanoparticles in single-cell analysis for surface enhanced Raman scattering. Molecules 2016, 21, 1617. [Google Scholar] [CrossRef] [PubMed]
- Mehta, B.; Chhajlani, M.; Shrivastava, B. Green synthesis of silver nanoparticles and their characterization by XRD. J. Phys. Conf. Ser. 2017, 836, 012050. [Google Scholar] [CrossRef]
- Shaligram, N.S.; Bule, M.; Bhambure, R.; Singhal, R.S.; Singh, S.K.; Szakacs, G.; Pandey, A. Biosynthesis of silver nanoparticles using aqueous extract from the compactin producing fungal strain. Process Biochem. 2009, 44, 939–943. [Google Scholar] [CrossRef]
- Binupriya, A.R.; Sathishkumar, M.; Yun, S.-I. Myco-crystallization of silver ions to nanosized particles by live and dead cell filtrates of Aspergillus oryzae var. viridis and its bactericidal activity toward Staphylococcus aureus KCCM 12256. Ind. Eng. Chem. Res. 2009, 49, 852–858. [Google Scholar] [CrossRef]
- Salvadori, M.R.; Lepre, L.F.; Ando, R.m.A.; do Nascimento, C.A.O.; Corrêa, B. Biosynthesis and uptake of copper nanoparticles by dead biomass of Hypocrea lixii isolated from the metal mine in the Brazilian Amazon region. PLoS ONE 2013, 8, e80519. [Google Scholar] [CrossRef]
- Salvadori, M.R.; Nascimento, C.A.O.; Corrêa, B. Nickel oxide nanoparticles film produced by dead biomass of filamentous fungus. Sci. Rep. 2014, 4, 6404. [Google Scholar] [CrossRef]
- Foldbjerg, R.; Dang, D.A.; Autrup, H. Cytotoxicity and genotoxicity of silver nanoparticles in the human lung cancer cell line, A549. Arch. Toxicol. 2011, 85, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Cheng, F.-Y.; Chiu, H.-W.; Tsai, J.-C.; Fang, C.-Y.; Chen, C.-W.; Wang, Y.-J. Cytotoxicity, oxidative stress, apoptosis and the autophagic effects of silver nanoparticles in mouse embryonic fibroblasts. Biomaterials 2014, 35, 4706–4715. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Degheidy, H.; Casey, B.J.; Goering, P.L. Flow cytometry evaluation of in vitro cellular necrosis and apoptosis induced by silver nanoparticles. Food Chem. Toxicol. 2015, 85, 45–51. [Google Scholar] [CrossRef] [PubMed]




| The Produced SNPs after Different Pretreatments | Z-Average (nm) | Zeta Potential (mV) |
|---|---|---|
| Pretreated in a hot air oven | 60 ± 2 | −21.23 ± 1.10 |
| Pretreated in a refrigerator | 62 ± 5 | −19.36 ± 0.54 |
| Pretreated at RT | 55 ± 4 | −21.34 ± 0.97 |
| Pretreated in an autoclave | 52 ± 4 | −21.35 ± 0.45 |
| Sample | Wavelength (nm): 328.068, Element: Ag (1 mg/mL) |
|---|---|
| Concentration (ppm) (Mean ± sd) | |
| Blank (ppm) | 0.00 ± 0.00 |
| Pretreated in a hot air oven | 3.20 ± 0.02 |
| Pretreated in a refrigerator | 2.63 ± 0.00 |
| Pretreated at RT | 3.52 ± 0.02 |
| Pretreated in an autoclave | 3.58 ± 0.05 |
| The Produced SNPs after Different Pretreatments | Inhibition Zones (mm) of the SNPs against the Tested Bacterial Strains (Mean ± sd) | ||
|---|---|---|---|
| E. coli | P. aeruginosa | S. aureus | |
| Hot air oven | 08.0 ± 0.4 | 11.0 ± 1.0 | 10.5 ± 0.9 |
| Refrigerator | 10.0 ± 0.7 | 07.0 ± 0.6 | 10.5 ± 0.2 |
| RT | 11.0 ± 0.8 | 11.0 ± 0.2 | 11.0 ± 0.3 |
| Autoclave | 10.0 ± 0.7 | 11.0 ± 0.6 | 11.0 ± 0.6 |
| Silver nitrate | 12.5 ± 0.6 | 12.0 ± 0.6 | 11.0 ± 0.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pourali, P.; Nouri, M.; Heidari, T.; Kheirkhahan, N.; Yahyaei, B. Comparison between the Nature and Activity of Silver Nanoparticles Produced by Active and Inactive Fungal Biomass Forms on Cervical Cancer Cells. Nanomanufacturing 2023, 3, 248-262. https://doi.org/10.3390/nanomanufacturing3020016
Pourali P, Nouri M, Heidari T, Kheirkhahan N, Yahyaei B. Comparison between the Nature and Activity of Silver Nanoparticles Produced by Active and Inactive Fungal Biomass Forms on Cervical Cancer Cells. Nanomanufacturing. 2023; 3(2):248-262. https://doi.org/10.3390/nanomanufacturing3020016
Chicago/Turabian StylePourali, Parastoo, Mahnaz Nouri, Tana Heidari, Niloufar Kheirkhahan, and Behrooz Yahyaei. 2023. "Comparison between the Nature and Activity of Silver Nanoparticles Produced by Active and Inactive Fungal Biomass Forms on Cervical Cancer Cells" Nanomanufacturing 3, no. 2: 248-262. https://doi.org/10.3390/nanomanufacturing3020016
APA StylePourali, P., Nouri, M., Heidari, T., Kheirkhahan, N., & Yahyaei, B. (2023). Comparison between the Nature and Activity of Silver Nanoparticles Produced by Active and Inactive Fungal Biomass Forms on Cervical Cancer Cells. Nanomanufacturing, 3(2), 248-262. https://doi.org/10.3390/nanomanufacturing3020016






