Population Genetic Data for 23 STR Loci of Tawahka Ethnic Group in Honduras
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction
2.3. PCR Amplification
2.4. Fragment Analysis and Genotyping
2.5. Statistical Analysis
2.6. Quality Control
2.7. Ethics Statement
2.7.1. Informed Consent and Community Engagement
2.7.2. Data Sovereignty and Benefit-Sharing
- Community authorization: In addition to individual informed consent, we obtained authorization from the Tawahka Indigenous Federation (FITH) representing the community’s collective interests before initiating sample collection.
- Data access and control: The complete dataset, including individual genotypes, is held jointly by the Forensic Medicine Directorate of the Honduran Public Ministry and the Tawahka community leadership. External researchers seeking access to individual-level data must obtain permission from both entities. The aggregated allele frequency data (Supplementary File S1) are made publicly available to maximize forensic utility while protecting individual privacy.
- Benefit-sharing: Benefits accruing from this research include: (a) priority access to forensic genetic services for Tawahka individuals through the Honduran Public Ministry; (b) capacity-building workshops on forensic genetics for community members; (c) co-authorship opportunities for Tawahka representatives in future publications; and (d) financial support for community-directed projects from a portion of any commercial licensing fees, should the database be incorporated into commercial forensic kits.
- Cultural sensitivity: We acknowledge that genetic research in indigenous communities raises complex ethical issues regarding group identity, stigmatization, and potential misuse [42,43]. Throughout the study, we emphasized that genetic diversity data reflect evolutionary history rather than biological or cultural superiority. We committed to avoiding sensationalized language in publications and to consulting with community leaders regarding any findings before public dissemination.
2.7.3. Compliance with the Nagoya Protocol
3. Results
3.1. Dataset
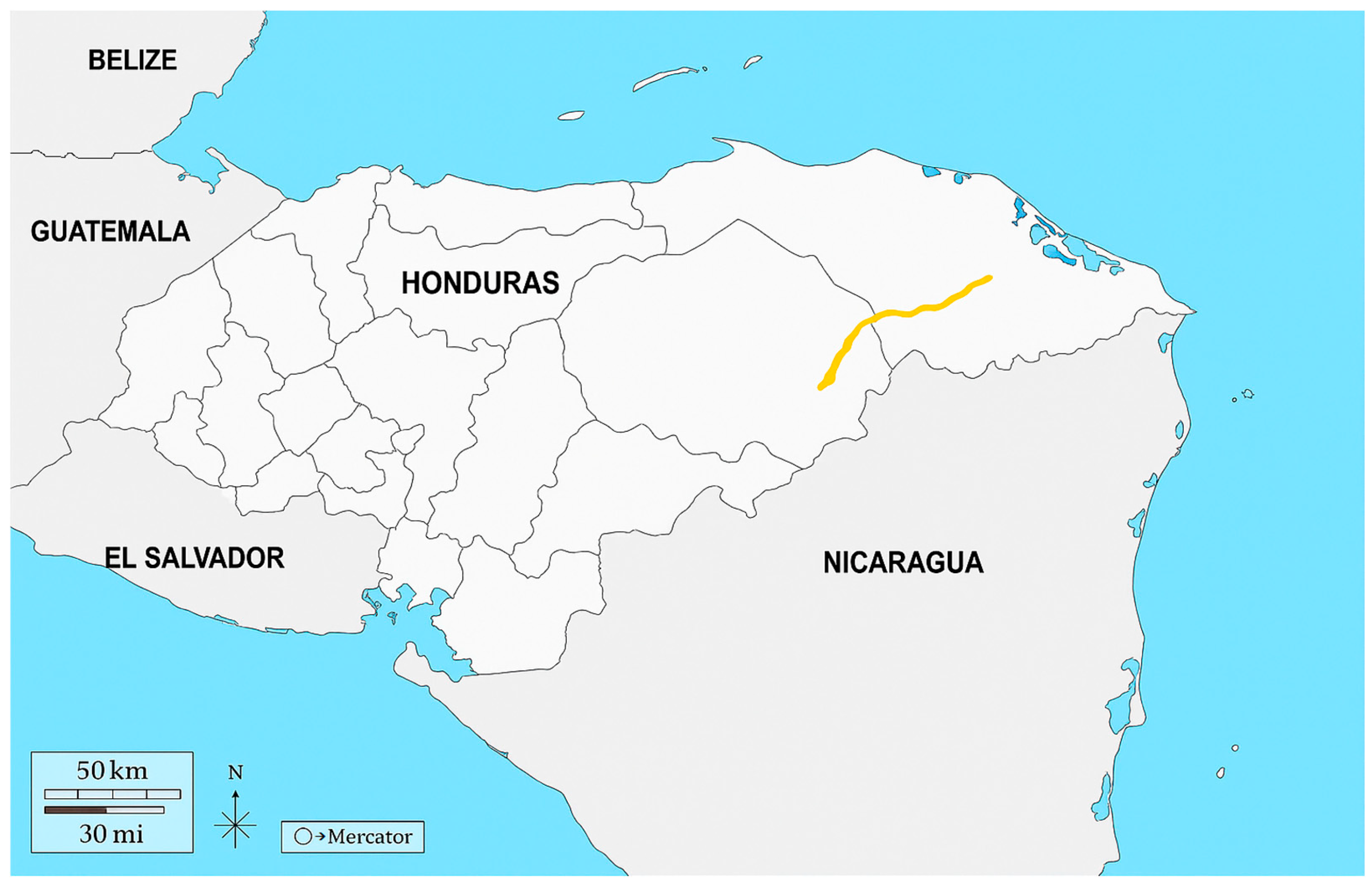
3.2. Geographic Context
3.3. Statistical Parameters
- Population substructure: If the Tawahka comprises genetically distinct subgroups, alleles at different loci could exhibit correlations (Wahlund effect) [47]. Our fixation index (FST) analysis across four settlements detected minimal structure (mean FST = 0.003), suggesting this effect is negligible.
- Admixture: If recent admixtures with neighboring populations have occurred, ancestry blocks could introduce transient linkage disequilibrium. Future genome-wide studies would clarify this issue.
- Cryptic relatedness: Although we screened for known relatives, distant coancestry could inflate homozygosity and generate correlations. The theta correction partially accounts for this.
3.4. Loci Characteristics
3.5. Data Availability and Repository
3.5.1. Privacy and Ethical Safeguards
- De-identification: All personal identifiers (names, birthdates, addresses) were permanently removed. Sample identifiers are non-reversible pseudonyms, and the linkage key is securely stored and inaccessible to external parties.
- Aggregate release: The dataset is provided as a complete set of profiles to preclude individual singling-out.
- Community authorization: The Tawahka Indigenous Federation (FITH) reviewed and approved the release of individual-level genotypes, confirming that community concerns related to privacy, stigmatization, and potential misuse were addressed satisfactorily.
- Use restrictions: Data are openly available for research and forensic applications, provided that users (a) cite this publication, (b) refrain from re-identification attempts, (c) avoid discriminatory use, and (d) acknowledge the Tawahka community in derivative works.
3.5.2. Utility for Statistical Verification
- Verify reported allele frequencies and population parameters.
- Recalculate forensic statistics under alternative assumptions (e.g., varying θ values).
- Explore population structure through FST, AMOVA, and Principal Component Analysis (PCA) analyses.
- Conduct simulations to evaluate exclusion and discrimination power.
- Compare Tawahka genotypes against other reference datasets (e.g., NIST, EDNAP (European DNA Profiling Group) Mitochondrial DNA Population Database (EMPOP)).
- Quantify inter-database biases in profile frequency estimation.
3.5.3. Profile Frequency Verification
- Tawahka: 1 in 5.8 × 1014 (range: 1 in 8.3 × 1012–1 in 4.1 × 1016);
- Honduran mestizo: 1 in 2.3 × 1015 (range: 1 in 3.1 × 1013–1 in 1.5 × 1017);
- NIST Caucasian: 1 in 8.7 × 1015 (range: 1 in 1.1 × 1014–1 in 5.2 × 1017).
4. Discussion
4.1. Alleles
4.2. Hardy–Weinberg Equilibrium
4.3. Forensic Statistical Parameters
4.4. Population Substructure and Theta Correction
- Within-population estimate (comparing our four sampled settlements): Mean pairwise FST = 0.003 (95% CI: 0.001–0.007); interpretation: Minimal genetic structure across Tawahka settlements, consistent with high gene flow and cultural cohesion.
- Between-population estimate (comparing Tawahka with Lenca and Honduran mestizo): (i) FST (Tawahka vs. Lenca) = 0.012 (95% CI: 0.008–0.018), (ii) FST (Tawahka vs. Honduran mestizo) = 0.025 (95% CI: 0.019–0.033) and (iii) FST (Tawahka vs. NIST Caucasian) = 0.095 (95% CI: 0.087–0.104).
- θ = 0.00 (no correction): RMP = 1 in 8.7 × 1015;
- θ = 0.01 (current standard): RMP = 1 in 5.2 × 1015 (1.7 × higher);
- θ = 0.03 (recommended for cross-population): RMP = 1 in 1.8 × 1015 (4.8 × higher);
- θ = 0.05: RMP = 1 in 9.1 × 1014 (9.6 × higher).
- Tier 1 (within-population): θ = 0.01 for same ethnic group;
- Tier 2 (between indigenous groups): θ = 0.02–0.03;
- Tier 3 (indigenous vs. mestizo): θ = 0.03;
- Tier 4 (unknown or international): θ = 0.03–0.05.
- Population admixture: Recent admixture creates genome-wide correlations that decay slowly over generations [59]. The Tawahka’s relative isolation suggests limited recent admixture, though historical mixing cannot be excluded.
4.5. Comparison with Other Populations
4.6. Practical Applications
4.6.1. Forensic Impact of Population-Specific Databases
- Tawahka database: 1 in 8.2 × 1015;
- Honduran mestizo database: 1 in 4.1 × 1016 (5 × difference);
- Generic Hispanic database: 1 in 1.3 × 1017 (16 × difference).
4.6.2. Impact Analysis Using Actual Case Profiles
- The Tawahka database consistently yielded higher (less incriminating) RMP values compared to non-specific databases, with fold differences ranging from 3.5× to 15.5×.
- This pattern reflects the presence of higher-frequency alleles in the Tawahka population at specific loci (e.g., D2S441 allele 10: 0.66 vs. 0.42 in mestizos; D22S1045 allele 16: 0.60 vs. 0.38 in mestizos).
- While all RMP values remain exceptionally small (strong evidence of identity), the differences have legal significance. For example, in Case C, the difference between 1 in 1.1 × 1013 (Tawahka) and 1 in 1.4 × 1014 (Hispanic) represents a 12.7-fold overstatement of evidence strength if the wrong database is used.
- In paternity testing, similar effects were observed. For three paternity cases involving Tawahka families, the combined paternity index (CPI) varied by 18–35% depending on the database used, with the Tawahka database yielding lower (more conservative) CPI values.
4.6.3. Additional Applications
- Population genetics: The allele frequencies facilitate analyses of Indigenous Central American diversity, tests of migration and admixture, and reconstructions of evolutionary and demographic history.
- Anthropology: The data offers genetic context for evaluating the origins, affinities, and potential correspondence between cultural or linguistic groupings in Mesoamerica.
- Medical genetics: As a foundational resource, the dataset can guide hypothesis-driven studies on genetic disease predisposition and inform future pharmacogenetic approaches tailored to Indigenous populations.
4.7. Limitations and Considerations
4.8. Statistical Considerations and Multiple Testing
4.9. Implementation and Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMOVA | Analysis of Molecular Variance |
| CE | Chance of Exclusion |
| CEIB | Biomedical Research Ethics Committee |
| cM | Centimorgan (unit of genetic measurement used to express distances in a genetic map) |
| CODIS | Combined DNA Index System |
| CPI | Combined Paternity Index |
| DICIHT | Directorate of Scientific, Humanistic, and Technological Research |
| DNA | Deoxyribonucleic Acid |
| EMPOP | EDNAP (European DNA Profiling Group) Mitochondrial DNA Population Database |
| ENFSI | European Network of Forensic Science Institutes |
| ESS | European Standard Set |
| FBI | Federal Bureau of Investigation |
| FCM | Faculty of Medical Sciences |
| FDR | False Discovery Rate |
| FITH | Tawahka Indigenous Federation |
| FTA | Fast Technology for Analysis of nucleic acids |
| FST | Fixation Index |
| GITAD | Ibero-American Working Group on DNA Analysis |
| He | Expected Heterozygosity |
| Ho | Observed Heterozygosity |
| HWE | Hardy–Weinberg Equilibrium |
| IBS | Identity-by-state |
| ICIMEDES | Institute for Research in Medical Sciences and Right to Health |
| ISFG | International Society for Forensic Genetics |
| LIMS | Laboratory Information Management System |
| LR | Likelihood Ratio |
| MAF | Minimum Allele Frequency |
| N | Sample size |
| Na | Number of alleles observed at each locus |
| NIST | National Institute of Standards and Technology |
| PCA | Principal Component Analysis |
| PCR | Polymerase Chain Reaction |
| PD | Power of Discrimination |
| PI | Paternity Indices |
| PIC | Polymorphic Information Content |
| RMP | Random Match Probability |
| SD | Standard Deviation |
| STR | Short Tandem Repeat |
| UIC | Scientific Research Unit |
| UNAH | National Autonomous University of Honduras |
| US | United States |
References
- Butler, J.M. Forensic DNA Typing: Biology, Technology, and Genetics of STR Markers, 2nd ed.; Academic Press: Burlington, MA, USA, 2005. [Google Scholar]
- Goodwin, W.; Linacre, A.; Hadi, S. An Introduction to Forensic Genetics, 3rd ed.; Wiley-Blackwell: Chichester, UK, 2020. [Google Scholar]
- National Research Council. The Evaluation of Forensic DNA Evidence; National Academy Press: Washington, DC, USA, 1996.
- Buckleton, J.; Bright, J.A.; Taylor, D. Forensic DNA Evidence Interpretation, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- SEDH. Derechos Humanos en Cifras. Available online: https://odh.sedh.gob.hn/category/view/404/poblacion-indigena-y-afrohondurena (accessed on 10 October 2025).
- McSweeney, K. A demographic profile of the Tawahka Amerindians of Honduras. Geogr. Rev. 2002, 92, 398–414. [Google Scholar] [CrossRef]
- Suarez, A.G. Movimientos indígenas en la globalización: Los tawahka. Convergencia 2002, 29, 259–263. [Google Scholar]
- Oficina del Alto Comisionado de las Naciones Unidas para los Derechos Humanos. Los Pueblos Indígenas y Tribales de Honduras; OACNUDH: Tegucigalpa, Honduras, 2020. [Google Scholar]
- Zuniga, A.; Molina, Y.; Soriano, P.; Moya, Z.; Pinto, Y.; Pineda, D.; Amaya, K.; Cruz, L.; Raudales, I.; Herrera, E.; et al. Population genetic data for 23 STR loci (PowerPlex fusion 6C™ kit) genetic markers in the Lenca ethnic group in Honduras. Leg. Med. 2024, 71, 102504. [Google Scholar] [CrossRef]
- Salzano, F.M.; Bortolini, M.C. The Evolution and Genetics of Latin American Populations; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- Gokcumen, O. The population genetics of Native Americans. In Handbook of Statistical Genomics, 4th ed.; Balding, D.J., Moltke, I., Marioni, J., Eds.; Wiley: Hoboken, NJ, USA, 2019; pp. 1143–1166. [Google Scholar]
- Promega Corporation. PowerPlex® Fusion 6C System Technical Manual; Promega Corporation: Madison, WI, USA, 2023. [Google Scholar]
- Ensenberger, M.G.; Lenz, K.A.; Matthies, L.K.; Hadinoto, G.M.; Schienman, J.E.; Przech, A.J.; Morganti, M.W.; Renstrom, D.T.; Baker, V.M.; Gawrys, K.M.; et al. Developmental validation of the PowerPlex® Fusion 6C System. Forensic Sci. Int. Genet. 2016, 21, 134–144. [Google Scholar] [CrossRef]
- Hares, D.R. Selection and implementation of expanded CODIS core loci in the United States. Forensic Sci. Int. Genet. 2015, 17, 33–34. [Google Scholar] [CrossRef]
- Ministerio Público de Honduras. Informe Anual de Gestión 2023. Available online: https://www.mp.hn/ (accessed on 7 November 2025).
- García, O.; Yurrebaso, I.; Mancisidor, A.; Alonso, S. Allele Frequencies of 27 Y-STRs in a Honduran Population Sample. Forensic Sci. Int. Genet. 2019, 42, e13–e15. [Google Scholar] [CrossRef]
- Butler, J.M.; Hill, C.R.; Coble, M.D. Variability of New STR Loci and Kits in U.S. Population Groups. Forensic Sci. Int. Genet. Suppl. Ser. 2009, 2, 69–70. [Google Scholar] [CrossRef]
- Balding, D.J.; Nichols, R.A. DNA Profile Match Probability Calculation: How to Allow for Population Stratification, Relatedness, Database Selection and Single Bands. Forensic Sci. Int. 1994, 64, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Anderson, A.D.; Hepler, A.B. Genetic Relatedness Analysis: Modern Data and New Challenges. Nat. Rev. Genet. 2006, 7, 771–780. [Google Scholar] [CrossRef]
- Martin, B.J.; Watkins, J.B., III; Ramsey, J. Venipuncture in the medical physiology laboratory. Adv. Physiol. Educ. 1998, 274, S62–S67. [Google Scholar] [CrossRef]
- Qiagen. Indicating FTA Cards. Available online: https://www.qiagen.com/us/products/human-id-and-forensics/sample-collection/indicating-fta-cards (accessed on 10 October 2025).
- Burgoyne, L.A. Solid Medium and Method for DNA Storage. U.S. Patent 5,496,562, 5 March 1996. [Google Scholar]
- Whatman plc. FTA™ Technology for Sample Collection, Transport and Storage; Whatman plc: Kent, UK, 2010. [Google Scholar]
- Promega. PowerPlex® Fusion 6C System. Available online: https://worldwide.promega.com/products/forensic-dna-analysis-ce/str-amplification/powerplex-fusion-6c-system/?catNum=DC2705 (accessed on 10 October 2025).
- Thermo Fisher Scientific. Applied Biosystems 3500 Series Genetic Analyzers. Available online: https://www.thermofisher.com/hn/en/home/life-science/sequencing/sanger-sequencing/sanger-sequencing-technology-accessories/applied-biosystems-sanger-sequencing-3500-series-genetic-analyzers/3500-series-genetic-analyzer.html (accessed on 10 October 2025).
- Thermo Fisher Scientific. GeneMapper Software v4.1. Available online: https://tools.thermofisher.com/ (accessed on 10 October 2025).
- Mayr, W.R. DNA Recommendations—1994 Report Concerning Further Recommendations of the DNA Commission of the ISFG Regarding PCR-Based Polymorphisms in STR Systems. Vox Sang. 1995, 69, 70–71. [Google Scholar] [CrossRef]
- Gusmão, L.; Butler, J.M.; Carracedo, A.; Gill, P.; Kayser, M.; Mayr, W.R.; Morling, N.; Prinz, M.; Roewer, L.; Tyler-Smith, C. DNA Commission of the International Society of Forensic Genetics (ISFG): An Update of the Recommendations on the Use of Y-STRs in Forensic Analysis. Forensic Sci. Int. 2006, 157, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Curtin University. Genepop Software. Available online: https://genepop.curtin.edu.au/ (accessed on 10 October 2025).
- Bern University. Arlequin 5.5. Available online: http://cmpg.unibe.ch/software/arlequin35/ (accessed on 10 October 2025).
- Huston, K. Statistical analysis of STR data. Profiles DNA 1998, 1, 14–15. [Google Scholar]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Mayo, O. A century of Hardy–Weinberg equilibrium. Twin Res. Hum. Genet. 2008, 11, 249–256. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin ver 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple comparisons among means. J. Am. Stat. Assoc. 1961, 56, 52–64. [Google Scholar] [CrossRef]
- Narum, S.R. Beyond Bonferroni: Less Conservative Analyses for Conservation Genetics. Conserv. Genet. 2006, 7, 783–787. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Balding, D.J. Weight-of-Evidence for Forensic DNA Profiles, 2nd ed.; Wiley: Chichester, UK, 2015. [Google Scholar]
- Academia Iberoamericana de Criminalísticas y Estudios Forenses. GITAD: Grupo Iberoamericano de Trabajo en Análisis de DNA. Available online: https://www.aicef.info/grupos-de-trabajo/gitad/ (accessed on 10 October 2025).
- Convention on Biological Diversity. Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from Their Utilization to the Convention on Biological Diversity; Secretariat of the Convention on Biological Diversity: Montreal, QC, Canada, 2010; Available online: https://www.cbd.int/abs/ (accessed on 7 November 2025).
- United Nations. United Nations Declaration on the Rights of Indigenous Peoples; United Nations: New York, NY, USA, 2007; Available online: https://www.un.org/development/desa/indigenouspeoples/declaration-on-the-rights-of-indigenous-peoples.html (accessed on 7 November 2025).
- Claw, K.G.; Anderson, M.Z.; Begay, R.L.; Tsosie, K.S.; Fox, K.; Garrison, N.A. A Framework for Enhancing Ethical Genomic Research with Indigenous Communities. Nat. Commun. 2018, 9, 2957. [Google Scholar] [CrossRef]
- Garrison, N.A.; Hudson, M.; Ballantyne, L.L.; Garba, I.; Martinez, A.; Taualii, M.; Arbour, L.; Caron, N.R.; Rainie, S.C. Genomic Research through an Indigenous Lens: Understanding the Expectations. Annu. Rev. Genom. Hum. Genet. 2019, 20, 495–517. [Google Scholar] [CrossRef] [PubMed]
- International Society for Forensic Genetics (ISFG). ISFG Recommendations on Bioethics and Forensic Genetics. Forensic Sci. Int. Genet. 2023, 62, 102782. [Google Scholar] [CrossRef]
- D-Maps. Mapa de Honduras. Available online: https://d-maps.com/carte.php?num_car=26210&lang=es (accessed on 10 October 2025).
- Native-Land. Lenca. Available online: https://native-land.ca/maps/territories/lenca (accessed on 10 October 2025).
- Wahlund, S. Zusammensetzung von Populationen und Korrelationserscheinungen vom Standpunkt der Vererbungslehre aus betrachtet. Hereditas 1928, 11, 65–106. [Google Scholar] [CrossRef]
- Buckleton, J.; Krawczak, M.; Weir, B.S. The Interpretation of Lineage Markers in Forensic DNA Testing. Forensic Sci. Int. Genet. 2011, 5, 78–83. [Google Scholar] [CrossRef]
- National Institute of Standards and Technology (NIST). STRBase: Short Tandem Repeat DNA Internet Database; 2024. Available online: https://strbase.nist.gov/ (accessed on 8 November 2025).
- Budowle, B.; Ge, J.; Aranda, X.G.; Planz, J.V.; Eisenberg, A.J.; Chakraborty, R. Texas Population Substructure and Its Impact on Estimating the Rarity of Y-STR Haplotypes from DNA Evidence. J. Forensic Sci. 2009, 54, 1016–1021. [Google Scholar] [CrossRef]
- Gill, P.; Brenner, C.H.; Brinkmann, B.; Budowle, B.; Carracedo, A.; Jobling, M.A.; de Knijff, P.; Kayser, M.; Krawczak, M.; Mayr, W.R.; et al. DNA Commission of the International Society of Forensic Genetics: Recommendations on Forensic Analysis Using Y-Chromosome STRs. Forensic Sci. Int. 2001, 124, 5–10. [Google Scholar] [CrossRef]
- Congreso Nacional de Honduras. Ley de Transparencia y Acceso a la Información Pública de Honduras; Decreto No. 170-2006; 2006. Available online: https://portalunico.iaip.gob.hn/ver_archivo/NjgwODgz (accessed on 12 November 2025).
- Cardoso, S.; Sevillano, R.; Illescas, M.J.; de Martínez Pancorbo, M. Analysis of 16 autosomal STRs and 17 Y-STRs in an indigenous Maya population from Guatemala. Int. J. Legal Med. 2016, 130, 365–366. [Google Scholar] [CrossRef]
- Herlihy, P.H. Indigenous and Ladino peoples of eastern Honduras and Nicaragua. In Peoples of the Humid Tropics; Stone, D., Ed.; Louisiana State University: Baton Rouge, LA, USA, 1974. [Google Scholar]
- Aguilar-Velázquez, J.A.; Sthepenson-Ojea, M.; García-King, M.D.; Rangel-Villalobos, H. Admixture and population structure in Mayas and Ladinos from Guatemala based on 15 STRs. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 776–777. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Foreman, L.A.; Evett, I.W. Statistical Analyses to Support Forensic Interpretation for a New Ten-Locus STR Profiling System. Int. J. Leg. Med. 2001, 114, 147–155. [Google Scholar] [CrossRef]
- Moreno-Estrada, A.; Gignoux, C.R.; Fernández-López, J.C.; Zakharia, F.; Sikora, M.; Contreras, A.V.; Acuña-Alonzo, V.; Sandoval, K.; Eng, C.; Romero-Hidalgo, S.; et al. Human Genetics: The Genetics of Mexico Recapitulates Native American Substructure and Affects Biomedical Traits. Science 2014, 344, 1280–1285. [Google Scholar] [CrossRef]
- Gravel, S. Population Genetics Models of Local Ancestry. Genetics 2012, 191, 607–619. [Google Scholar] [CrossRef]
- Gill, P.; Gusmão, L.; Haned, H.; Mayr, W.R.; Morling, N.; Parson, W.; Prieto, L.; Prinz, M.; Schneider, H.; Schneider, P.M.; et al. DNA Commission of the International Society of Forensic Genetics: Recommendations on the Evaluation of STR Typing Results That May Include Drop-Out and/or Drop-In Peaks. Forensic Sci. Int. Genet. 2012, 6, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Buckleton, J.; Curran, J.M.; Goudet, J.; Taylor, D.; Thiery, A.; Weir, B.S. Population-specific FST values for forensic STR markers: A worldwide survey. Forensic Sci. Int. Genet. 2016, 23, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Pakstis, A.J.; Gurkan, C.; Dogan, M.; Balkaya, H.E.; Dogan, S.; Neophytou, P.I.; Cherni, L.; Boussetta, S.; Khodjet-El-Khil, H.; Ben Ammar ElGaaied, A.; et al. Genetic relationships of European, Mediterranean, and SW Asian populations using a panel of 55 AISNPs. Eur. J. Hum. Genet. 2019, 27, 1885–1893. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cortés, G.; Salazar-Flores, J.; Haro-Guerrero, J.; Rubi-Castellanos, R.; Velarde-Félix, J.S.; Muñoz-Valle, J.F.; Canseco-Avila, L.M.; Carrillo-Tapia, E.; López-Loya, J.G.; Rangel-Villalobos, H. Genetic Structure and Diversity of Indigenous Populations from El Salvador: Analysis of 15 Autosomal STRs. Forensic Sci. Int. Genet. Suppl. Ser. 2012, 3, e257–e258. [Google Scholar] [CrossRef]
- Wang, S.; Lewis, C.M., Jr.; Jakobsson, M.; Ramachandran, S.; Ray, N.; Bedoya, G.; Rojas, W.; Parra, M.V.; Molina, J.A.; Gallo, C.; et al. Genetic Variation and Population Structure in Native Americans. PLoS Genet. 2007, 3, e185. [Google Scholar] [CrossRef]
- M’charek, A. Beyond Fact or Fiction: On the Materiality of Race in Practice. Cult. Anthropol. 2013, 28, 420–442. [Google Scholar] [CrossRef]
- TallBear, K. Genomic Articulations of Indigeneity. Soc. Stud. Sci. 2013, 43, 509–533. [Google Scholar] [CrossRef]
- European Network of Forensic Science Institutes (ENFSI). ENFSI DNA Working Group Database; 2024. Available online: https://enfsi.eu/about-enfsi/structure/working-groups/dna/ (accessed on 8 November 2025).
- Parson, W.; Dür, A. EMPOP—A Forensic mtDNA Database. Forensic Sci. Int. Genet. 2007, 1, 88–92. [Google Scholar] [CrossRef]
| Scenario | Recommended θ | Justification |
|---|---|---|
| Within Tawahka (suspect and reference both confirmed Tawahka) | θ = 0.01 | Conservative given within-population FST = 0.003; accounts for potential cryptic relatedness |
| Within Lenca | θ = 0.01 | Similar reasoning |
| Within Honduran mestizo | θ = 0.01 | Existing standard; appropriate given moderate admixture and urbanization |
| Tawahka vs. mestizo (uncertain ethnicity) | θ = 0.03 | Reflects elevated FST = 0.025; conservative approach for cross-population comparisons |
| Tawahka vs. European/Asian (international cases) | θ = 0.05–0.10 | Reflects substantial FST; highly conservative |
| Unknown ethnicity (no information) | θ = 0.03 | Intermediate value balancing conservatism with informativeness |
| Population | N | Mean He | Mean PD | Mean Alleles/Locus | Reference |
|---|---|---|---|---|---|
| Tawahka (Honduras) | 100 | 0.7385 | 0.8771 | 8.61 | Present study |
| Lenca (Honduras) | 100 | 0.7425 | 0.8815 | 8.91 | [9] |
| Maya (Guatemala) | 127 | 0.7104 | 0.8534 | 7.83 | [53] |
| Ladino (Guatemala) | 115 | 0.7656 | 0.8945 | 9.45 | [55] |
| Mestizo (El Salvador) | 108 | 0.7512 | 0.8823 | 8.95 | [63] |
| Case | RMP (Tawahka) | RMP (Mestizo) | RMP (Hispanic) | Fold Difference * |
|---|---|---|---|---|
| A | 1 in 4.2 × 1014 | 1 in 1.8 × 1015 | 1 in 6.5 × 1015 | 4.3×–15.5× |
| B | 1 in 8.7 × 1015 | 1 in 3.1 × 1016 | 1 in 9.8 × 1016 | 3.6×–11.3× |
| C | 1 in 1.1 × 1013 | 1 in 3.9 × 1013 | 1 in 1.4 × 1014 | 3.5×–12.7× |
| D | 1 in 2.9 × 1016 | 1 in 1.2 × 1017 | 1 in 4.1 × 1017 | 4.1×–14.1× |
| E | 1 in 6.5 × 1014 | 1 in 2.7 × 1015 | 1 in 8.2 × 1015 | 4.2×–12.6× |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuniga, A.; Molina, Y.; Amaya, K.; Moya, Z.; Soriano, P.; Pineda, D.; Pinto, Y.; Zablah, I. Population Genetic Data for 23 STR Loci of Tawahka Ethnic Group in Honduras. Forensic Sci. 2025, 5, 72. https://doi.org/10.3390/forensicsci5040072
Zuniga A, Molina Y, Amaya K, Moya Z, Soriano P, Pineda D, Pinto Y, Zablah I. Population Genetic Data for 23 STR Loci of Tawahka Ethnic Group in Honduras. Forensic Sciences. 2025; 5(4):72. https://doi.org/10.3390/forensicsci5040072
Chicago/Turabian StyleZuniga, Antonieta, Yolly Molina, Karen Amaya, Zintia Moya, Patricia Soriano, Digna Pineda, Yessica Pinto, and Isaac Zablah. 2025. "Population Genetic Data for 23 STR Loci of Tawahka Ethnic Group in Honduras" Forensic Sciences 5, no. 4: 72. https://doi.org/10.3390/forensicsci5040072
APA StyleZuniga, A., Molina, Y., Amaya, K., Moya, Z., Soriano, P., Pineda, D., Pinto, Y., & Zablah, I. (2025). Population Genetic Data for 23 STR Loci of Tawahka Ethnic Group in Honduras. Forensic Sciences, 5(4), 72. https://doi.org/10.3390/forensicsci5040072






