Advances in Personalized Cancer Vaccine Development: AI Applications from Neoantigen Discovery to mRNA Formulation
Abstract
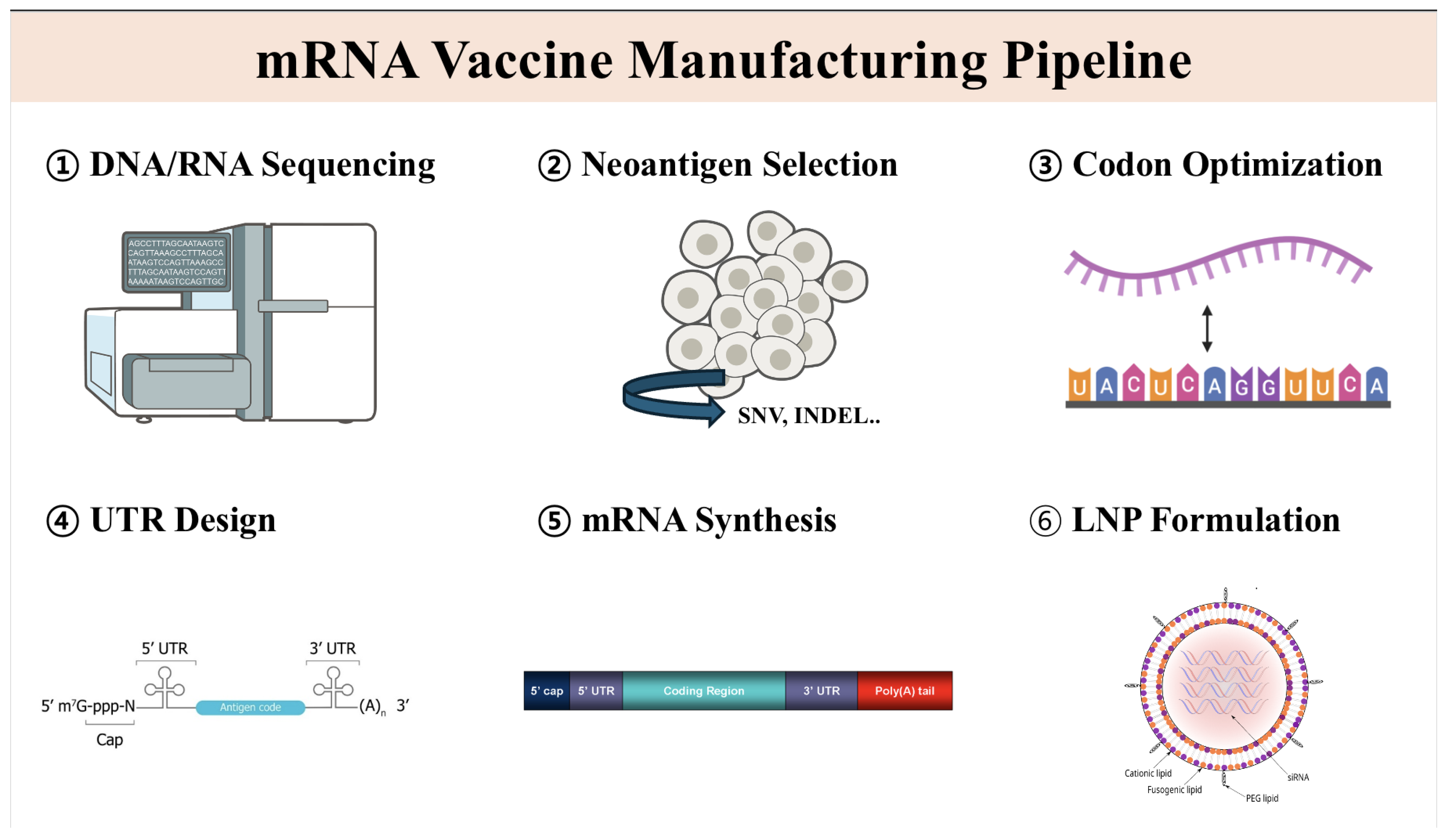
1. Introduction
2. AI in Neoantigen Discovery
2.1. Enhancing Neoantigen Prediction with Machine Learning
2.2. Improved Pipelines and Web Services
3. AI in Codon Optimization for Vaccine Antigens
3.1. Deep Learning Models for Codon Optimization
3.2. Experimental Validation of AI-Optimized Codon Sequences
4. AI in UTR Sequence Generation and mRNA Design
4.1. AI-Driven 5′ UTR Optimization
4.2. AI-Driven 3′ UTR Optimization
4.3. Implications for mRNA Vaccine Design
5. AI in mRNA Vaccine Formulation and Delivery
5.1. AI-Guided Lipid Nanoparticle (LNP) Optimization
5.2. AI in Adjuvant and Immune-Stimulatory Element Design
5.3. AI in Personalized mRNA Vaccine Formulation
6. Challenges and Future Perspectives
7. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Blass, E.; Ott, P.A. Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat. Rev. Clin. Oncol. 2021, 18, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Ott, P.A.; Hu, Z.; Keskin, D.B.; Shukla, S.A.; Sun, J.; Bozym, D.J.; Zhang, W.; Luoma, A.; Giobbie-Hurder, A.; Peter, L.; et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017, 547, 217–221. [Google Scholar] [CrossRef]
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.P.; Simon, P.; Löwer, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrörs, B. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Keskin, D.B.; Anandappa, A.J.; Sun, J.; Tirosh, I.; Mathewson, N.D.; Li, S.; Oliveira, G.; Giobbie-Hurder, A.; Felt, K.; Gjini, E.; et al. Neoantigen vaccine generates intratumoral T cell responses in glioblastoma. Nature 2019, 565, 234–239. [Google Scholar] [CrossRef]
- Sahin, U.; Türeci, Ö. Personalized vaccines for cancer immunotherapy. Science 2018, 359, 1355–1360. [Google Scholar] [CrossRef]
- Liu, M.A. A comparison of plasmid DNA and mRNA as vaccine technologies. Vaccines 2019, 7, 37. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Nguyen, T.B.Q.; Pham, Q.T.M.; Tran, L.S. 84P T cell receptor repertoire profiles of tumor-infiltrating lymphocytes improves neoantigen prioritization for personalized cancer immunotherapy. Ann. Oncol. 2023, 34, S1499. [Google Scholar] [CrossRef]
- Hong, W.; Chen, C.; Zhu, Z.; Tang, K. An Elite Archive-Assisted Multi-Objective Evolutionary Algorithm for mRNA Design. In Proceedings of the 2024 IEEE Congress on Evolutionary Computation (CEC), Yokohama, Japan, 30 June–5 July 2024; pp. 1–8. [Google Scholar]
- Kaushik, R.; Kant, R.; Christodoulides, M. Artificial intelligence in accelerating vaccine development-current and future perspectives. Front. Bacteriol. 2023, 2, 1258159. [Google Scholar] [CrossRef]
- Imani, S.; Li, X.; Chen, K.; Maghsoudloo, M.; Kaboli, P.J.; Hashemi, M.; Khoushab, S.; Li, X. Computational biology and artificial intelligence in mRNA vaccine design for cancer immunotherapy. Front. Cell. Infect. Microbiol. 2025, 14, 1501010. [Google Scholar] [CrossRef]
- Xin, P.; Yang, D.; Zhou, Y.; Peng, S. TlcMHCpan: A Novel Deep Learning Model for Enhanced Pan-Specific Prediction of Peptide-HLA Binding. IEEE Access 2024, 12, 184644–184656. [Google Scholar] [CrossRef]
- Pu, T.; Peddle, A.; Zhu, J.; Tejpar, S.; Verbandt, S. Neoantigen identification: Technological advances and challenges. Methods Cell Biol. 2024, 183, 265–302. [Google Scholar] [PubMed]
- Springer, I.; Besser, H.; Tickotsky-Moskovitz, N.; Dvorkin, S.; Louzoun, Y. Prediction of specific TCR-peptide binding from large dictionaries of TCR-peptide pairs. Front. Immunol. 2020, 11, 1803. [Google Scholar]
- Montemurro, A.; Schuster, V.; Povlsen, H.R.; Bentzen, A.K.; Jurtz, V.; Chronister, W.D.; Crinklaw, A.; Hadrup, S.R.; Winther, O.; Peters, B.; et al. NetTCR-2.0 enables accurate prediction of TCR-peptide binding by using paired TCRα and β sequence data. Commun. Biol. 2021, 4, 1060. [Google Scholar]
- Lu, T.; Zhang, Z.; Zhu, J.; Wang, Y.; Jiang, P.; Xiao, X.; Bernatchez, C.; Heymach, J.V.; Gibbons, D.L.; Wang, J.; et al. Deep learning-based prediction of the T cell receptor–antigen binding specificity. Nat. Mach. Intell. 2021, 3, 864–875. [Google Scholar] [CrossRef]
- Fu, H.; Liang, Y.; Zhong, X.; Pan, Z.; Huang, L.; Zhang, H.; Xu, Y.; Zhou, W.; Liu, Z. Codon optimization with deep learning to enhance protein expression. Sci. Rep. 2020, 10, 17617. [Google Scholar]
- Kumar, A.; Dixit, S.; Srinivasan, K.; M, D.; Vincent, P.M.D.R. Personalized cancer vaccine design using AI-powered technologies. Front. Immunol. 2024, 15, 1357217. [Google Scholar]
- Li, S.; Moayedpour, S.; Li, R.; Bailey, M.; Riahi, S.; Kogler-Anele, L.; Miladi, M.; Miner, J.; Zheng, D.; Wang, J.; et al. CodonBERT: Large language models for mRNA design and optimization. bioRxiv 2023. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, R.; Gao, S.; Li, W.; Liu, Y.; Su, G.; Song, M.; Jiang, M.; Jiang, C.; Zhang, X. Artificial intelligence applied in neoantigen identification facilitates personalized cancer immunotherapy. Front. Oncol. 2023, 12, 1054231. [Google Scholar]
- Castillo-Hair, S.; Fedak, S.; Wang, B.; Linder, J.; Havens, K.; Certo, M.; Seelig, G. Optimizing 5′UTRs for mRNA-delivered gene editing using deep learning. Nat. Commun. 2024, 15, 5284. [Google Scholar] [CrossRef]
- Chu, Y.; Yu, D.; Li, Y.; Huang, K.; Shen, Y.; Cong, L.; Zhang, J.; Wang, M. A 5′UTR Language Model for Decoding Untranslated Regions of mRNA and Function Predictions. bioRxiv 2023. [Google Scholar] [CrossRef]
- Cafri, G.; Gartner, J.J.; Zaks, T.; Hopson, K.; Levin, N.; Paria, B.C.; Parkhurst, M.R.; Yossef, R.; Lowery, F.J.; Jafferji, M.S.; et al. mRNA vaccine–induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J. Clin. Investig. 2020, 130, 5976–5988. [Google Scholar] [PubMed]
- Mekki-Berrada, F.; Ren, Z.; Huang, T.; Wong, W.K.; Zheng, F.; Xie, J.; Tian, I.P.S.; Jayavelu, S.; Mahfoud, Z.; Bash, D.; et al. Two-step machine learning enables optimized nanoparticle synthesis. Npj Comput. Mater. 2021, 7, 55. [Google Scholar]
- He, S.; Gao, B.; Sabnis, R.; Sun, Q. RNAdegformer: Accurate prediction of mRNA degradation at nucleotide resolution with deep learning. Brief. Bioinform. 2023, 24, bbac581. [Google Scholar]
- Laumont, C.M.; Vincent, K.; Hesnard, L.; Audemard, É.; Bonneil, É.; Laverdure, J.-P.; Gendron, P.; Courcelles, M.; Hardy, M.-P.; Côté, C.; et al. Noncoding regions are the main source of tumor-specific antigens. Sci. Transl. Med. 2018, 10, eaau5516. [Google Scholar] [CrossRef]
- Wells, D.K.; van Buuren, M.M.; Dang, K.K.; Hubbard-Lucey, V.M.; Sheehan, K.C.; Campbell, K.M.; Lamb, A.; Ward, J.P.; Sidney, J.; Blazquez, A.B.; et al. Key parameters of tumor epitope immunogenicity revealed through a consortium approach improve neoantigen prediction. Cell 2020, 183, 818–834.e13. [Google Scholar] [CrossRef]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef]
- Hilf, N.; Kuttruff-Coqui, S.; Frenzel, L.P.; Bukur, T.; Stevanović, S.; Gouttefangeas, C.; Platten, M.; Tabatabai, J.; Dutoit, V.; van der Burg, S.H.; et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature 2019, 565, 240–245. [Google Scholar] [CrossRef]
- Yadav, M.; Jhunjhunwala, S.; Phung, Q.; Lupardus, P.; Tanguay, J.; Bumbaca, S.; Franci, C.; Cheung, T.K.; Fritsche, F.; Weinschenk, T.; et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014, 515, 572–576. [Google Scholar] [CrossRef]
- Hundal, J.; Kiwala, S.; McMichael, J.; Miller, C.A.; Xia, H.; Wollam, A.T.; Liu, C.J.; Zhao, S.; Feng, Y.-Y.; Graubert, A.P.; et al. pVACtools: A computational toolkit to identify and visualize cancer neoantigens. Cancer Immunol. Res. 2020, 8, 409–420. [Google Scholar] [CrossRef]
- Laumont, C.M.; Wouters, M.C.A.; Smazynski, J.; Gierc, N.S. The landscape of tumor antigens and neoantigens in cancer immunotherapy. Nat. Rev. Cancer 2022, 22, 682–696. [Google Scholar] [CrossRef]
- Vitiello, A.; Zanetti, M. Neoantigen prediction and the need for validation. Nat. Biotech. 2017, 35, 815–817. [Google Scholar]
- Jurtz, V.; Paul, S.; Andreatta, M.; Marcatili, P.; Peters, B.; Nielsen, M. NetMHCpan-4.0: Improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 2017, 199, 3360–3368. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, X.; Li, X.; Zhang, L.; Wang, Y. Advances in deep learning for neoantigen prediction. Front. Immunol. 2021, 12, 705096. [Google Scholar] [CrossRef]
- Zhao, W.; Li, S.; Liu, Y.; Yang, H. NeoaPred: A structure-based deep learning approach for predicting peptide-MHC binding and immunogenicity. Bioinformatics 2022, 38, 3792–3800. [Google Scholar] [CrossRef]
- Abella, J.R.; Brown, J.R.; Pal, M.; Silvestri, G. DeepHLApan: A convolutional neural network for MHC-peptide binding prediction. PLoS Comput. Biol. 2020, 16, e1007869. [Google Scholar] [CrossRef]
- Racle, J.; Michaux, J. Machine learning-based approaches for neoantigen prediction. Trends Cancer 2022, 8, 45–57. [Google Scholar] [CrossRef]
- Carri, I.; Schwab, E.; Podaza, E.; Alvarez, H.M.G.; Mordoh, J.; Nielsen, M.; Barrio, M.M. Beyond MHC binding: Immunogenicity prediction tools to refine neoantigen selection in cancer patients. Explor. Immunol. 2023, 3, 82–103. [Google Scholar]
- Xin, K.; Wei, X.; Shao, J.; Chen, F.; Liu, Q.; Liu, B. Establishment of a novel tumor neoantigen prediction tool for personalized vaccine design. Hum. Vaccines Immunother. 2024, 20, 2300881. [Google Scholar]
- Kim, J.Y.; Bang, H.; Noh, S.J.; Choi, J.K. DeepNeo: A webserver for predicting immunogenic neoantigens. Nucleic Acids Res. 2023, 51, W134–W140. [Google Scholar]
- Yang, Z.; Bogdan, P.; Nazarian, S. An in silico deep learning approach to multi-epitope vaccine design: A SARS-CoV-2 case study. Sci. Rep. 2021, 11, 3238. [Google Scholar]
- Bulashevska, A.; Nacsa, Z.; Lang, F.; Braun, M.; Machyna, M.; Diken, M.; Childs, L.; König, R. Artificial intelligence and neoantigens: Paving the path for precision cancer immunotherapy. Front. Immunol. 2024, 15, 1394003. [Google Scholar]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [PubMed]
- Sharp, P.M.; Li, W.H. The codon adaptation index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar]
- Li, Y.; Wang, F.; Yang, J.; Han, Z.; Chen, L.; Jiang, W.; Zhou, H.; Li, T.; Tang, Z.; Deng, J.; et al. Deep Generative Optimization of mRNA Codon Sequences for Enhanced Protein Production and Therapeutic Efficacy. bioRxiv 2024. [Google Scholar] [CrossRef]
- Joshi, M.; Wang, A.; Myong, S. 5′UTR G-quadruplex structure enhances translation in size dependent manner. Nat. Commun. 2024, 15, 3963. [Google Scholar] [CrossRef]
- Hong, D.; Jeong, S. 3′UTR Diversity: Expanding Repertoire of RNA Alterations in Human mRNAs. Mol. Cells 2023, 46, 48–56. [Google Scholar] [CrossRef]
- Tang, X.; Huo, M.; Chen, Y.; Huang, H.; Qin, S.; Luo, J.; Qin, Z.; Jiang, X.; Liu, Y.; Duan, X.; et al. A novel deep generative model for mRNA vaccine development: Designing 5′ UTRs with N1-methyl-pseudouridine modification. Acta Pharm. Sin. B 2024, 14, 1814–1826. [Google Scholar]
- Morrow, A.; Thornal, A.; Flynn, E.D.; Hoelzli, E.; Shan, M.; Garipler, G.; Kirchner, R.; Reddy, A.J.; Tabchouri, S.; Gupta, A.S.; et al. ML-driven design of 3′ UTRs for mRNA stability. bioRxiv 2024. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, J.; Zhang, X.; Fang, X. Joint Design of 5′Untranslated Region and Coding Sequence of mRNA. arXiv 2024, arXiv:2410.20781. [Google Scholar]
- Castillo-Hair, S.M.; Seelig, G. Machine learning for designing next-generation mRNA therapeutics. Acc. Chem. Res. 2021, 55, 24–34. [Google Scholar] [PubMed]
- Shi, R.; Liu, X.; Wang, Y.; Pan, M.; Wang, S.; Shi, L.; Ni, B. Long-term stability and immunogenicity of lipid nanoparticle COVID-19 mRNA vaccine is affected by particle size. Hum. Vaccines Immunother. 2024, 20, 2342592. [Google Scholar]
- Xie, C.; Yao, R.; Xia, X. The advances of adjuvants in mRNA vaccines. npj Vaccines 2023, 8, 162. [Google Scholar] [PubMed]
- Li, X.; Qi, J.; Wang, J.; Hu, W.; Zhou, W.; Wang, Y.; Li, T. Nanoparticle technology for mRNA: Delivery strategy, clinical application and developmental landscape. Theranostics 2024, 14, 738. [Google Scholar]
- Xu, Y.; Ma, S.; Cui, H.; Chen, J.; Xu, S.; Gong, F.; Golubovic, A.; Zhou, M.; Wang, K.C.; Varley, A.; et al. AGILE platform: A deep learning powered approach to accelerate LNP development for mRNA delivery. Nat. Commun. 2024, 15, 6305. [Google Scholar]
- Wu, K.; Yang, X.; Wang, Z.; Li, N.; Zhang, J.; Liu, L. Data-balanced transformer for accelerated ionizable lipid nanoparticles screening in mRNA delivery. Brief. Bioinform. 2024, 25, bbae186. [Google Scholar]
- Zhang, W.Y.; Zheng, X.L.; Coghi, P.S.; Chen, J.H.; Dong, B.J.; Fan, X.X. Revolutionizing adjuvant development: Harnessing AI for next-generation cancer vaccines. Front. Immunol. 2024, 15, 1438030. [Google Scholar]
- Chaudhury, S.; Duncan, E.H.; Atre, T.; Storme, C.K.; Beck, K.; Kaba, S.A.; Lanar, D.E.; Bergmann-Leitner, E.S. Identification of immune signatures of novel adjuvant formulations using machine learning. Sci. Rep. 2018, 8, 17508. [Google Scholar]
- Ma, J.; Wang, S.; Zhao, C.; Yan, X.; Ren, Q.; Dong, Z.; Qiu, J.; Liu, Y.; Shan, Q.; Xu, M.; et al. Computer-Aided Discovery of Potent Broad-Spectrum Vaccine Adjuvants. Angew. Chem. Int. Ed. 2023, 62, e202301059. [Google Scholar]
- Gude, S.; Abburi, S.K.; Gali, P.K.; Gorlagunta, S. Advancing single-shot vaccine design through AI and computational models. Transl. Regul. Sci. 2025. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 2017, 168, 613–628. [Google Scholar]
- Sugiyama, N.; Terry, F.E.; Gutierrez, A.H.; Hirano, T.; Hoshi, M.; Mizuno, Y.; Martin, W.; Yasunaga, S.; Niiro, H.; Fujio, K.; et al. Individual and population-level variability in HLA-DR associated immunogenicity risk of biologics used for the treatment of rheumatoid arthritis. Front. Immunol. 2024, 15, 1377911. [Google Scholar] [CrossRef]
| Aspect | Reference | AI Model | Data and Inputs | Key Outcome |
|---|---|---|---|---|
| Neoantigen discovery | Springer et al. (2020) [14] | LSTM neural network for TCR–peptide binding prediction | >170,000 TCR–peptide pairs | Learned TCR–peptide specificity; achieved performance on par with state-of-the-art methods |
| Montemurro et al. (2021) [15] | CNN model for paired TCRα and TCRβ sequences | Known TCRαβ sequences and cognate peptides (HLA-A*02:01, 9-mer) | Improved prediction of TCR–peptide binding (79% specificity at a 2% false-positive rate) | |
| Lu et al. (2021) [16] | Transfer learning deep learning model for pMHC–TCR binding | Sequence data: mutated peptide, patient’s MHC class I, and TCR sequence | Achieved high accuracy (AUC 0.827 on independent testing) in predicting TCR binding | |
| Codon optimization | Fu et al. (2020) [17] | BiLSTM-CRF deep learning model using “codon boxes” | E. coli expression of multiple genes (e.g., vaccine antigen FALVAC-1, PTP4A3) | AI-optimized genes showed a higher protein expression than industry-optimized sequences |
| Costa/Absci (2024) [18] | Transformer-based language model (“CO-BERT”) for codon choice | Large-scale coding sequence dataset (multiple organisms) | Predicted optimal synonymous codons for maximal protein expression | |
| Wang et al. (2023) [19] | Multi-species transformer (“CodonTransformer”) for codon optimization | >1 million DNA–protein sequence pairs from 164 species | Learned codon usage across species; enabled cross-species gene optimization | |
| UTR sequence generation | Song et al. (2024) [20] | Deep generative model (“Smart5UTR”)—multitask autoencoder with a CNN encoder | 5′ UTR library with >200,000 sequences tested via MPRA | Generated optimized 5′ UTRs for N1-methyl-pseudouridine mRNA, improving vaccine efficacy |
| Castillo-Hair et al. (2024) [21] | Deep learning regression + generative design for 5′UTR (CNN models and optimization) | Polysome profiling data from random 5′UTR libraries in 3 human cell types | Designed synthetic 5′ UTRs that enhanced translation of a gene editor enzyme | |
| Chu et al. (2024) [22] | Pretrained transformer language model (“UTR-LM”) for 5′UTRs | Endogenous 5′ UTR sequences from multiple species (unsupervised pretraining) | Learned a language of 5′ UTRs enabling improved translation initiation efficiency | |
| mRNA vaccine design | Cafri et al. (2020) [23] | Personalized neoantigen mRNA vaccine (clinical trial) | 13 patients with gastrointestinal tumors; 5–20 neoantigens per mRNA vaccine | First-in-human phase I trial of an individualized mRNA neoantigen vaccine |
| Mekki-Berrada et al. (2021) [24] | Two-step ML for lipid nanoparticle formulation optimization | Data from combinatorial synthesis of polymeric nanoparticles for mRNA delivery | AI-guided nanoparticle formulation improved mRNA delivery efficiency | |
| He et al. (2023) [25] | CNN with self-attention (“RNAdeformer”) for mRNA degradation prediction | Public mRNA stability datasets (e.g., OpenVaccine COVID-19 mRNA data) | Achieved state-of-the-art accuracy in predicting mRNA half-life and degradation sites |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, H. Advances in Personalized Cancer Vaccine Development: AI Applications from Neoantigen Discovery to mRNA Formulation. BioChem 2025, 5, 5. https://doi.org/10.3390/biochem5020005
Kong H. Advances in Personalized Cancer Vaccine Development: AI Applications from Neoantigen Discovery to mRNA Formulation. BioChem. 2025; 5(2):5. https://doi.org/10.3390/biochem5020005
Chicago/Turabian StyleKong, Hyunseung. 2025. "Advances in Personalized Cancer Vaccine Development: AI Applications from Neoantigen Discovery to mRNA Formulation" BioChem 5, no. 2: 5. https://doi.org/10.3390/biochem5020005
APA StyleKong, H. (2025). Advances in Personalized Cancer Vaccine Development: AI Applications from Neoantigen Discovery to mRNA Formulation. BioChem, 5(2), 5. https://doi.org/10.3390/biochem5020005







