Mammarenavirus Z Protein Myristoylation and Oligomerization Are Not Required for Its Dose-Dependent Inhibitory Effect on vRNP Activity
Abstract
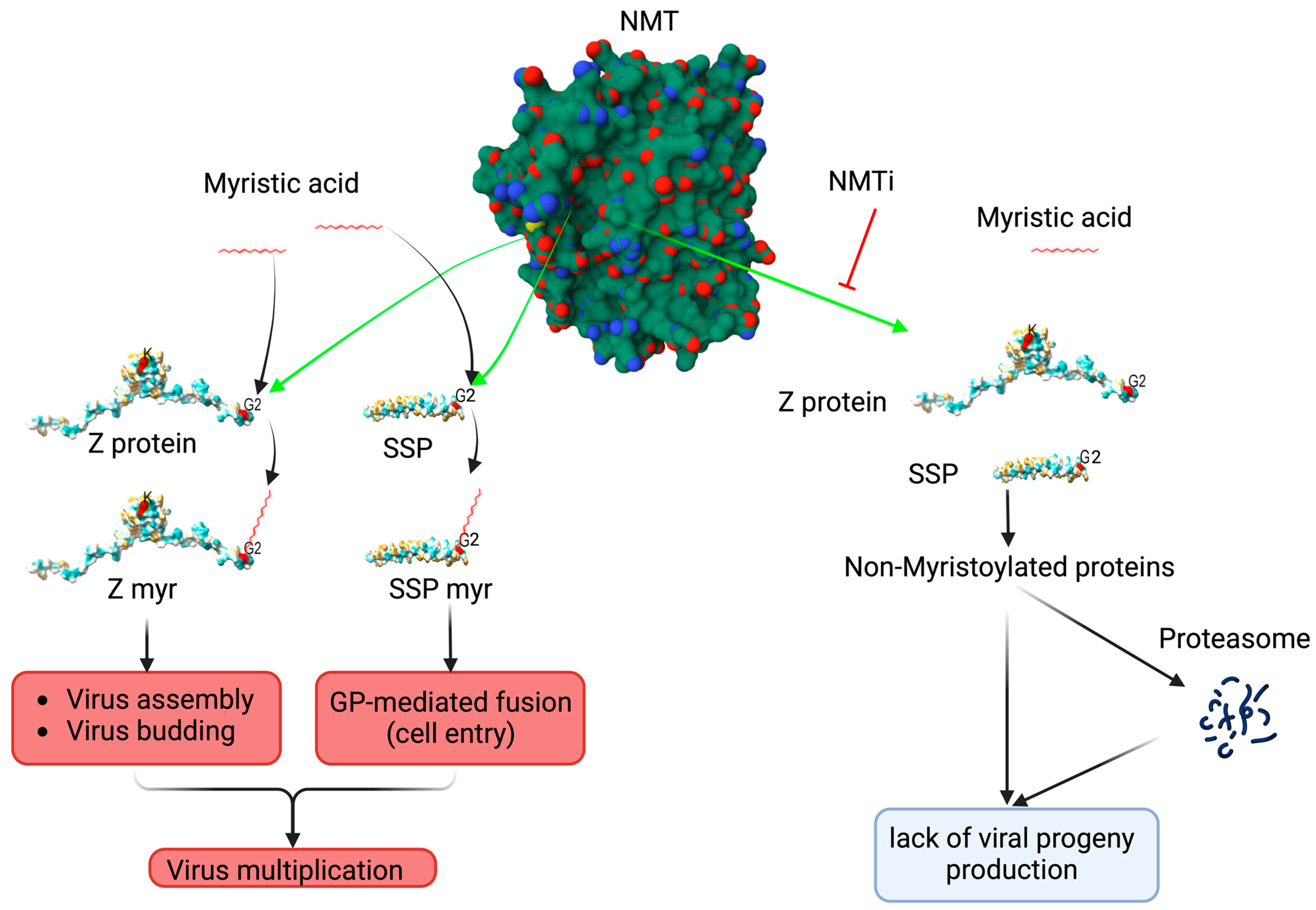
1. Introduction
2. Materials and Methods
2.1. Cell Line and Compounds
2.2. Plasmids
2.3. Minigenome Assay
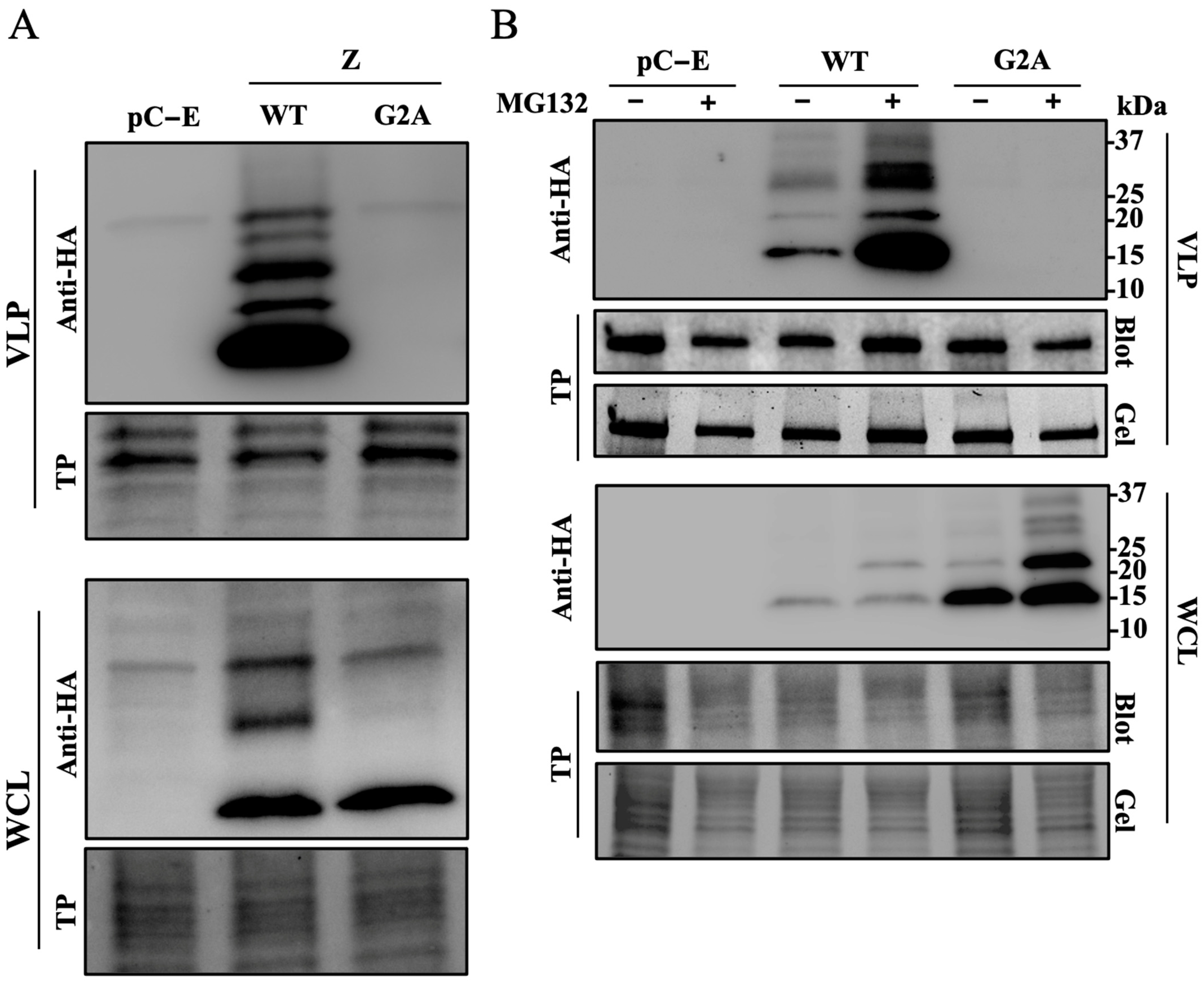
2.4. VLP Assay
2.5. Immunoblotting
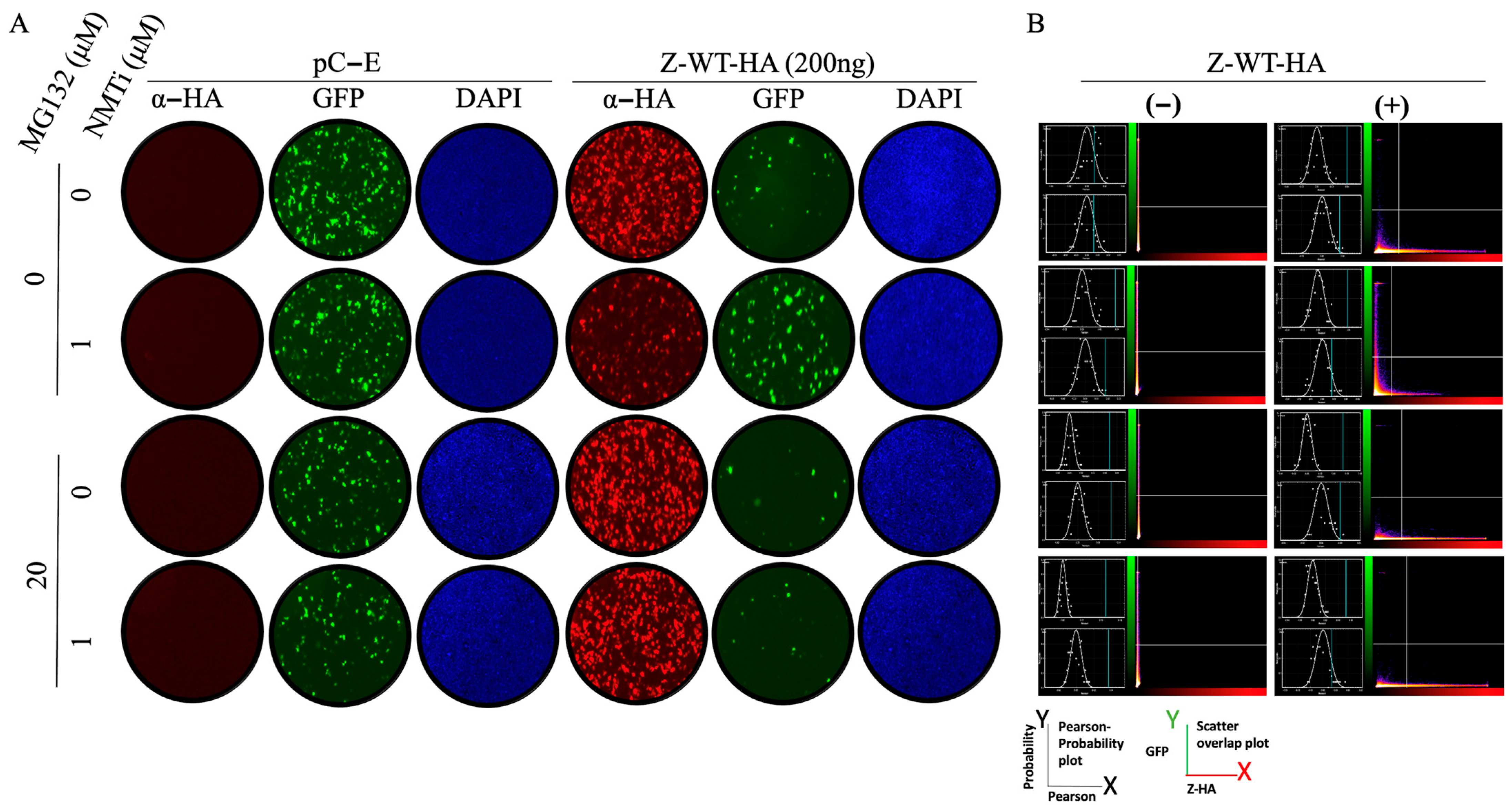
2.6. Immunofluorescence Imaging
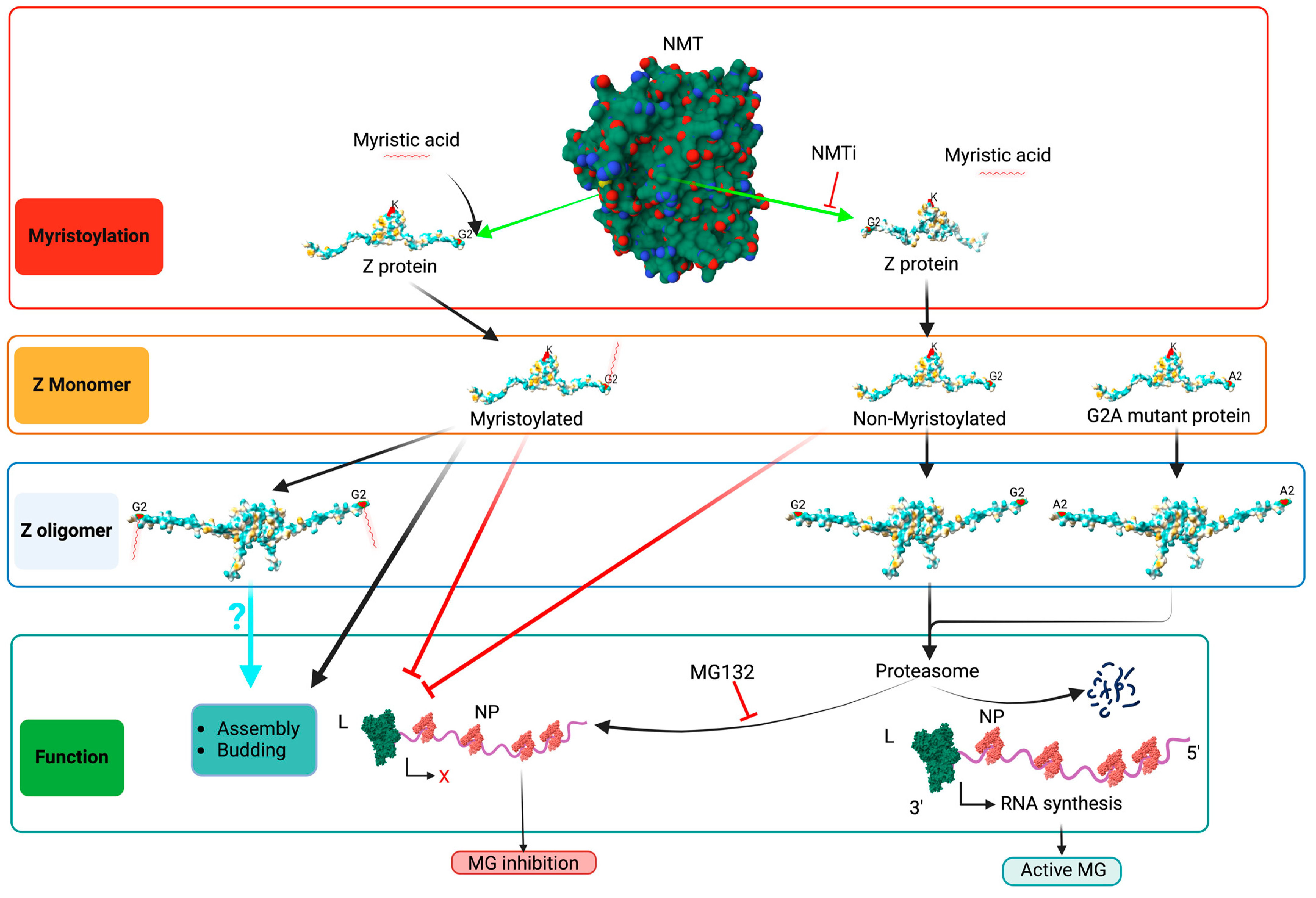
3. Results
3.1. Effect of NMTi on Z-Mediated Inhibition of vRNP Activity
3.2. Role of N-Myristoylation on Z-Mediated Inhibition of vRNP Activity
3.3. Role of Z Oligomerization on Z-Mediated Inhibition of vRNP Activity and Z Budding Activity
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yadav, K.; Mathur, G.; Ford, B.; Miller, R.; Group, C.W. A Case Cluster of Lymphocytic Choriomeningitis Virus Transmitted Via Organ Transplantation.: Abstract# D2381. Transplantation 2014, 98, 768. [Google Scholar]
- Sayyad, L.E.; Smith, K.L.; Sadigh, K.S.; Cossaboom, C.M.; Choi, M.J.; Whitmer, S.; Cannon, D.; Krapiunaya, I.; Morales-Betoulle, M.; Annambhotla, P.; et al. Severe Non–Donor-Derived Lymphocytic Choriomeningitis Virus Infection in 2 Solid Organ Transplant Recipients. Open Forum Infect. Dis. 2025, 12, ofaf002. [Google Scholar] [CrossRef]
- Schafer, I.J.; Miller, R.; Ströher, U.; Knust, B.; Nichol, S.T.; Rollin, P.E.; Centers for Disease Control and Prevention (CDC). Notes from the Field: A Cluster of Lymphocytic Choriomeningitis Virus Infections Transmitted Through Organ Transplantation—Iowa, 2013. MMWR Morb. Mortal. Wkly. Rep. 2014, 63, 249. [Google Scholar] [CrossRef]
- Carrillo-Bustamante, P.; Nguyen, T.H.T.; Oestereich, L.; Günther, S.; Guedj, J.; Graw, F. Determining Ribavirin’s Mechanism of Action against Lassa Virus Infection. Sci. Rep. 2017, 7, 11693. [Google Scholar] [CrossRef]
- Radoshitzky, S.R.; Buchmeier, M.; de la Torre, J.C. Emerging Viruses: Arenaviridae. In Fields Virology, 7th ed.; Knipe, D., Howley, P., Whelan, S., Eds.; Wolters Kluwer: Philadelphia, PA, USA, 2021; Volume I, ISBN 978-1-975112-54-7. [Google Scholar]
- Kunz, S.; Edelmann, K.H.; de la Torre, J.-C.; Gorney, R.; Oldstone, M.B.A. Mechanisms for Lymphocytic Choriomeningitis Virus Glycoprotein Cleavage, Transport, and Incorporation into Virions. Virology 2003, 314, 168–178. [Google Scholar] [CrossRef]
- Rojek, J.M.; Lee, A.M.; Nguyen, N.; Spiropoulou, C.F.; Kunz, S. Site 1 Protease Is Required for Proteolytic Processing of the Glycoproteins of the South American Hemorrhagic Fever Viruses Junin, Machupo, and Guanarito. J. Virol. 2008, 82, 6045–6051. [Google Scholar] [CrossRef]
- Fedeli, C.; Moreno, H.; Kunz, S. Novel Insights into Cell Entry of Emerging Human Pathogenic Arenaviruses. J. Mol. Biol. 2018, 430, 1839–1852. [Google Scholar] [CrossRef]
- York, J.; Nunberg, J.H. Role of the Stable Signal Peptide of Junín Arenavirus Envelope Glycoprotein in pH-Dependent Membrane Fusion. J. Virol. 2006, 80, 7775–7780. [Google Scholar] [CrossRef]
- York, J.; Nunberg, J.H. Myristoylation of the Arenavirus Envelope Glycoprotein Stable Signal Peptide Is Critical for Membrane Fusion but Dispensable for Virion Morphogenesis. J. Virol. 2016, 90, 8341–8350. [Google Scholar] [CrossRef]
- Perez, M.; Greenwald, D.L.; de La Torre, J.C. Myristoylation of the RING Finger Z Protein Is Essential for Arenavirus Budding. J. Virol. 2004, 78, 11443–11448. [Google Scholar] [CrossRef]
- Capul, A.A.; Perez, M.; Burke, E.; Kunz, S.; Buchmeier, M.J.; de la Torre, J.C. Arenavirus Z-Glycoprotein Association Requires Z Myristoylation but Not Functional RING or Late Domains. J. Virol. 2007, 81, 9451–9460. [Google Scholar] [CrossRef]
- Cordo, S.M.; Candurra, N.A.; Damonte, E.B. Myristic Acid Analogs Are Inhibitors of Junin Virus Replication. Microbes Infect. 1999, 1, 609–614. [Google Scholar] [CrossRef]
- Kallemeijn, W.W.; Lueg, G.A.; Faronato, M.; Hadavizadeh, K.; Grocin, A.G.; Song, O.-R.; Howell, M.; Calado, D.P.; Tate, E.W. Validation and Invalidation of Chemical Probes for the Human N-Myristoyltransferases. Cell Chem. Biol. 2019, 26, 892–900.e4. [Google Scholar] [CrossRef]
- Ramljak, I.C.; Stanger, J.; Real-Hohn, A.; Dreier, D.; Wimmer, L.; Redlberger-Fritz, M.; Fischl, W.; Klingel, K.; Mihovilovic, M.D.; Blaas, D.; et al. Cellular N-Myristoyltransferases Play a Crucial Picornavirus Genus-Specific Role in Viral Assembly, Virion Maturation, and Infectivity. PLoS Pathog. 2018, 14, e1007203. [Google Scholar] [CrossRef]
- Witwit, H.; Betancourt, C.A.; Cubitt, B.; Khafaji, R.; Kowalski, H.; Jackson, N.; Ye, C.; Martinez-Sobrido, L.; de la Torre, J.C. Cellular N-Myristoyl Transferases Are Required for Mammarenavirus Multiplication. Viruses 2024, 16, 1362. [Google Scholar] [CrossRef]
- Cornu, T.I.; de la Torre, J.C. Characterization of the Arenavirus RING Finger Z Protein Regions Required for Z-Mediated Inhibition of Viral RNA Synthesis. J. Virol. 2002, 76, 6678–6688. [Google Scholar] [CrossRef]
- Jácamo, R.; López, N.; Wilda, M.; Franze-Fernández, M.T. Tacaribe Virus Z Protein Interacts with the L Polymerase Protein To Inhibit Viral RNA Synthesis. J. Virol. 2003, 77, 10383–10393. [Google Scholar] [CrossRef]
- López, N.; Jácamo, R.; Franze-Fernández, M.T. Transcription and RNA Replication of Tacaribe Virus Genome and Antigenome Analogs Require N and L Proteins: Z Protein Is an Inhibitor of These Processes. J. Virol. 2001, 75, 12241–12251. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Liu, A.; Zhang, L.; Yan, L.; Guo, Y.; Xiao, G.; Rao, Z.; Lou, Z. Structure Basis for Allosteric Regulation of Lymphocytic Choriomeningitis Virus Polymerase Function by Z Matrix Protein. Protein Cell 2023, 14, 703–707. [Google Scholar] [CrossRef]
- Cornu, T.I.; de la Torre, J.C. RING Finger Z Protein of Lymphocytic Choriomeningitis Virus (LCMV) Inhibits Transcription and RNA Replication of an LCMV S-Segment Minigenome. J. Virol. 2001, 75, 9415–9426. [Google Scholar] [CrossRef]
- Iwasaki, M.; de la Torre, J.C. A Highly Conserved Leucine in Mammarenavirus Matrix Z Protein Is Required for Z Interaction with the Virus L Polymerase and Z Stability in Cells Harboring an Active Viral Ribonucleoprotein. J. Virol. 2018, 92, e02256-17. [Google Scholar] [CrossRef] [PubMed]
- Hastie, K.M.; Zandonatti, M.; Liu, T.; Li, S.; Woods, V.L.; Saphire, E.O. Crystal Structure of the Oligomeric Form of Lassa Virus Matrix Protein Z. J. Virol. 2016, 90, 4556–4562. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, M.E.; Wilda, M.; Levingston Macleod, J.M.; D’Antuono, A.; Foscaldi, S.; Buslje, C.M.; Lopez, N. Molecular Determinants of Arenavirus Z Protein Homo-Oligomerization and L Polymerase Binding. J. Virol. 2011, 85, 12304–12314. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Hutchinson, A.; Wernimont, A.; Lin, Y.-H.; Kania, A.; Ravichandran, M.; Kozieradzki, I.; Cossar, D.; Schapira, M.; Arrowsmith, C.H.; et al. Crystal Structure of Human Type-I N-Myristoyltransferase with Bound Myristoyl-CoA and Inhibitor DDD85646. 2010; To be published. [Google Scholar] [CrossRef]
- Volpon, L.; Osborne, M.J.; Capul, A.A.; de la Torre, J.C.; Borden, K.L.B. Structural Characterization of the Z RING-eIF4E Complex Reveals a Distinct Mode of Control for eIF4E. Proc. Natl. Acad. Sci. USA 2010, 107, 5441–5446. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Perez, M.; Craven, R.C.; De La Torre, J.C. The Small RING Finger Protein Z Drives Arenavirus Budding: Implications for Antiviral Strategies. Proc. Natl. Acad. Sci. USA 2003, 100, 12978–12983. [Google Scholar] [CrossRef]
- Baudin, F.; Petit, I.; Weissenhorn, W.; Ruigrok, R.W. In Vitro Dissection of the Membrane and RNP Binding Activities of Influenza Virus M1 Protein. Virology 2001, 281, 102–108. [Google Scholar] [CrossRef]
- Cornu, T.I.; Feldmann, H.; de la Torre, J.C. Cells Expressing the RING Finger Z Protein Are Resistant to Arenavirus Infection. J. Virol. 2004, 78, 2979–2983. [Google Scholar] [CrossRef]
- Kranzusch, P.J.; Whelan, S.P.J. Arenavirus Z Protein Controls Viral RNA Synthesis by Locking a Polymerase–Promoter Complex. Proc. Natl. Acad. Sci. USA 2011, 108, 19743–19748. [Google Scholar] [CrossRef]
- Witwit, H.; Cubitt, B.; Khafaji, R.; Castro, E.M.; Goicoechea, M.; Lorenzo, M.M.; Blasco, R.; Martinez-Sobrido, L.; de la Torre, J.C. Repurposing Drugs for Synergistic Combination Therapies to Counteract Monkeypox Virus Tecovirimat Resistance. Viruses 2025, 17, 92. [Google Scholar] [CrossRef]
- Sangha, R.; Davies, N.M.; Namdar, A.; Chu, M.; Spratlin, J.; Beauchamp, E.; Berthiaume, L.G.; Mackey, J.R. Novel, First-in-Human, Oral PCLX-001 Treatment in a Patient with Relapsed Diffuse Large B-Cell Lymphoma. Curr. Oncol. 2022, 29, 1939–1946. [Google Scholar] [CrossRef] [PubMed]
- Sangha, R.S.; Jamal, R.; Spratlin, J.L.; Kuruvilla, J.; Sehn, L.H.; Weickert, M.; Berthiaume, L.G.; Mackey, J.R. A First-in-Human, Open-Label, Phase I Trial of Daily Oral PCLX-001, an NMT Inhibitor, in Patients with Relapsed/Refractory B-Cell Lymphomas and Advanced Solid Tumors. JCO 2023, 41, e15094. [Google Scholar] [CrossRef]
- Priyamvada, L.; Kallemeijn, W.W.; Faronato, M.; Wilkins, K.; Goldsmith, C.S.; Cotter, C.A.; Ojeda, S.; Solari, R.; Moss, B.; Tate, E.W.; et al. Inhibition of Vaccinia Virus L1 N-Myristoylation by the Host N-Myristoyltransferase Inhibitor IMP-1088 Generates Non-Infectious Virions Defective in Cell Entry. PLoS Pathog. 2022, 18, e1010662. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Witwit, H.; de la Torre, J.C. Mammarenavirus Z Protein Myristoylation and Oligomerization Are Not Required for Its Dose-Dependent Inhibitory Effect on vRNP Activity. BioChem 2025, 5, 10. https://doi.org/10.3390/biochem5020010
Witwit H, de la Torre JC. Mammarenavirus Z Protein Myristoylation and Oligomerization Are Not Required for Its Dose-Dependent Inhibitory Effect on vRNP Activity. BioChem. 2025; 5(2):10. https://doi.org/10.3390/biochem5020010
Chicago/Turabian StyleWitwit, Haydar, and Juan C. de la Torre. 2025. "Mammarenavirus Z Protein Myristoylation and Oligomerization Are Not Required for Its Dose-Dependent Inhibitory Effect on vRNP Activity" BioChem 5, no. 2: 10. https://doi.org/10.3390/biochem5020010
APA StyleWitwit, H., & de la Torre, J. C. (2025). Mammarenavirus Z Protein Myristoylation and Oligomerization Are Not Required for Its Dose-Dependent Inhibitory Effect on vRNP Activity. BioChem, 5(2), 10. https://doi.org/10.3390/biochem5020010







