Primer Binding Site (PBS) Profiling of Genetic Diversity of Natural Populations of Endemic Species Allium ledebourianum Schult.
Abstract
1. Introduction
2. Materials and Methods
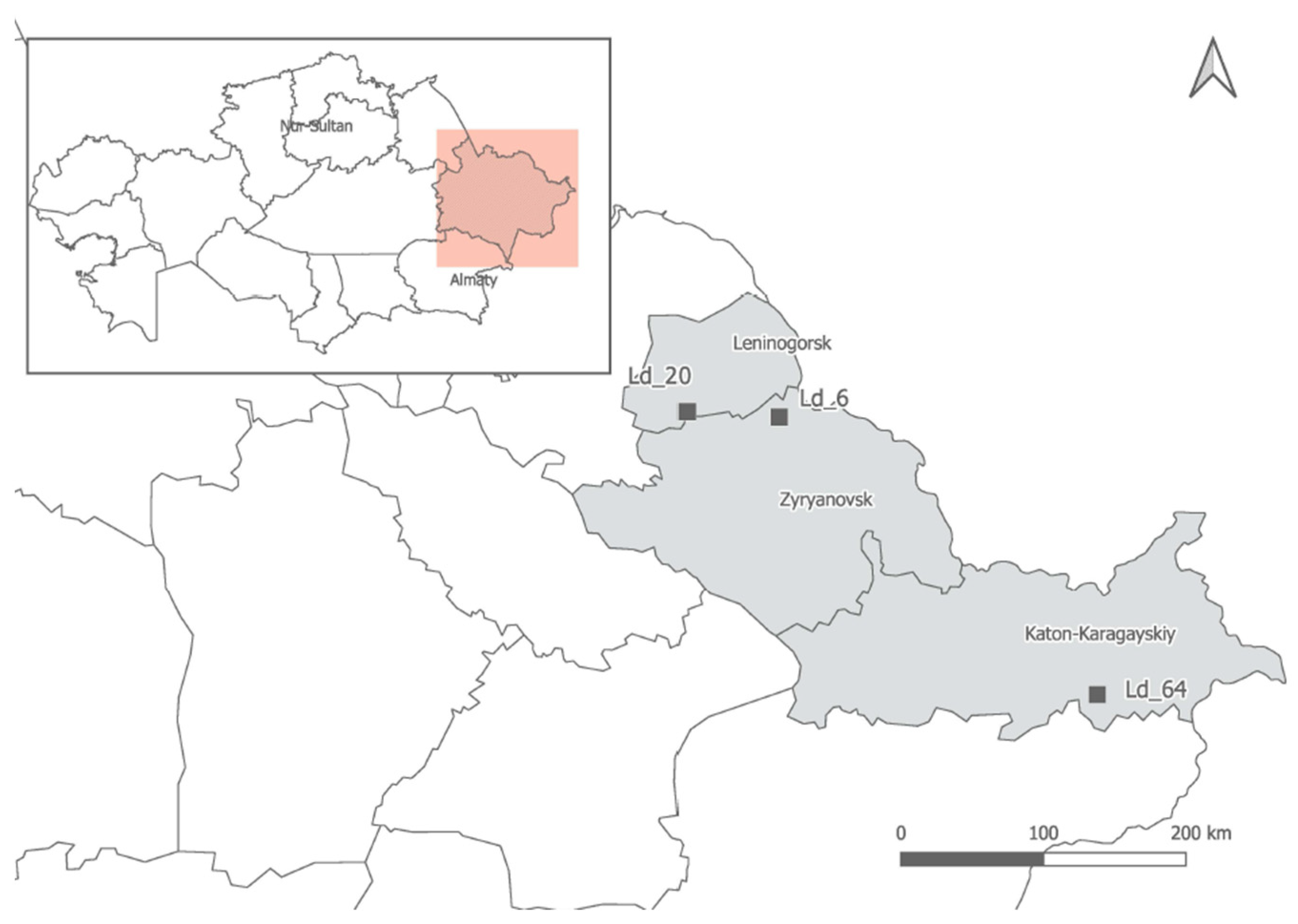
2.1. Plant Material
2.2. DNA Isolation and PCR Amplification
2.3. Data Scoring and Analysis
2.4. Allium ledebourianum Sample Collection and Characteristics
3. Results
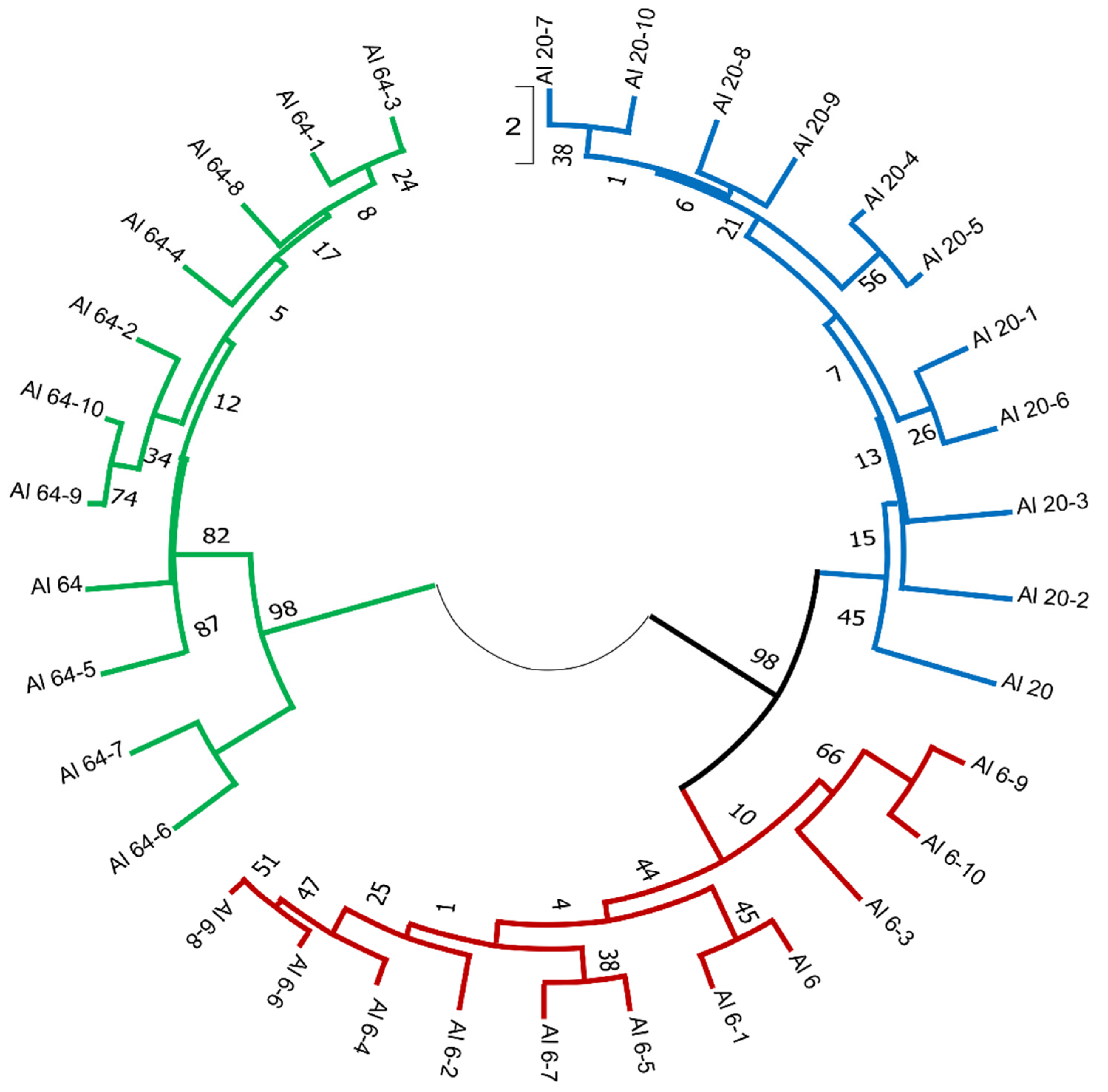
3.1. iPBS Loci Variability Polymorphism Analysis of Natural A. ledebourianum Populations
3.2. Analysis of the Genetic Differences between A. ledebourianum Populations Based on iPBS Amplification
3.3. Relationship between the Analyzed Parameters of A. ledebourianum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Newbold, T.; Hudson, L.N.; Contu, S.; Hill, S.L.; Beck, J.; Liu, Y.; Meyer, C.; Phillips, H.R.; Scharlemann, J.P.; Purvis, A. Widespread winners and narrow-ranged losers: Land use homogenizes biodiversity in local assemblages worldwide. PLoS Biol. 2018, 16, e2006841. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, D.; Li, W.; Li, Y.; Zhang, C.; Guan, K.; Pan, B. Characteristics and utilization of plant diversity and resources in central asia. Reg. Sustain. 2020, 1, 1–10. [Google Scholar] [CrossRef]
- Brugière, D.; Scholte, P. Biodiversity gap analysis of the protected area system in poorly-documented chad. J. Nat. Conserv. 2013, 21, 286–293. [Google Scholar] [CrossRef]
- Monastersky, R. Biodiversity: Life—A status report. Nature 2014, 516, 158–161. [Google Scholar] [CrossRef]
- Pyak, A.I.; Shaw, S.C.; Ebel, A.L.; Zverev, A.A.; Hodgson, J.G.; Wheeler, B.D.; Gaston, K.J.; Morenko, M.O.; Revushkin, A.S.; Kotukhov, Y.A.; et al. Endemic Plants of the Altai Mountain Country; WILDGuides Ltd.: Hampshire, UK, 2008; Volume 368. [Google Scholar]
- Gemejiyeva, N.G.; Grudzinskaya, L.M. Current state and prospects for studies on the diversity of medicinal flora in kazakhstan. In Vegetation of Central Asia and Environs; Springer International Publishing: Berlin, Gemany, 2018; pp. 239–262. [Google Scholar] [CrossRef]
- Baitulin, I.O. Red Book of Kazakhstan; Public-Welfare: Astana, Kazakhstan, 2014; Volume 2, p. 452. [Google Scholar]
- Ivachenko, A.; Grudzinskaya, L.; Gemedzhievo, N.; Da Silva, J.; Ryabushkina, N. Genetic resources of kazakhstan flora: Experience, basic targets and methods for conservation of flowering plants. Floric. Ornam. Plant Biotechnol. 2006, 12, 583–584. [Google Scholar]
- Kaulfuß, F.; Reisch, C. Reintroduction of the endangered and endemic plant species cochlearia bavarica—Implications from conservation genetics. Ecol. Evol. 2017, 7, 11100–11112. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Ji, Y.; Liu, J.; Gong, X. Genetic characterization of the entire range of Cycas panzhihuaensis (cycadaceae). Plant Divers. 2020, 42, 7–18. [Google Scholar] [CrossRef]
- Kotukhov, Y.; Danilova, A.; Anufrieva, O. Abstract of bows (Allium L.) of Kazakhstan altai, sauro-manrak and zaisan depression. Anniv. Editor. 2011, 1–171. [Google Scholar]
- Kruckeberg, A.R.; Rabinowitz, D. Biological aspects of endemism in higher plants. Annu. Rev. Ecol. Syst. 1985, 16, 447–479. [Google Scholar] [CrossRef]
- Kramina, T.E.; Degtjareva, G.V.; Samigullin, T.H.; Valiejo-Roman, C.M.; Kirkbride, J.; Joseph, H.; Volis, S.; Deng, T.; Sokoloff, D.D. Phylogeny of lotus (leguminosae: Loteae): Partial incongruence between nrits, nrets and plastid markers and biogeographic implications. Taxon 2016, 65, 997–1018. [Google Scholar] [CrossRef]
- Pérez-Vargas, I.; Portero Álvarez, A.M.; Pérez de Paz, P.L.; PÉrez, J.A. Retrotransposon-based molecular markers as a tool in delimiting species in section ryncholotus, a recent radiation group of macaronesian lotus. Syst. Biodivers. 2020, 19, 110–120. [Google Scholar] [CrossRef]
- Martínez, S.D.; Boedeker, C.; Zuccarello, G. Genetic data support reproductively isolated species in the endemic cladophoraceae (chlorophyta) of lake baikal, russia. Phycologia 2020, 60, 120–130. [Google Scholar] [CrossRef]
- Auvinen, A.-P.; Kemppainen, E.; Von Weissenberg, M. Fourth National Report on the Implementation of the Convention on Biological Diversity of Finland; Ministry of the Environment: Helsinki, Finland, 2010. [Google Scholar]
- McGlaughlin, M.E.; Riley, L.; Brandsrud, M.; Arcibal, E.; Helenurm, M.K.; Helenurm, K. How much is enough? Minimum sampling intensity required to capture extant genetic diversity in ex situ seed collections: Examples from the endangered plant Sibara filifolia (brassicaceae). Conserv. Genet. 2015, 16, 253–266. [Google Scholar] [CrossRef]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by simple sequence repeat (ssr)-anchored polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. Aflp: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Shustov, A.; Schulman, A. Palindromic sequence-targeted (pst) pcr, version 2: An advanced method for high-throughput targeted gene characterization and transposon display. Front. Plant Sci. 2021, 12, 691940. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Shustov, A.V.; Seppanen, M.M.; Schulman, A.H.; Stoddard, F.L. Palindromic sequence-targeted (pst) pcr: A rapid and efficient method for high-throughput gene characterization and genome walking. Sci. Rep. 2019, 9, 17707. [Google Scholar] [CrossRef]
- Kalendar, R.; Amenov, A.; Daniyarov, A. Use of retrotransposon-derived genetic markers to analyse genomic variability in plants. Funct. Plant Biol. 2019, 46, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Kospanova, D.; Schulman, A.H. Transposon-based tagging in silico using fastpcr software. Methods Mol. Biol. 2021, 2250, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Muterko, A.; Boronnikova, S. Retrotransposable elements: DNA fingerprinting and the assessment of genetic diversity. Methods Mol. Biol. 2021, 2222, 263–286. [Google Scholar] [CrossRef]
- Kaul, S.; Koo, H.L.; Jenkins, J.; Rizzo, M.; Rooney, T.; Tallon, L.J.; Feldblyum, T.; Nierman, W.; Benito, M.I.; Lin, X. Analysis of the genome sequence of the flowering plant arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Sabot, F.; Schulman, A.H. Parasitism and the retrotransposon life cycle in plants: A hitchhiker’s guide to the genome. Heredity 2006, 97, 381–388. [Google Scholar] [CrossRef]
- Fu, J.; Zhang, H.; Guo, F.; Ma, L.; Wu, J.; Yue, M.; Zheng, X.; Qiu, Z.; Li, L. Identification and characterization of abundant repetitive sequences in allium cepa. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef]
- Peška, V.; Mandáková, T.; Ihradská, V.; Fajkus, J. Comparative dissection of three giant genomes: Allium cepa, allium sativum, and allium ursinum. Int. J. Mol. Sci. 2019, 20, 733. [Google Scholar] [CrossRef] [PubMed]
- Kirov, I.V.; Kiseleva, A.V.; Van Laere, K.; Van Roy, N.; Khrustaleva, L.I. Tandem repeats of allium fistulosum associated with major chromosomal landmarks. Mol. Genet. Genom. 2017, 292, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Khapilina, O.; Raiser, O.; Danilova, A.; Shevtsov, V.; Turzhanova, A.; Kalendar, R. DNA profiling and assessment of genetic diversity of relict species allium altaicum pall. On the territory of altai. PeerJ 2021, 9, e10674. [Google Scholar] [CrossRef] [PubMed]
- Belyayev, A.; Josefiova, J.; Jandova, M.; Kalendar, R.; Krak, K.; Mandak, B. Natural history of a satellite DNA family: From the ancestral genome component to species-specific sequences, concerted and non-concerted evolution. Int. J. Mol. Sci. 2019, 20, 1201. [Google Scholar] [CrossRef]
- Dorogina, O.; Zhmud, E. Molecular-genetic methods in plant ecology. Contemp. Probl. Ecol. 2020, 13, 333–345. [Google Scholar] [CrossRef]
- Erper, I.; Ozer, G.; Kalendar, R.; Avci, S.; Yildirim, E.; Alkan, M.; Turkkan, M. Genetic diversity and pathogenicity of rhizoctonia spp. Isolates associated with red cabbage in samsun (turkey). J. Fungi 2021, 7, 234. [Google Scholar] [CrossRef]
- Turzhanova, A.; Khapilina, O.N.; Tumenbayeva, A.; Shevtsov, V.; Raiser, O.; Kalendar, R. Genetic diversity of alternaria species associated with black point in wheat grains. PeerJ 2020, 8, e9097. [Google Scholar] [CrossRef]
- Hosid, E.; Brodsky, L.; Kalendar, R.; Raskina, O.; Belyayev, A. Diversity of long terminal repeat retrotransposon genome distribution in natural populations of the wild diploid wheat aegilops speltoides. Genetics 2012, 190, 263–274. [Google Scholar] [CrossRef]
- Domingues, D.S.; Cruz, G.M.; Metcalfe, C.J.; Nogueira, F.T.; Vicentini, R.; de S Alves, C.; Van Sluys, M.-A. Analysis of plant ltr-retrotransposons at the fine-scale family level reveals individual molecular patterns. BMC Genom. 2012, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Poczai, P.; Varga, I.; Laos, M.; Cseh, A.; Bell, N.; Valkonen, J.P.; Hyvönen, J. Advances in plant gene-targeted and functional markers: A review. Plant Methods 2013, 9, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Gurlen, A.; Gundogdu, M.; Ozer, G.; Ercisli, S.; Duralija, B. Primary, secondary metabolites and molecular characterization of hawthorn (Crataegus spp.) genotypes. Agronomy 2020, 10, 1731. [Google Scholar] [CrossRef]
- Vanijajiva, O.; Pornpongrungrueng, P. Inter-primer binding site (ipbs) markers reveal the population genetic diversity and structure of tropical climbing cissampelopsis (asteraceae) in thailand. Biodiversitas J. Biol. Divers. 2020, 21. [Google Scholar] [CrossRef]
- Pakhrou, O.; Medraoui, L.; Yatrib, C.; Alami, M.; Filali-Maltouf, A.; Belkadi, B. Assessment of genetic diversity and population structure of an endemic moroccan tree (Argania spinosa L.) based in irap and issr markers and implications for conservation. Physiol. Mol. Biol. Plants 2017, 23, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Beguiristain, T.; Grandbastien, M.-A.; Puigdomènech, P.; Casacuberta, J.M. Three tnt1 subfamilies show different stress-associated patterns of expression in tobacco. Consequences for retrotransposon control and evolution in plants. Plant Physiol. 2001, 127, 212–221. [Google Scholar] [CrossRef]
- Kalendar, R.; Tanskanen, J.; Immonen, S.; Nevo, E.; Schulman, A.H. Genome evolution of wild barley (Hordeum spontaneum) by bare-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc. Natl. Acad. Sci. USA 2000, 97, 6603–6607. [Google Scholar] [CrossRef] [PubMed]
- Boyko, A.; Kovalchuk, I. Epigenetic control of plant stress response. Environ. Mol. Mutagenesis 2008, 49, 61–72. [Google Scholar] [CrossRef]
- Cavrak, V.V.; Lettner, N.; Jamge, S.; Kosarewicz, A.; Bayer, L.M.; Scheid, O.M. How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 2014, 10, e1004115. [Google Scholar] [CrossRef]
- Dufoo-Hurtado, M.D.; Zavala-Gutiérrez, K.G.; Cao, C.-M.; Cisneros-Zevallos, L.; Guevara-González, R.n.G.; Torres-Pacheco, I.; Vázquez-Barrios, M.E.; Rivera-Pastrana, D.M.; Mercado-Silva, E.M. Low-temperature conditioning of “seed” cloves enhances the expression of phenolic metabolism related genes and anthocyanin content in ‘coreano’garlic (Allium sativum) during plant development. J. Agric. Food Chem. 2013, 61, 10439–10446. [Google Scholar] [CrossRef]
- Belyayev, A.; Kalendar, R.; Brodsky, L.; Nevo, E.; Schulman, A.H.; Raskina, O. Transposable elements in a marginal plant population: Temporal fluctuations provide new insights into genome evolution of wild diploid wheat. Mob. DNA 2010, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Grandbastien, M.-A. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998, 3, 181–187. [Google Scholar] [CrossRef]
- Vicient, C.M.; Jääskeläinen, M.J.; Kalendar, R.; Schulman, A.H. Active retrotransposons are a common feature of grass genomes. Plant Physiol. 2001, 125, 1283–1292. [Google Scholar] [CrossRef]
- Capy, P.; Gasperi, G.; Biémont, C.; Bazin, C. Stress and transposable elements: Co-evolution or useful parasites? Heredity 2000, 85, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Horbowicz, M.; Kotlinska, T. Level of flavonols in wild and cultivated allium species. Acta Hortic. 2000, 375–380. [Google Scholar] [CrossRef]
- Uranov, A.A.; Smirnova, O.V. Classification and main features of development of populations of perennial plants. Bull. Mosc. Soc. Nat. Dep. Biol. 1969, 2, 119–134. [Google Scholar]
- Gatsuk, L.E.; Smirnova, O.V.; Vorontzova, L.I.; Zaugolnova, L.B.; Zhukova, L.A. Age states of plants of various growth forms: A review. J. Ecol. 1980, 68, 675. [Google Scholar] [CrossRef]
- Zhivotovsky, L. Ontogenetic states, effective density, and classification of plant populations. Russ. J. Ecol. 2001, 32, 1–5. [Google Scholar] [CrossRef]
- Mirkin, B.; Rozenberg, G. Phytocenology: Principles and Methods; Nauka: Moscow, Russia, 1978; Volume 212. [Google Scholar]
- Kalendar, R.; Boronnikova, S.; Seppanen, M. Isolation and purification of DNA from complicated biological samples. Methods Mol. Biol. 2021, 2222, 57–67. [Google Scholar] [CrossRef]
- Kalendar, R.; Antonius, K.; Smykal, P.; Schulman, A.H. Ipbs: A universal method for DNA fingerprinting and retrotransposon isolation. Theor. Appl. Genet. 2010, 121, 1419–1430. [Google Scholar] [CrossRef]
- Kalendar, R.; Khassenov, B.; Ramankulov, Y.; Samuilova, O.; Ivanov, K.I. Fastpcr: An in silico tool for fast primer and probe design and advanced sequence analysis. Genomics 2017, 109, 312–319. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. Genalex 6: Genetic analysis in excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega x: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- RC Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Meirmans, P.G. The trouble with isolation by distance. Mol. Ecol. 2012, 21, 2839–2846. [Google Scholar] [CrossRef] [PubMed]
- Bezemer, N.; Krauss, S.L.; Roberts, D.G.; Hopper, S.D. Conservation of old individual trees and small populations is integral to maintain species’ genetic diversity of a historically fragmented woody perennial. Mol. Ecol. 2019, 28, 3339–3357. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.J.; Bradburd, G.S. Isolation by environment. Mol. Ecol. 2014, 23, 5649–5662. [Google Scholar] [CrossRef]
- Klaas, M. Applications and impact of molecular markers on evolutionary and diversity studies in the genus allium. Plant Breed. 1998, 117, 297–308. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Driscoll, H.E.; Specht, C.D. A molecular phylogeny of the wild onions (allium; alliaceae) with a focus on the western north american center of diversity. Mol. Phylogenet. Evol. 2008, 47, 1157–1172. [Google Scholar] [CrossRef] [PubMed]
- Duminil, J.; Di Michele, M. Plant species delimitation: A comparison of morphological and molecular markers. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2009, 143, 528–542. [Google Scholar] [CrossRef]
- Havey, M.J. Onion breeding. Plant Breed. Rev. 2018, 42, 39–85. [Google Scholar]
- Khosa, J.S.; McCallum, J.; Dhatt, A.S.; Macknight, R.C. Enhancing onion breeding using molecular tools. Plant Breed. 2015, 135, 9–20. [Google Scholar] [CrossRef]
- Karic, L.; Golzardi, M.; Glamoclija, P.; Sutkovic, J. Genetic diversity assessment of allium cepa l. Cultivars from bosnia and herzegovina using ssr makers. Genet. Mol. Res. 2018, 17, 16039870. [Google Scholar] [CrossRef]
- Abugalieva, S.; Volkova, L.; Genievskaya, Y.; Ivaschenko, A.; Kotukhov, Y.; Sakauova, G.; Turuspekov, Y. Taxonomic assessment of allium species from kazakhstan based on its and matk markers. BMC Plant Biol. 2017, 17, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Vasfilova, E.S.; Vorob’eva, T.y.A. Peculiarities of accumulation of glucofructans in Allium L. (Amaryllidaceae) species. Vestn. Tomsk. Gos. Universiteta. Biol. 2018, 42, 160–175. [Google Scholar] [CrossRef] [PubMed]
- Kotukhov, Y.; Danilova, A.; Anufrieva, O. Current State of Rare and Endangered Plants in East Kazakhstan; Tethys: Almaty, Kazakhstan, 2009. [Google Scholar]
- Żuraw, B. Biological value and morphological traits of pollen of selected garlic species Allium L. Acta Agrobot. 2012, 60, 67–71. [Google Scholar] [CrossRef][Green Version]
- Friesen, N.; Borisjuk, N.; Mes, T.H.; Klaas, M.; Hanelt, P. Allotetraploid origin of Allium altyncolicum (Alliaceae, Allium sect. Schoenoprasum) as investigated by karyological and molecular markers. Plant Syst. Evol. 1997, 206, 317–335. [Google Scholar] [CrossRef]
- Friesen, N.; Fritsch, R.; Bachmann, K. Hybrid origin of some ornamentals of allium subgenus melanocrommyum verified with gish and rapd. Theor. Appl. Genet. 1997, 95, 1229–1238. [Google Scholar] [CrossRef]
- Friesen, N.; Fritsch, R.M.; Blattner, F.R. Phylogeny and new intrageneric classification of allium (Alliaceae) based on nuclear ribosomal DNA its sequences. Aliso A J. Syst. Evol. Bot. 2006, 22, 372–395. [Google Scholar] [CrossRef]
- Buso, G.; Paiva, M.; Torres, A.; Resende, F.; Ferreira, M.; Buso, J.; Dusi, A. Genetic diversity studies of brazilian garlic cultivars and quality control of garlic-clover production. Genet. Mol. Res. 2008, 7, 534–541. [Google Scholar] [CrossRef]
- Chen, S.; Chen, W.; Shen, X.; Yang, Y.; Qi, F.; Liu, Y.; Meng, H. Analysis of the genetic diversity of garlic (Allium sativum L.) by simple sequence repeat and inter simple sequence repeat analysis and agro-morphological traits. Biochem. Syst. Ecol. 2014, 55, 260–267. [Google Scholar] [CrossRef]
- Dyachenko, E.; Filyushin, M.; Seredin, T. Nuclear and chloroplast genome variability in leek (Allium porrum L.). Vavilovskij Žurnal Genet. I Sel. 2019, 23, 902–909. [Google Scholar] [CrossRef]
- Sudha, G.S.; Ramesh, P.; Sekhar, A.C.; Krishna, T.S.; Bramhachari, P.; Riazunnisa, K. Genetic diversity analysis of selected onion (Allium cepa L.) germplasm using specific rapd and issr polymorphism markers. Biocatal. Agric. Biotechnol. 2019, 17, 110–118. [Google Scholar] [CrossRef]
- Villano, C.; Esposito, S.; Carucci, F.; Iorizzo, M.; Frusciante, L.; Carputo, D.; Aversano, R. High-throughput genotyping in onion reveals structure of genetic diversity and informative snps useful for molecular breeding. Mol. Breed. 2019, 39, 1–11. [Google Scholar] [CrossRef]
- Filyushin, M.A.; Beletsky, A.V.; Mazur, A.M.; Kochieva, E.Z. Characterization of the complete plastid genome of lop-sided onion Allium obliquum L. (Amaryllidaceae). Mitochondrial DNA Part B 2018, 3, 393–394. [Google Scholar] [CrossRef] [PubMed]
- Nault, A.; Gagnon, D. Ramet demography of allium tricoccum, a spring ephemeral, perennial forest herb. J. Ecol. 1993, 81, 101–119. [Google Scholar] [CrossRef]
- Ellstrand, N.C.; Elam, D.R. Population genetic consequences of small population size: Implications for plant conservation. Annu. Rev. Ecol. Syst. 1993, 24, 217–242. [Google Scholar] [CrossRef]



| Primer ID | Sequence 5′–3′ | Tm (°C) * | Total Bands | Polymorphism Loci (%) | Polymorphism Information Content |
|---|---|---|---|---|---|
| 2228 | CATTGGCTCTTGATACCA | 50.2 | 258 | 48.7 | 0.763 |
| 2240 | AACCTGGCTCAGATGCCA | 54.7 | 195 | 38.1 | 0.965 |
| 2395 | TCCCCAGCGGAGTCGCCA | 53.0 | 175 | 41.6 | 0.886 |
| Mean | 209 | 42.8 |
| Population | |||
|---|---|---|---|
| LD20 | LD6 | LD64 | |
| Geographical Coordinates | Western Altai. Northwestern foot of the Ivanovsky ridge, the Gray Lug tract, northwestern slope | Western Altai, southern part of Lineisky ridge, top of the Barsuk river | Southern Altai, Sarymsakty ridge, southeastern Foothills, Tautekeli river valley, park larch forest |
| 50°21′27″ N 83°53′54″ E 1170 m above sea level | 50°19′12″ N 84°11′49″ E 1925 m above sea level | 49°05′37″ N 86°12′36″ E 1925 m above sea level | |
| Soil moistening | Moderate | Excessive | Moderate |
| Soil | Mountain-meadow | Wetland meadows | Mountain meadow |
| Subsoil | Moderately developed, 5–7 cm | Not developed, represented by litter | Intensively developed, more than 10 cm |
| Spatial structure of the population | Diffuse scattered | Diffused | Densely diffuse, spots of 3–5 individuals |
| Projective cover | 75% | 75% | 90% |
| Phytocenosis | Calamagrostis purpurea, Filipendula ulmaria, Veratrum lobelianum Bernh., Allium ledebourianum, Poa remota, Alopecurus pratensis, Veratrum lobelianum. | Filipendula ulmaria (L.) Maxim., Phalaroides arundinaceae (L.) Rauschert, Calamagrostis purpurea (Trin.) Trin., Allium ledebourianum. Lathyrus pratensis L., Cerastium davuricum Fisch. ex Spreng., Carex rostrata Stokes, Alopecurus pratensis L., Poa sibirica Roshev., P. remota Forsell. | Calamagrostis epigeios (L.) Roth, Elymus mutabilis (Drob.) Tzvel. |
| Signs | LD20 | LD6 | LD64 |
|---|---|---|---|
| Shoot height during the flowering phase (min/max), cm | 93/108 | 32/53 | 65/84 |
| Average height, cm | 99.6 ± 1.2 | 44.8 ± 2.9 | 68.2 ± 3.4 |
| Number of inflorescences per bush | 6 | 5 | 4 |
| Number of flowers per inflorescence | 80.8 | 69.1 | 87.2 |
| Number of formed bolls per inflorescence | 33.9 | 36.6 | 64.6 |
| Fruit formation coefficient, % | 41.9 | 52.9 | 74.1 |
| Number of bolls per bush | 995.0 | 647.0 | 565.0 |
| Boll formations per bush, % | 98.0 | 95.0 | 93.0 |
| Potential seed production of one generative shoot, pcs. | 484.8 | 414.6 | 522.0 |
| Actual seed production of one generative shoot, pcs. | 140.1 | 91.5 | 161.5 |
| Productivity rate, % | 28.9 | 22.1 | 30.9 |
| Number of seeds per inflorescence | 1194.0 | 725.0 | 678.0 |
| Weight 1000 pcs, g | 1.31 | 1.22 | 1.11 |
| Seed length, mm | 3.46 ± 0.25 | 3.08 ± 0.31 | 3.19 ± 0.24 |
| Seed width, mm | 2.01 ± 0.25 | 2.32 ± 0.24 | 1.98 ± 0.55 |
| Ground germination, % | 28 | 48 | 61.2 |
| Laboratory germination, % | 73 | 71.5 | 83.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khapilina, O.; Turzhanova, A.; Danilova, A.; Tumenbayeva, A.; Shevtsov, V.; Kotukhov, Y.; Kalendar, R. Primer Binding Site (PBS) Profiling of Genetic Diversity of Natural Populations of Endemic Species Allium ledebourianum Schult. BioTech 2021, 10, 23. https://doi.org/10.3390/biotech10040023
Khapilina O, Turzhanova A, Danilova A, Tumenbayeva A, Shevtsov V, Kotukhov Y, Kalendar R. Primer Binding Site (PBS) Profiling of Genetic Diversity of Natural Populations of Endemic Species Allium ledebourianum Schult. BioTech. 2021; 10(4):23. https://doi.org/10.3390/biotech10040023
Chicago/Turabian StyleKhapilina, Oxana, Ainur Turzhanova, Alevtina Danilova, Asem Tumenbayeva, Vladislav Shevtsov, Yuri Kotukhov, and Ruslan Kalendar. 2021. "Primer Binding Site (PBS) Profiling of Genetic Diversity of Natural Populations of Endemic Species Allium ledebourianum Schult." BioTech 10, no. 4: 23. https://doi.org/10.3390/biotech10040023
APA StyleKhapilina, O., Turzhanova, A., Danilova, A., Tumenbayeva, A., Shevtsov, V., Kotukhov, Y., & Kalendar, R. (2021). Primer Binding Site (PBS) Profiling of Genetic Diversity of Natural Populations of Endemic Species Allium ledebourianum Schult. BioTech, 10(4), 23. https://doi.org/10.3390/biotech10040023







