Current Understanding of Novel Coronavirus: Molecular Pathogenesis, Diagnosis, and Treatment Approaches
Abstract
:1. Introduction
2. Classification, Origin, Primary Reservoirs, and Hosts of Coronavirus
3. Transmission Modes of Coronavirus
4. Genome Structure and Life Cycle
5. Risk Factors of Coronavirus
6. The Clinical Manifestations of COVID-19
7. Effect of ACE-2 on SARS-CoV-2 Infection
8. Immunopathological Mechanisms of SARS-CoV-2 Infection
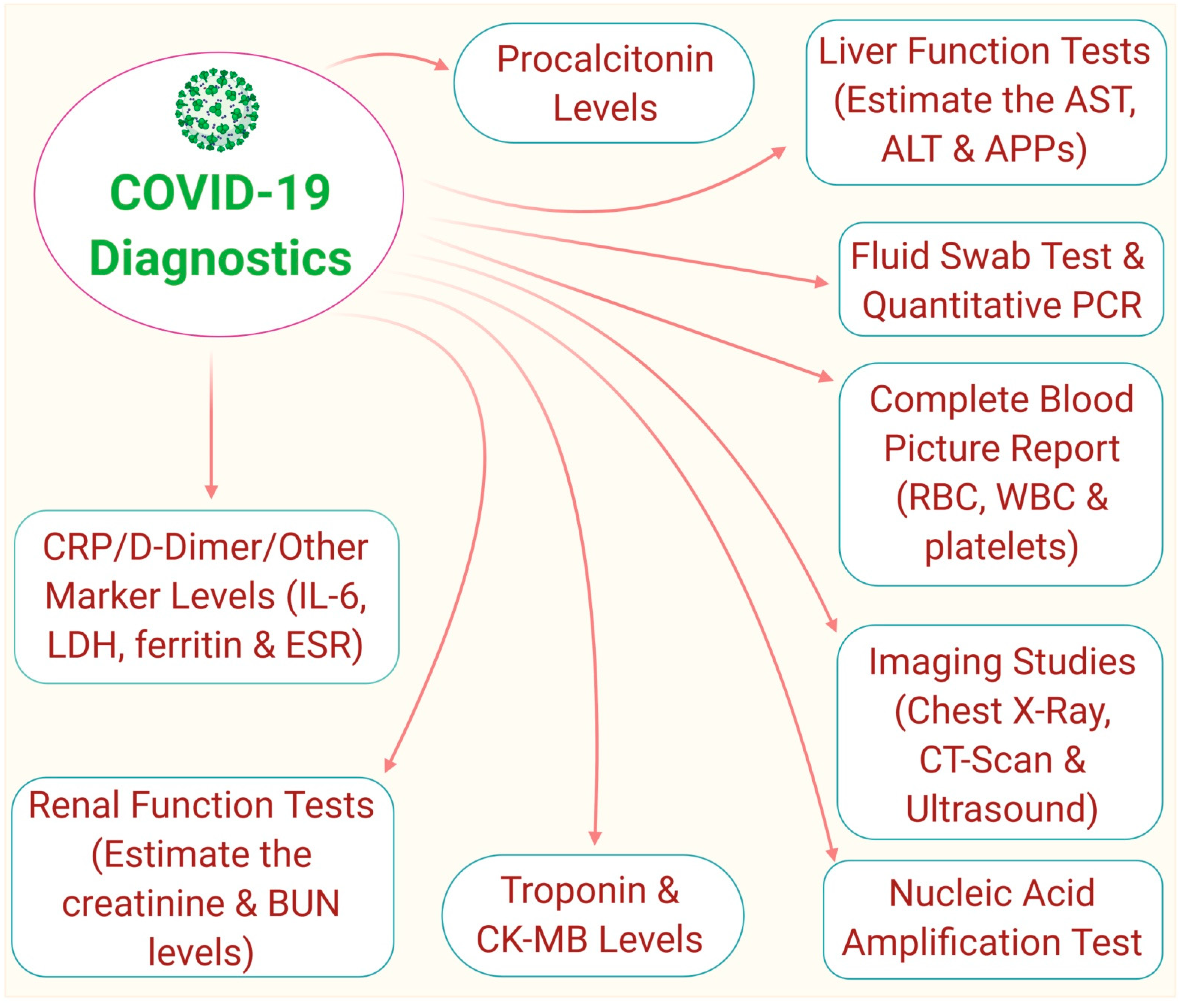
9. Diagnosis of COVID-19
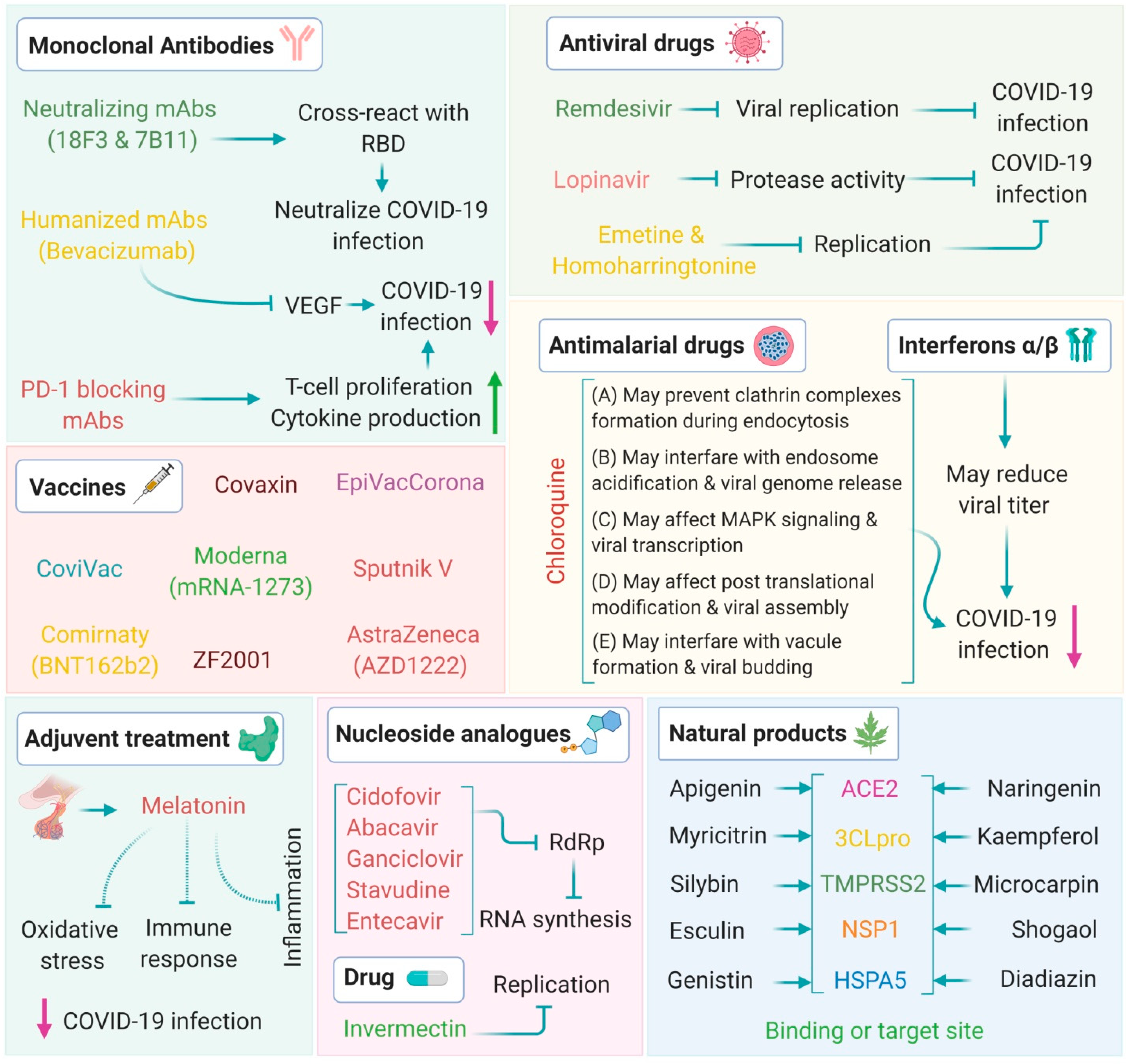
10. Therapeutic Strategies
10.1. Antiviral Drugs
10.2. Vaccines
11. Other Promising Therapeutics
12. Obstacles to Research on COVID-19 Pathogenesis
13. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Liu, S.-M.; Yu, X.-H.; Tang, S.-L.; Tang, C.-K. Coronavirus disease 2019 (COVID-19): Current status and future perspectives. Int. J. Antimicrob. Agents 2020, 55, 105951. [Google Scholar] [CrossRef]
- Guo, Y.-R.; Cao, Q.-D.; Hong, Z.-S.; Tan, Y.-Y.; Chen, S.-D.; Jin, H.-J.; Tan, K.-S.; Wang, D.-Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Mil. Med Res. 2020, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.-Y.; Ma, Y.-T.; Zhang, J.-Y.; Xie, X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020, 17, 259–260. [Google Scholar] [CrossRef] [Green Version]
- Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H. Treatment of SARS with human interferons. Lancet 2003, 362, 293–294. [Google Scholar] [CrossRef]
- Florindo, H.F.; Kleiner, R.; Vaskovich-Koubi, D.; Acúrcio, R.C.; Carreira, B.; Yeini, E.; Tiram, G.; Liubomirski, Y.; Satchi-Fainaro, R. Immune-mediated approaches against COVID-19. Nat. Nanotechnol. 2020, 15, 630–645. [Google Scholar] [CrossRef]
- Chan, J.F.; Chan, K.-H.; Kao, R.Y.; To, K.K.; Zheng, B.-J.; Li, C.P.; Li, P.T.; Dai, J.; Mok, F.K.; Chen, H.; et al. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013, 67, 606–616. [Google Scholar] [CrossRef] [Green Version]
- Cheng, K.-W.; Cheng, S.-C.; Chen, W.-Y.; Lin, M.-H.; Chuang, S.-J.; Cheng, I.-H.; Sun, C.-Y.; Chou, C.-Y. Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antivir. Res. 2015, 115, 9–16. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y.; Wu, A.; Xu, S.; Pan, R.; Zeng, C.; Jin, X.; Ge, X.; Shi, Z.; Ahola, T.; et al. Coronavirus nsp10/nsp16 methyltransferase can be targeted by nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J. Virol. 2015, 89, 8416–8427. [Google Scholar] [CrossRef] [Green Version]
- Mair-Jenkins, J.; Saavedra-Campos, M.; Baillie, J.K.; Cleary, P.; Khaw, F.M.; Lim, W.S.; Makki, S.; Rooney, K.D.; Nguyen-Van-Tam, J.S.; Beck, C.R. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: A systematic review and exploratory meta-analysis. J. Infect. Dis. 2015, 211, 80–90. [Google Scholar] [CrossRef] [Green Version]
- Graham, R.L.; Donaldson, E.F.; Baric, R.S. A decade after SARS: Strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013, 11, 836–848. [Google Scholar] [CrossRef] [Green Version]
- De Wit, E.; Feldmann, F.; Cronin, J.; Jordan, R.; Okumura, A.; Thomas, T.; Scott, D.; Cihlar, T.; Feldmann, H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA 2020, 3, 2019–2208. [Google Scholar] [CrossRef] [Green Version]
- Burrell, C.J.; Howard, C.R.; Murphy, F.A. Fenner and White’s Medical Virology; Elsevier BV: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Burrell, A.J.; Pellegrini, B.; Salimi, F.; Begum, H.; Broadley, T.; Campbell, L.T.; Cheng, A.C.; Cheung, W.; Cooper, D.J.; Earnest, A.; et al. Outcomes for patients with COVID-19 admitted to Australian intensive care units during the first four months of the pandemic. Med. J. Aust. 2021, 214, 23–30. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Hughes, J.; Wilson, M.; Luby, S.; Gurley, E.; Hossain, M. Transmission of human infection with Nipah virus. Clin. Infect. Dis. 2009, 49, 1743–1748. [Google Scholar]
- Kan, B.; Wang, M.; Jing, H.; Xu, H.; Jiang, X.; Yan, M.; Liang, W.; Zheng, H.; Wan, K.; Liu, Q.; et al. Molecular Evolution Analysis and Geographic Investigation of Severe Acute Respiratory Syndrome Coronavirus-Like Virus in Palm Civets at an Animal Market and on Farms. J. Virol. 2005, 79, 11892–11900. [Google Scholar] [CrossRef] [Green Version]
- Zheng, B.J.; Guan, Y.; Wong, K.H.; Zhou, J.; Wong, K.L.; Young, B.W.Y.; Lu, L.W.; Lee, S.S. SARS-related Virus Predating SARS Outbreak, Hong Kong. Emerg. Infect. Dis. 2004, 10, 176–178. [Google Scholar] [CrossRef]
- Shi, Z.; Hu, Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008, 133, 74–87. [Google Scholar] [CrossRef]
- Memish, Z.A.; Zumla, A.I.; Al-Hakeem, R.F.; Al-Rabeeah, A.A.; Stephens, G.M. Family Cluster of Middle East Respiratory Syndrome Coronavirus Infections. N. Engl. J. Med. 2013, 368, 2487–2494. [Google Scholar] [CrossRef]
- Annan, A.; Baldwin, H.J.; Corman, V.M.; Klose, S.M.; Owusu, M.; Nkrumah, E.E.; Badu, E.K.; Anti, P.; Agbenyega, O.; Meyer, B. Human betacorona virus 2c EMC/2012–related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013, 19, 456. [Google Scholar] [CrossRef] [PubMed]
- Huynh, J.; Li, S.; Yount, B.; Smith, A.; Sturges, L.; Olsen, J.C.; Nagel, J.; Johnson, J.B.; Agnihothram, S.; Gates, J.E.; et al. Evidence Supporting a Zoonotic Origin of Human Coronavirus Strain NL63. J. Virol. 2012, 86, 12816–12825. [Google Scholar] [CrossRef] [Green Version]
- Lau, S.K.P.; Li, K.S.M.; Tsang, A.K.L.; Lam, C.S.F.; Ahmed, S.; Chen, H.; Chan, K.-H.; Woo, P.C.Y.; Yuen, K.-Y. Genetic Characterization of Betacoronavirus Lineage C Viruses in Bats Reveals Marked Sequence Divergence in the Spike Protein of Pipistrellus Bat Coronavirus HKU5 in Japanese Pipistrelle: Implications for the Origin of the Novel Middle East Respiratory Syndrome Coronavirus. J. Virol. 2013, 87, 8638–8650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Stratton, C.W.; Tang, Y. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J. Med. Virol. 2020, 92, 401–402. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Zhao, K.; Shi, Z.-L.; Zhou, P. Bat Coronaviruses in China. Viruses 2019, 11, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahase, E. China coronavirus: What do we know so far? BMJ 2020, 368, m308. [Google Scholar] [CrossRef] [Green Version]
- Hui, D.S.; Azhar, E.I.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Morales, A.J.; Bonilla-Aldana, D.K.; Balbin-Ramon, G.J.; Rabaan, A.A.; Sah, R.; Paniz-Mondolfi, A.; Pagliano, P.; Esposito, S. History is repeating itself: Probable zoonotic spillover as the cause of the 2019 novel coronavirus epidemic. Infez. Med. 2020, 28, 3–5. [Google Scholar]
- Xu, X.-W.; Wu, X.-X.; Jiang, X.-G.; Xu, K.-J.; Ying, L.-J.; Ma, C.-L.; Li, S.-B.; Wang, H.-Y.; Zhang, S.; Gao, H.-N.; et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: Retrospective case series. BMJ 2020, 368, m606. [Google Scholar] [CrossRef] [Green Version]
- Ison, M.G.; Hirsch, H.H. Community-acquired respiratory viruses in transplant patients: Diversity, impact, unmet clinicalneeds. Clin. Microbiol. Rev. 2019, 32, e00042-19. [Google Scholar]
- Kramer, A.; Schwebke, I.; Kampf, G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 2006, 6, 130. [Google Scholar] [CrossRef] [Green Version]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Rothe, C.; Schunk, M.; Sothmann, P.; Bretzel, G.; Froeschl, G.; Wallrauch, C.; Zimmer, T.; Thiel, V.; Janke, C.; Guggemos, W. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020, 382, 970–971. [Google Scholar] [CrossRef] [Green Version]
- Kupferschmidt, K. Study claiming new coronavirus can be transmitted by people without symptoms was flawed. Science 2020. [Google Scholar] [CrossRef]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Guan, X.; Wu, P.; Qun, L.; Xuhua, G.; Peng, W.; Xiaoye, W.; Lei, Z.; Yeqing, T.; Ruiqi, R.; et al. Early transmission dynamics in Wuhan, China, of Novel Coronavirus—Infected pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- WHO (World Health Organization). Clinical Management of Severe Acute Respiratory Infection When Novel Coronavirus (2019-nCoV) Infection Is Suspected: Interim Guidance; WHO: New York, NY, USA, 2020; pp. 1–19. [Google Scholar]
- Bauch, C.; Oraby, T. Assessing the pandemic potential of MERS-CoV. Lancet 2013, 382, 662–664. [Google Scholar] [CrossRef] [Green Version]
- Riley, S.; Fraser, C.; Donnelly, C.A.; Ghani, A.C.; Abu-Raddad, L.J.; Hedley, A.J.; Leung, G.M.; Ho, L.-M.; Lam, T.-H.; Thach, T.Q.; et al. Transmission Dynamics of the Etiological Agent of SARS in Hong Kong: Impact of Public Health Interventions. Science 2003, 300, 1961–1966. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Sugimoto, J.D.; Halloran, M.E.; Basta, N.E.; Chao, D.L.; Matrajt, L.; Potter, G.; Kenah, E.; Longini, I.M. The Transmissibility and Control of Pandemic Influenza A (H1N1) Virus. Science 2009, 326, 729–733. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Gayle, A.A.; Wilder-Smith, A.; Rocklöv, J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020, 27, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kucharski, A.; Althaus, C. The role of superspreading in Middle East respiratory syndrome coronavirus (MERS-CoV) transmission. Euro Surveill. 2015, 20, 21167. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Guo, J.; Wang, C.; Luo, F.; Yu, X.; Zhang, W.; Li, J.; Zhao, D.; Xu, D.; Gong, Q.; et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: A retrospective review of medical records. Lancet 2020, 395, 809–815. [Google Scholar] [CrossRef] [Green Version]
- McBride, R.; van Zyl, M.; Fielding, B.C. The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of 350 the same clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef]
- Klausegger, A.; Strobl, B.; Regl, G.; Kaser, A.; Luytjes, W.; Vlasak, R. Identification of a Coronavirus Hemagglutinin-Esterase with a Substrate Specificity Different from Those of Influenza C Virus and Bovine Coronavirus. J. Virol. 1999, 73, 3737–3743. [Google Scholar] [CrossRef] [Green Version]
- Bárcena, M.; Oostergetel, G.T.; Bartelink, W.; Faas, F.G.; Verkleij, A.; Rottier, P.J.; Koster, A.J.; Bosch, B.J. Cryo-electron tomography of mouse hepatitis virus: Insight to the structure of the coronavirion. Proc. Natl. Acad. Sci. USA 2009, 106, 582–587. [Google Scholar] [CrossRef] [Green Version]
- Mortola, E.; Roy, P. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 2004, 576, 174–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beniac, D.R.; Andonov, A.; Grudeski, E.; Booth, T.F. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006, 13, 751–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delmas, B.; Laude, H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 1990, 64, 5367–5375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nal, B. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005, 86, 1423–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuman, B.W.; Kiss, G.; Kunding, A.H.; Bhella, D.; Baksh, M.F.; Connelly, S.; Droese, B.; Klaus, J.P.; Makino, S.; Sawicki, S.G.; et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011, 174, 11–22. [Google Scholar] [CrossRef]
- De Diego, M.L.; Álvarez, E.; Almazán, F.; Rejas, M.T.; Lamirande, E.; Roberts, A.; Shieh, W.-J.; Zaki, S.R.; Subbarao, K.; Enjuanes, L. A Severe Acute Respiratory Syndrome Coronavirus That Lacks the E Gene Is Attenuated In Vitro and In Vivo. J. Virol. 2006, 81, 1701–1713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieto-Torres, J.L.; de Diego, M.L.; Verdiá-Báguena, C.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Fernandez-Delgado, R.; Castaño-Rodriguez, C.; Alcaraz, A.; Torres, J.; Aguilella, V.M. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014, 10, e1004077. [Google Scholar] [CrossRef] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar]
- Chang, C.-K.; Sue, S.-C.; Yu, T.-H.; Hsieh, C.-M.; Tsai, C.-K.; Chiang, Y.-C.; Lee, S.-J.; Hsiao, H.-H.; Wu, W.-J.; Chang, W.-L.; et al. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2005, 13, 59–72. [Google Scholar] [CrossRef] [Green Version]
- Hurst, K.R.; Koetzner, C.A.; Masters, P.S. Identification of In Vivo-Interacting Domains of the Murine Coronavirus Nucleocapsid Protein. J. Virol. 2009, 83, 7221–7234. [Google Scholar] [CrossRef] [Green Version]
- Cui, L.; Wang, H.; Ji, Y.; Yang, J.; Xu, S.; Huang, X.; Wang, Z.; Qin, L.; Tien, P.; Zhou, X.; et al. The Nucleocapsid Protein of Coronaviruses Acts as a Viral Suppressor of RNA Silencing in Mammalian Cells. J. Virol. 2015, 89, 9029–9043. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.J.; Goh, P.Y.; Fielding, B.C.; Shen, S.; Chou, C.F.; Fu, J.L.; Leong, H.N.; Leo, Y.S.; Ooi, E.E.; Ling, A.E.; et al. Profiles of Antibody Responses against Severe Acute Respiratory Syndrome Coronavirus Recombinant Proteins and Their Potential Use as Diagnostic Markers. Clin. Diagn. Lab. Immunol. 2004, 11, 362–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, X.; Stjohn, S.E.; Osswald, H.L.; O'Brien, A.; Banach, B.S.; Sleeman, K.; Ghosh, A.K.; Mesecar, A.D.; Baker, S.C. Coronaviruses Resistant to a 3C-Like Protease Inhibitor Are Attenuated for Replication and Pathogenesis, Revealing a Low Genetic Barrier but High Fitness Cost of Resistance. J. Virol. 2014, 88, 1186–11898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Götte, B.; Liu, L.; McInerney, G.M. The enigmatic alphavirus non-structural protein 3 (nsP3) revealing its secretsat last. Viruses 2018, 10, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirchdoerfer, R.N.; Ward, A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co365 factors. Nat. Commun. 2019, 10, 2342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Lokugamage, K.G.; Rozovics, J.M.; Narayanan, K.; Semler, B.L.; Makino, S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: Viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011, 7, e1002433. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Kamitani, W.; de Diego, M.L.; Enjuanes, L.; Matsuura, Y. Severe Acute Respiratory Syndrome Coronavirus nsp1 Facilitates Efficient Propagation in Cells through a Specific Translational Shutoff of Host mRNA. J. Virol. 2012, 86, 11128–11137. [Google Scholar] [CrossRef] [Green Version]
- Graham, R.L.; Sims, A.C.; Brockway, S.M.; Baric, R.S.; Denison, M.R. The nsp2 Replicase Proteins of Murine Hepatitis Virus and Severe Acute Respiratory Syndrome Coronavirus Are Dispensable for Viral Replication. J. Virol. 2005, 79, 13399–13411. [Google Scholar] [CrossRef] [Green Version]
- Gadlage, M.J.; Graham, R.L.; Denison, M.R. Murine Coronaviruses Encoding nsp2 at Different Genomic Loci Have Altered Replication, Protein Expression, and Localization. J. Virol. 2008, 82, 11964–11969. [Google Scholar] [CrossRef] [Green Version]
- Lei, J.; Kusov, Y.; Hilgenfeld, R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antivir. Res. 2018, 149, 58–74. [Google Scholar] [CrossRef]
- Serrano, P.; Johnson, M.A.; Chatterjee, A.; Neuman, B.W.; Joseph, J.S.; Buchmeier, M.J.; Kuhn, P.; Wüthrich, K. Nuclear Magnetic Resonance Structure of the Nucleic Acid-Binding Domain of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein 3. J. Virol. 2009, 83, 12998–13008. [Google Scholar] [CrossRef] [Green Version]
- Beachboard, D.C.; Anderson-Daniels, J.M.; Denison, M.R. Mutations across Murine Hepatitis Virus nsp4 Alter Virus Fitness and Membrane Modifications. J. Virol. 2014, 89, 2080–2089. [Google Scholar] [CrossRef] [Green Version]
- Gadlage, M.J.; Sparks, J.S.; Beachboard, D.C.; Cox, R.G.; Doyle, J.D.; Stobart, C.C.; Denison, M.R. Murine Hepatitis Virus Nonstructural Protein 4 Regulates Virus-Induced Membrane Modifications and Replication Complex Function. J. Virol. 2009, 84, 280–290. [Google Scholar] [CrossRef] [Green Version]
- Stobart, C.C.; Sexton, N.R.; Munjal, H.; Lu, X.; Molland, K.L.; Tomar, S.; Mesecar, A.D.; Denison, M.R. Chimeric Exchange of Coronavirus nsp5 Proteases (3CLpro) Identifies Common and Divergent Regulatory Determinants of Protease Activity. J. Virol. 2013, 87, 12611–12618. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Fang, L.; Wang, D.; Yang, Y.; Chen, J.; Ye, X.; Foda, M.F.; Xiao, S. Porcine delta coronavirus nsp5 inhibits interferon-beta production through the cleavage of NEMO. Virology 2017, 502, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, D.; Zhou, J.; Pan, T.; Chen, J.; Yang, Y.; Lv, M.; Ye, X.; Peng, G.; Fang, L.; et al. Porcine Deltacoronavirus nsp5 Antagonizes Type I Interferon Signaling by Cleaving STAT2. J. Virol. 2017, 91, e00003-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelini, M.M.; Akhlaghpour, M.; Neuman, B.W.; Buchmeier, M.J. Severe Acute Respiratory Syndrome Coronavirus Nonstructural Proteins 3, 4, and 6 Induce Double-Membrane Vesicles. mBio 2013, 4, 00524-13. [Google Scholar] [CrossRef] [Green Version]
- Cottam, E.M.; Whelband, M.C.; Wileman, T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy 2014, 10, 1426–1441. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Y.; Sun, F.; Li, X.; Pang, H.; Xu, X.; Bartlam, M.; Rao, Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7–nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005, 12, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Velthuis, A.J.T.; Worm, S.H.E.V.D.; Snijder, E.J. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2011, 40, 1737–1747. [Google Scholar] [CrossRef] [Green Version]
- Egloff, M.-P.; Ferron, F.; Campanacci, V.; Longhi, S.; Rancurel, C.; Dutartre, H.; Snijder, E.J.; Gorbalenya, A.E.; Cambillau, C.; Canard, B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. USA 2004, 101, 3792–3796. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Z.; Deng, F.; Shi, K.; Ye, G.; Wang, G.; Fang, L.; Xiao, S.; Fu, Z.; Peng, G. Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity. J. Virol. 2018, 92, e00692-18. [Google Scholar] [CrossRef] [Green Version]
- Bouvet, M.; Lugari, A.; Posthuma, C.C.; Zevenhoven, J.C.; Bernard, S.; Betzi, S.; Imbert, I.; Canard, B.; Guillemot, J.-C.; Lécine, P.; et al. Coronavirus Nsp10, a Critical Co-factor for Activation of Multiple Replicative Enzymes. J. Biol. Chem. 2014, 289, 25783–25796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Su, C.; Ke, M.; Jin, X.; Xu, L.; Zhang, Z.; Wu, A.; Sun, Y.; Yang, Z.; Tien, P.; et al. Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by nsp16/nsp10 Protein Complex. PLoS Pathog. 2011, 7, e1002294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decroly, E.; Debarnot, C.; Ferron, F.; Bouvet, M.; Coutard, B.; Imbert, I.; Gluais, L.; Papageorgiou, N.; Sharff, A.; Bricogne, G.; et al. Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase nsp10/nsp16 Complex. PLoS Pathog. 2011, 7, e1002059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Wu, L.; Shaw, N.; Gao, Y.; Wang, J.; Sun, Y.; Lou, Z.; Yan, L.; Zhang, R.; Rao, Z. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc. Natl. Acad. Sci. USA 2015, 112, 9436–9441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, S.G.; Shen, H.; Wang, J.; Tay, F.P.L.; Liu, D.X. Proteolytic processing of polyproteins 1a and 1ab between non-structural proteins 10 and 11/ 12 of coronavirus infectious bronchitis virus is dispensable for viral replication in cultured cells. Virology, 2008; 379, 175–180. [Google Scholar]
- Ahn, D.-G.; Choi, J.-K.; Taylor, D.R.; Oh, J.-W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012, 157, 2095–2104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velthuis, A.J.W.T.; Arnold, J.J.; Cameron, C.E.; Worm, S.H.E.V.D.; Snijder, E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2009, 38, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Adedeji, A.O.; Lazarus, H. Biochemical Characterization of Middle East Respiratory Syndrome Coronavirus Helicase. mSphere 2016, 1, e00235-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, W.; Wojdyla, J.A.; Zhao, R.; Han, R.; Das, R.; Zlatev, I.; Manoharan, M.; Wang, M.; Cui, S. Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog. 2017, 13, e1006474. [Google Scholar] [CrossRef]
- Jia, Z.; Yan, L.; Ren, Z.; Wu, L.; Wang, J.; Guo, J.; Zheng, L.; Ming, Z.; Zhang, L.; Lou, Z.; et al. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019, 47, 6538–6550. [Google Scholar] [CrossRef] [Green Version]
- Bouvet, M.; Imbert, I.; Subissi, L.; Gluais, L.; Canard, B.; Decroly, E. RNA 3’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. USA 2012, 109, 9372–9377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Cai, H.; Pan, J.; Xiang, N.; Tien, P.; Ahola, T.; Guo, D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. USA 2009, 106, 3484–3489. [Google Scholar] [CrossRef] [Green Version]
- Minskaia, E.; Hertzig, T.; Gorbalenya, A.E.; Campanacci, V.; Cambillau, C.; Canard, B.; Ziebuhr, J. Discovery of an RNA virus 3’→5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5108–5113. [Google Scholar] [CrossRef] [Green Version]
- Eckerle, L.D.; Becker, M.M.; Halpin, R.A.; Li, K.; Venter, E.; Lu, X.; Scherbakova, S.; Graham, R.L.; Baric, R.S.; Stockwell, T.B.; et al. Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing. PLoS Pathog. 2010, 6, e1000896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhardwaj, K.; Sun, J.; Holzenburg, A.; Guarino, L.A.; Kao, C.C. RNA Recognition and Cleavage by the SARS Coronavirus Endoribonuclease. J. Mol. Biol. 2006, 361, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Hackbart, M.; Mettelman, R.C.; O’Brien, A.; Mielech, A.M.; Yi, G.; Kao, C.C.; Baker, S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. USA 2017, 114, E4251–E4260. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Li, L.; Yan, L.; Ming, Z.; Jia, Z.; Lou, Z.; Rao, Z. Structural and Biochemical Characterization of Endoribonuclease Nsp15 Encoded by Middle East Respiratory Syndrome Coronavirus. J. Virol. 2018, 92, e00893-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, P.; Su, Y.; Li, R.; Liang, Z.; Dong, S.; Huang, J. PEDV nsp16 negatively regulates innate immunity to promote viral proliferation. Virus Res. 2019, 265, 57–66. [Google Scholar]
- Van Boheemen, S.; de Graaf, M.; Lauber, C.; Bestebroer, T.; Raj, V.; Zaki, A.; Osterhaus, A.; Haagmans, B.; Gorbalenya, A.; Snijder, E.; et al. Genomic Characterization of a Newly Discovered Coronavirus Associated with Acute Respiratory Distress Syndrome in Humans. mBio 2012, 3, 00473-12. [Google Scholar] [CrossRef] [Green Version]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.W.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.A.; Zaki, A.; Fouchier, R.A.M.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [Green Version]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Genet. 2009, 7, 439–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Shi, X.; Jiang, L.; Zhang, S.; Wang, D.; Tong, P.; Guo, D.; Fu, L.; Cui, Y.; Liu, X.; et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013, 23, 986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertram, S.; Glowacka, I.; Müller, M.A.; Lavender, H.; Gnirss, K.; Nehlmeier, I.; Niemeyer, D.; He, Y.; Simmons, G.; Drosten, C.; et al. Cleavage and Activation of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein by Human Airway Trypsin-Like Protease. J. Virol. 2011, 85, 13363–13372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glowacka, I.; Bertram, S.; Müller, M.A.; Allen, P.D.; Soilleux, E.J.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence that TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J. Virol. 2011, 85, 4122–4134. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Chen, P.; Wang, J.; Feng, J.; Zhou, H.; Li, X.; Zhong, W.; Hao, P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020, 63, 457–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by novel coro-navirus from Wuhan: An analysis based on decade-long structural studies of SARS. J Virol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus—Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Chang, D.; Lin, M.; Wei, L.; Xie, L.; Zhu, G.; Cruz, C.S.D.; Sharma, L. Epidemiologic and Clinical Characteristics of Novel Coronavirus Infections Involving 13 Patients Outside Wuhan, China. JAMA 2020, 323, 1092. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Zhang, Q.; Chen, J.; Xiang, R.; Song, H.; Shu, S.; Chen, L.; Liang, L.; Zhou, J.; You, L.; et al. Detection of Covid-19 in Children in Early January 2020 in Wuhan, China. N. Engl. J. Med. 2020, 382, 1370–1371. [Google Scholar] [CrossRef]
- Shen, K.L.; Yang, Y.H. Diagnosis and treatment of 2019 novel coronavirus infectionin children: A pressing issue. World J. Pediatr. 2020, 16, 219–221. [Google Scholar]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv 2020, 1–10. [Google Scholar] [CrossRef]
- Zhang, J.-J.; Dong, X.; Cao, Y.-Y.; Yuan, Y.-D.; Yang, Y.-B.; Yan, Y.-Q.; Akdis, C.A.; Gao, Y.-D. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy 2020, 75, 1730–1741. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Hui, D.S.; Perlman, S. Middle East respiratory syndrome. Lancet 2015, 386, 995–1007. [Google Scholar] [CrossRef] [Green Version]
- Al-Omari, A.; Rabaan, A.A.; Salih, S.; Al-Tawfiq, J.A.; Memish, Z.A. MERS coronavirus outbreak: Implications for emerging viral infections. Diagn. Microbiol. Infect. Dis. 2019, 93, 265–285. [Google Scholar] [CrossRef] [PubMed]
- Yeung, M.-L.; Yao, Y.; Jia, L.; Chan, J.F.W.; Chan, K.-H.; Cheung, K.-F.; Chen, H.; Poon, V.K.M.; Tsang, A.K.L.; To, K.K.; et al. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat. Microbiol. 2016, 1, 16004. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Guo, Y.; Pan, Y.; Zhao, Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020, 525, 135–140. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 180, 281–292. [Google Scholar] [CrossRef]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [Green Version]
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020, 14, 185–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan-Yeung, M.; Xu, R.-H. SARS: Epidemiology. Respirology 2003, 8, S9–S14. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yang, J.; Zhao, F.; Zhi, L.; Wang, X.; Liu, L.; Bi, Z.; Zhao, Y. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin. Res. Cardiol. 2020, 109, 531–538. [Google Scholar] [CrossRef]
- Saavedra, J.M. Angiotensin receptor blockers and COVID-19. Pharmacol. Res. 2020, 156, 104832. [Google Scholar] [CrossRef] [PubMed]
- Belouzard, S.; Chu, V.C.; Whittaker, G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA 2009, 106, 5871–5876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millet, J.K.; Whittaker, G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA 2014, 111, 15214–15219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [Green Version]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Totura, A.L.; Baric, R.S. SARS coronavirus pathogenesis: Host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012, 2, 264–275. [Google Scholar] [CrossRef]
- Hamming, I.; Timens, W.; Bulthuis, M.L.; Lely, A.T.; Navis, G.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A firststep in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.K.; Lam, C.W.K.; Wu, A.K.L.; Ip, W.K.; Lee, N.L.S.; Chan, I.H.S.; Lit, L.C.W.; Hui, D.S.C.; Chan, M.H.M.; Chung, S.S.C. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. ExpImmunol. 2004, 136, 95–103. [Google Scholar] [CrossRef] [Green Version]
- Smits, S.L.; de Lang, A.; van den Brand, J.M.; Leijten, L.M.; van Ijcken, W.F.; Eijkemans, M.J.; van Amerongen, G.; Kuiken, T.; Andeweg, A.C.; Osterhaus, A.D. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. 2010, 6, e1000756. [Google Scholar]
- Jin, Y.H.; Cai, L.; Cheng, Z.S.; Cheng, H.; Deng, T.; Fan, Y.P.; Fang, C.; Huang, D.; Huang, L.Q.; Huang, Q. A rapid advice guideline forthe diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil. Med. Res. 2020, 7, 4. [Google Scholar] [PubMed] [Green Version]
- Lei, J.; Li, J.; Li, X.; Qi, X. CT Imaging of the 2019 Novel Coronavirus (2019-nCoV) Pneumonia. Radiology 2020, 295, 18. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.; Bernheim, A.; Mei, X.; Zhang, N.; Huang, M.; Zeng, X.; Cui, J.; Xu, W.; Yang, Y.; Fayad, Z.A. CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology 2020, 295, 200–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habibzadeh, P.; Stoneman, E.K. The novel coronavirus: A bird’s eye view. Int. J. Occup. Environ. Med. 2020, 11, 65–71. [Google Scholar] [CrossRef] [Green Version]
- WHO (World Health Organization). Global Surveillance for Human Infection with Novel Coronavirus (2019-nCoV); WHO: New York, NY, USA, 2020. [Google Scholar]
- Russell, C.C.; Millar, J.K.; Baillie, J.E. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 2020, 395, 473–475. [Google Scholar]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.S.; Kasumba, D.M.; Fujita, T.; Luo, H. Spatio-temporal characterization of the antiviral activity of the XRN1-DCP1/2 aggregation against cytoplasmic RNA viruses to prevent cell death. Cell Death Differ 2020, 8, 1–20, Correction in 2020, 27, 2363–2382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.X.; Fish, E.N. Global virus outbreaks: Interferons as 1st responders. Semin. Immunol. 2019, 43, 101300. [Google Scholar] [CrossRef]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 2018, 9, e00221-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paules, C.I.; Marston, H.D.; Fauci, A.S. Coronavirus infections—More than just the common cold. JAMA 2020, 323, 707–708. [Google Scholar] [CrossRef] [Green Version]
- Holshue, M.L.; de Bolt, C.; Lindquist, S.; Lofy, K.H.; Wiesman, J.; Bruce, H.; Spitters, C.; Ericson, K.; Wilkerson, S.; Tural, A.; et al. First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020, 382, 929–936. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Leist, S.R.; Schäfer, A.; Won, J.; Brown, A.J.; Montgomery, S.A.; Hogg, A.; Babusis, D.; Clarke, M.O.; et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020, 11, 222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, P.; Griffin, I.; Tucker, C.; Smith, D.; Oechsle, O.; Phelan, A.; Rawling, M.; Savory, E.; Stebbing, J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet 2020, 395, e30–e31. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wang, X.-J. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J. Genet. Genom. 2020, 47, 119–121. [Google Scholar] [CrossRef] [PubMed]
- Lu, H. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci. Trends 2020, 14, 69–71. [Google Scholar] [CrossRef] [Green Version]
- Gordon, C.J.; Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Götte, M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020, 295, 4773–4779. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Chan, K.; Jiang, Y.; Kao, R.; Lu, H.; Fan, K.; Cheng, V.; Tsui, W.; Hung, I.; Lee, T. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004, 31, 69–75. [Google Scholar] [CrossRef]
- Yao, T.; Qian, J.; Zhu, W.; Wang, Y.; Wang, G. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus—A possible reference for coronavirus disease-19 treatment option. J. Med Virol. 2020, 92, 556–563. [Google Scholar] [CrossRef] [Green Version]
- Riva, A.; Conti, F.; Bernacchia, D.; Pezzati, L.; Sollima, S.; Merli, S.; Siano, M.; Lupo, A.; Rusconi, S.; Cattaneo, D. Darunavir does not prevent SARS-CoV-2 infection in HIV patients. Pharmacol. Res. 2020, 157, 104826. [Google Scholar] [CrossRef] [PubMed]
- Furuta, Y.; Gowen, B.B.; Takahashi, K.; Shiraki, K.; Smee, D.F.; Barnard, D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 2013, 100, 446–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stockman, L.J.; Bellamy, R.; Garner, P. SARS: Systematic Review of Treatment Effects. PLoS Med. 2006, 3, e343. [Google Scholar] [CrossRef] [Green Version]
- Kadam, R.U.; Wilson, I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. USA 2017, 114, 206–214. [Google Scholar] [CrossRef] [Green Version]
- Khamitov, R.A.; Loginova, S.; Shchukina, V.N.; Borisevich, S.V.; Maksimov, V.A.; Shuster, A.M. Antiviral activity of arbidoland its derivatives against the pathogen of severe acute respiratory syndromein the cell cultures. Vopr. Virusol. 2008, 53, 9. [Google Scholar]
- Al-Bari, M.A.A. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 2017, 5, e00293. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Ye, F.; Zhang, M.; Cui, C.; Huang, B.; Niu, P.; Liu, X.; Zhao, L.; Dong, E.; Song, C.; et al. In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2020, 71, 732–739. [Google Scholar] [CrossRef] [Green Version]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA—Approved Drug Ivermectin inhibits the replication ofSARS-CoV-2 in vitro. J Antivir. 2020, 178, 1047–1087. [Google Scholar]
- Rossignol, J.-F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J. Infect. Public Heal. 2016, 9, 227–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Zhang, F.; Xu, M.; Huang, K.; Zhong, W.; Cai, W.; Yin, Z.; Huang, S.; Deng, Z.; Wei, M.; et al. Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J. Med. Microbiol. 2003, 52, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Gautret, P.; Lagier, J.C.; Parola, P.; Meddeb, L.; Mailhe, M.; Doudier, B.; Courjon, J.; Giordanengo, V.; Vieira, V.E.; Dupont, H.T.; et al. Hydroxychloroquine and Azithryomycin as a treatment of COVID-results of an openlabel non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020, 20, 105949. [Google Scholar] [CrossRef]
- Pan, L.; Mu, M.; Yang, P.; Sun, Y.; Wang, R.; Yan, J.; Li, P.; Hu, B.; Wang, J.; Hu, C.; et al. Clinical Characteristics of COVID-19 Patients with Digestive Symptoms in Hubei, China: A Descriptive, Cross-Sectional, Multicenter Study. Am. J. Gastroenterol. 2020, 115, 766–773. [Google Scholar] [CrossRef]
- Vaduganathan, M.; Vardeny, O.; Michel, T.; McMurray, J.J.V.; Pfeffer, M.A.; Solomon, S.D. Renin-Angiotensin-Aldosterone System Inhibitors in Patients with Covid-19. N. Engl. J. Med. 2020, 382, 1653–1659. [Google Scholar] [CrossRef]
- Berry, D.J.; Hesketh, K.; Power, C.; Hyppönen, E. Vitamin D status has a linear association with seasonal infections and lung function in British adults. Br. J. Nutr. 2011, 106, 1433–1440. [Google Scholar] [CrossRef] [Green Version]
- Derbyshire, E.; Delange, J. COVID-19: Is there a role for immunonutrition, particularly in the over 65s? BMJ Nutr. Prev. Heal. 2020, 3, 100–105. [Google Scholar] [CrossRef]
- Kandimalla, R. SARS-CoV-2 pathophysiology and assessment of coronaviruses in CNS diseases with a focus on therapeutic targets. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165889. [Google Scholar] [CrossRef]
- Atyeo, C.; Slein, M.D.; Fischinger, S.; Burke, J.; Schäfer, A.; Leist, S.R.; Kuzmina, N.A.; Mire, C.; Honko, A.; Johnson, R.; et al. Dissecting strategies to tune the therapeutic potential of SARS-CoV-2–specific monoclonal antibody CR3022. JCI Insight 2021, 6. [Google Scholar] [CrossRef]
- Choy, K.T.; Wong, A.Y.; Kaewpreedee, P.; Sia, S.F.; Chen, D.; Hui, K.P.Y.; Chu, D.K.W.; Chan, M.C.W.; Cheung, P.P.; Huang, X. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020, 178, 104786. [Google Scholar] [CrossRef] [PubMed]
- Mantlo, E.; Bukreyeva, N.; Maruyama, J.; Paessler, S.; Huanga, C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020, 179, 104811. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; Zhang, X.; He, Y.; Jiang, S.; Dua, L. Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2. Antivir. Res. 2020, 179, 104820. [Google Scholar] [CrossRef] [PubMed]
- Jockusch, S.; Tao, C.; Li, X.; Anderson, T.K.; Chien, M.; Kumar, S.; Russo, J.J.; Kirchdoerfer, R.N.; Ju, J. A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antivir. Res. 2020, 180, 104857. [Google Scholar] [CrossRef]
- Sallard, E.; Lescure, F.-X.; Yazdanpanah, Y.; Mentre, F.; Peiffer-Smadja, N. Type 1 interferons as a potential treatment against COVID-19. Antivir. Res. 2020, 178, 104791. [Google Scholar] [CrossRef]
- Jeyaraman, M.; Ranjan, R.; Kumar, R.; Arora, A.; Chaudhary, D.; Ajay, S.S.; Jain, R. Cellular Therapy: Shafts of Light Emerging for COVID-19. Stem Cell Investig. 2020, 7, 11. [Google Scholar] [CrossRef]
- Yang, Y.; Islam, M.S.; Wang, J.; Li, Y.; Chen, X. Traditional Chinese Medicine in the Treatment of Patients Infected with 2019-New Coronavirus (SARS-CoV-2): A Review and Perspective. Int. J. Biol. Sci. 2020, 16, 1708–1717. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Karakiulakis, G.; Roth, M. Antihypertensive drugs and risk of COVID-19? Authors’ reply. Lancet Respir. Med. 2020, 8, e32–e33. [Google Scholar] [CrossRef] [Green Version]
- Rahman, N.; Basharat, Z.; Yousuf, M.; Castaldo, G.; Rastrelli, L.; Khan, H. Virtual Screening of Natural Products against Type II Transmembrane Serine Protease (TMPRSS2), the Priming Agent of Coronavirus 2 (SARS-CoV-2). Molecules 2020, 25, 2271. [Google Scholar] [CrossRef]
- Oo, A.; Teoh, B.T.; Sam, S.S.; Abu Bakar, S.; Zandi, K. Baicalein and baicalin as Zika virus inhibitors. Arch. Virol. 2019, 164, 585–593. [Google Scholar] [CrossRef]
- Gurung, A.B.; Ali, M.A.; Lee, J.; Farah, M.A.; Al-Anazi, K.M. Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in silico approach. Life Sci. 2020, 255, 117831. [Google Scholar] [CrossRef] [PubMed]
- Gentile, D.; Patamia, V.; Scala, A.; Sciortino, M.T.; Piperno, A.; Rescina, A. Putative Inhibitors of SARS-CoV-2 Main Protease from A Library of Marine Natural Products: A Virtual Screening and Molecular Modeling Study. Mar. Drugs 2020, 18, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Q.; Huang, C.; Ji, X.; Zhang, W.; Zhang, F.; Wang, L. Possible Inhibitors of ACE2, the Receptor of 2019-nCoV. Preprints. 2020. Available online: https://www.preprints.org/manuscript/202002.0047/v1 (accessed on 14 March 2021).
- Cheng, J.; Tang, Y.; Bao, B.; Zhang, P. Exploring the Active Compounds of Traditional Mongolian Medicine Agsirga in Intervention of Novel Coronavirus (2019-nCoV) Based on HPLC-Q-Exactive-MS/MS and Molecular Docking Method. ChemRxiv. 2020. Available online: https://chemrxiv.org/articles/preprint/Exploring_the_Active_Compounds_of_Traditional_Mongolian_Medicine_Agsirga_in_Intervention_of_Novel_Coronavirus_2019-nCoV_Based_on_HPLC-Q-Exactive-MS_MS_and_Molecular_Docking_Method/11955273/2 (accessed on 14 March 2021).
- Meneguzzo, F.; Ciriminna, R.; Zabini, F.; Pagliaro, M. Review of Evidence Available on Hesperidin-Rich Products as Potential Tools against COVID-19 and Hydrodynamic Cavitation-Based Extraction as a Method of Increasing Their Production. Processes 2020, 8, 549. [Google Scholar] [CrossRef]
- Pandit, M.; Latha, N. In silico studies reveal potential antiviral activity of phytochemicals from medicinal plants for the treatment of COVID-19 infection. Res. Sq. 2020, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.S.; Jagdale, S.S.; Bansode, S.B.; Shankar, S.S.; Tellis, M.B.; Pandya, V.K.; Chugh, A.; Giri, A.P.; Kulkarni, M.J. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. 2020, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khaerunnisa, S.; Kurniawan, H.; Awaluddin, R.; Suhartati, S.; Soetjipto, S. Potential Inhibitor of COVID-19 Main Protease (Mpro) From Several Medicinal Plant Compounds by Molecular Docking Study. Preprints. 2020. Available online: https://www.preprints.org/manuscript/202003.0226/v1 (accessed on 14 March 2021).
- Da Silva, J.K.R.; Figueiredo, P.L.B.; Byler, K.G.; Setzer, W.N. Essential Oils as Antiviral Agents, Potential of Essential Oils to Treat SARS-CoV-2 Infection: An In-Silico Investigation. Int. J. Mol. Sci. 2020, 21, 3426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-H.; Wu, K.-L.; Zhang, X.; Deng, S.-Q.; Peng, B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J. Integr. Med. 2020, 18, 152–158. [Google Scholar] [CrossRef]
- Qamar, M.T.U.; Alqahtani, S.M.; Alamri, M.A.; Chen, L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020, 10, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Olubiyi, O.O.; Olagunju, M.; Keutmann, M.; Strodel, B. High Throughput Virtual Screening to Discover Inhibitors of the Main Protease of the Coronavirus SARS-CoV-2. Preprints. 2020. Available online: https://www.preprints.org/manuscript/202004.0161/v2 (accessed on 14 March 2021).
- Chen, C.N.; Lin, C.P.C.; Huang, K.K.; Chen, W.C.; Hsieh, H.P.; Liang, P.H.; Hsu, J.T.A. Inhibition of SARS-CoV 3C-like protease activity by Theaflavin-3,30-digallate (TF3). Evid. Based Complementary Altern. Med. 2005, 2, 209–215. [Google Scholar] [CrossRef] [Green Version]
- El Fiky, A.A. Natural products may interfere with SARS-CoV-2 attachment to the host cell. J. Biomol. Struct. Dyn. 2020, 1–10. [Google Scholar] [CrossRef]
- Sharma, A.; Tiwari, V.; Sowdhamini, R. Computational search for potential COVID-19 drugs from FDA-approved drugs and small molecules of natural origin identifies several anti-virals and plant products. J. Biosci. 2020, 45, 1–18. [Google Scholar] [CrossRef]
- Gretebeck, L.M.; Subbarao, K. Animal models for SARS and MERS coronaviruses. Curr. Opin. Virol. 2015, 13, 123–129. [Google Scholar] [CrossRef]
- Cockrell, A.S.; Peck, K.M.; Yount, B.L.; Agnihothram, S.S.; Scobey, T.; Curnes, N.R.; Baric, R.S.; Heise, M.T. Mouse Dipeptidyl Peptidase 4 Is Not a Functional Receptor for Middle East Respiratory Syndrome Coronavirus Infection. J. Virol. 2014, 88, 5195–5199. [Google Scholar] [CrossRef] [Green Version]
- Nagata, N.; Iwata, N.; Hasegawa, H.; Fukushi, S.; Yokoyama, M.; Harashima, A.; Sato, Y.; Saijo, M.; Morikawa, S.; Sata, T. Participation of both Host and Virus Factors in Induction of Severe Acute Respiratory Syndrome (SARS) in F344 Rats Infected with SARS Coronavirus. J. Virol. 2006, 81, 1848–1857. [Google Scholar] [CrossRef] [Green Version]
- Roychoudhury, S.; Das, A.; Sengupta, P.; Dutta, S.; Roychoudhury, S.; Choudhury, A.P.; Ahmed, A.B.F.; Bhattacharjee, S.; Slama, P. Viral Pandemics of the Last Four Decades: Pathophysiology, Health Impacts and Perspectives. In Int. J. Environ. Res. Public Health; 2020; Volume 17, p. 9411. [Google Scholar]
- Roychoudhury, S.; Das, A.; Jha, N.K.; Kesari, K.K.; Roychoudhury, S.; Jha, S.K.; Kosgi, R.; Choudhury, A.P.; Lukac, N.; Madhu, N.R.; et al. Viral pathogenesis of SARS-CoV-2 infection and male reproductive health. Open Biol. 2021, 11, 200347. [Google Scholar]
- Roychoudhury, S.; Das, A.; Sengupta, P.; Dutta, S.; Roychoudhury, S.; Kolesarova, A.; Hleba, L.; Massanyi, P.; Slama, P. Viral pandemics of twenty-first century. J. Microbiol. Biotech. Food Sci. 2021, 10, 711–716. [Google Scholar]
- Jha, N.K.; Ojha, S.; Jha, S.K.; Dureja, H.; Singh, S.K.; Shukla, S.D.; Chellappan, D.K.; Gupta, G.; Bhardwaj, S.; Kumar, N.; et al. Evidence of Coronavirus (CoV) Pathogenesis and Emerging Pathogen SARS-CoV-2 in the Nervous System: A Review on Neurological Impairments and Manifestations. J Mol Neurosci. 2021, 19, 1–18. [Google Scholar]
- Javed, H.; Meeran, M.F.N.; Jha, N.K.; Ojha, S. Carvacrol, a Plant Metabolite Targeting Viral Protease (M pro) and ACE2 in Host Cells Can Be a Possible Candidate for COVID-19. Front Plant Sci. 2021, 11, 601335. [Google Scholar] [CrossRef] [PubMed]
- Jeyaraman, M.; John, A.; Koshy, S.; Ranjan, R.; Anudeep, T.C.; Jain, R.; Swati, K.; Jha, N.K.; Sharma, A.; Kesari, K.K.; et al. Fostering mesenchymal stem cell therapy to halt cytokine storm in COVID-19. Biochim Biophys Acta MolBasis Dis. 2021, 1867, 166014. [Google Scholar] [CrossRef] [PubMed]
- Nagoor Meeran, M.F.; Javed, H.; Sharma, C.; Goyal, S.N.; Kumar, S.; Jha, N.K.; Ojha, S. Can Echinacea be a potential candidate to target immunity, inflammation, and infection—The trinity of coronavirus disease 2019. Heliyon. 2021, 7, e05990. [Google Scholar] [CrossRef] [PubMed]
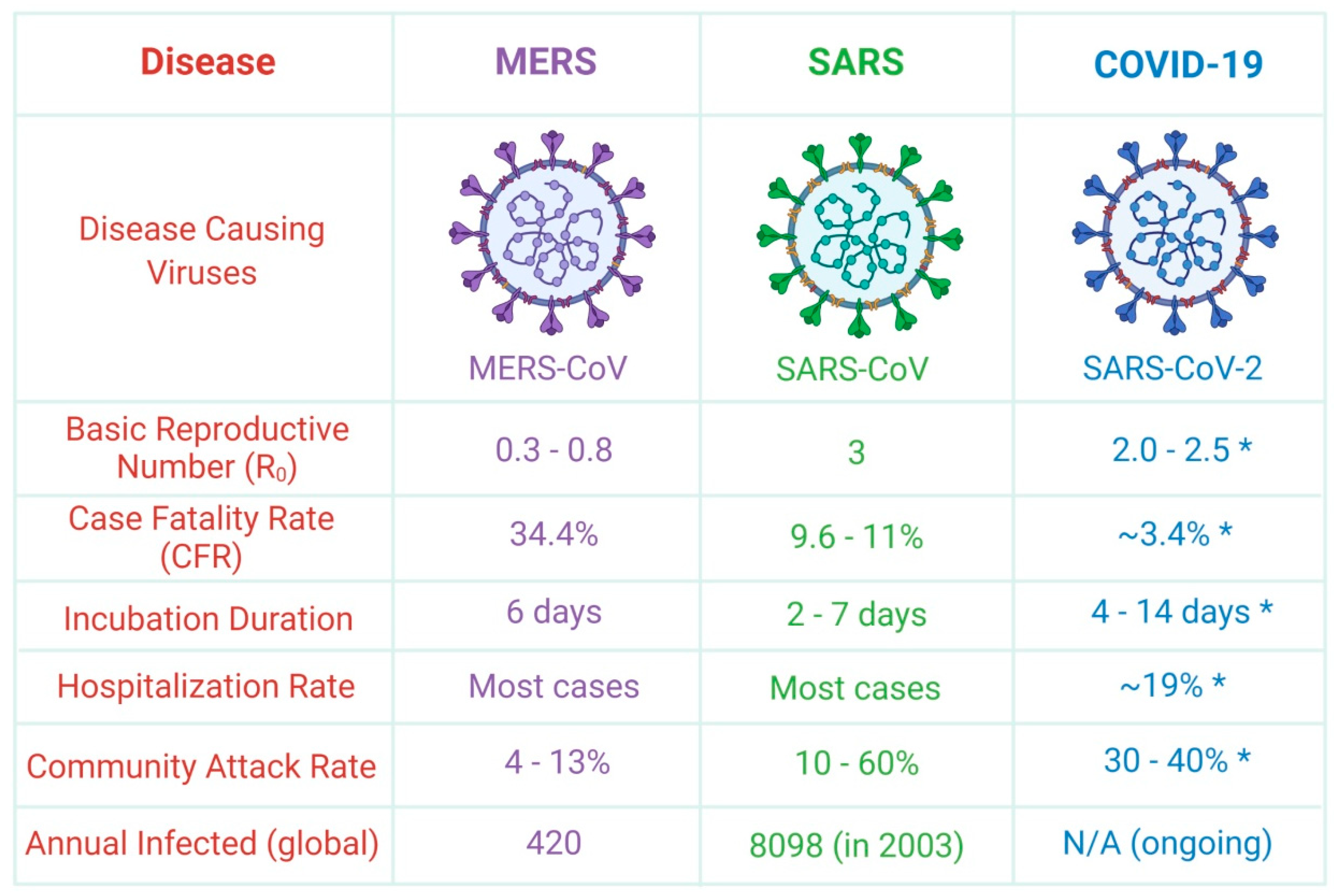
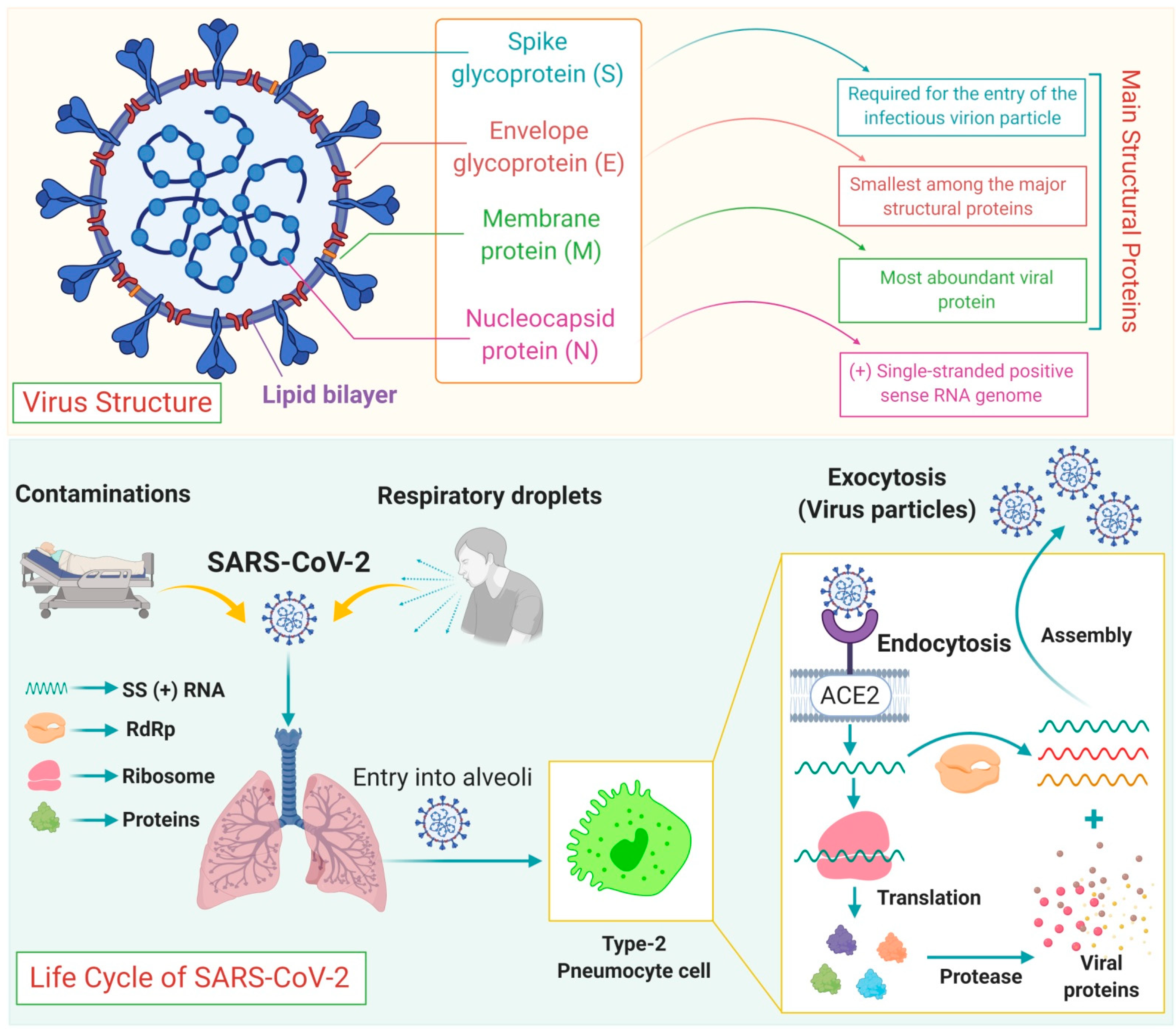
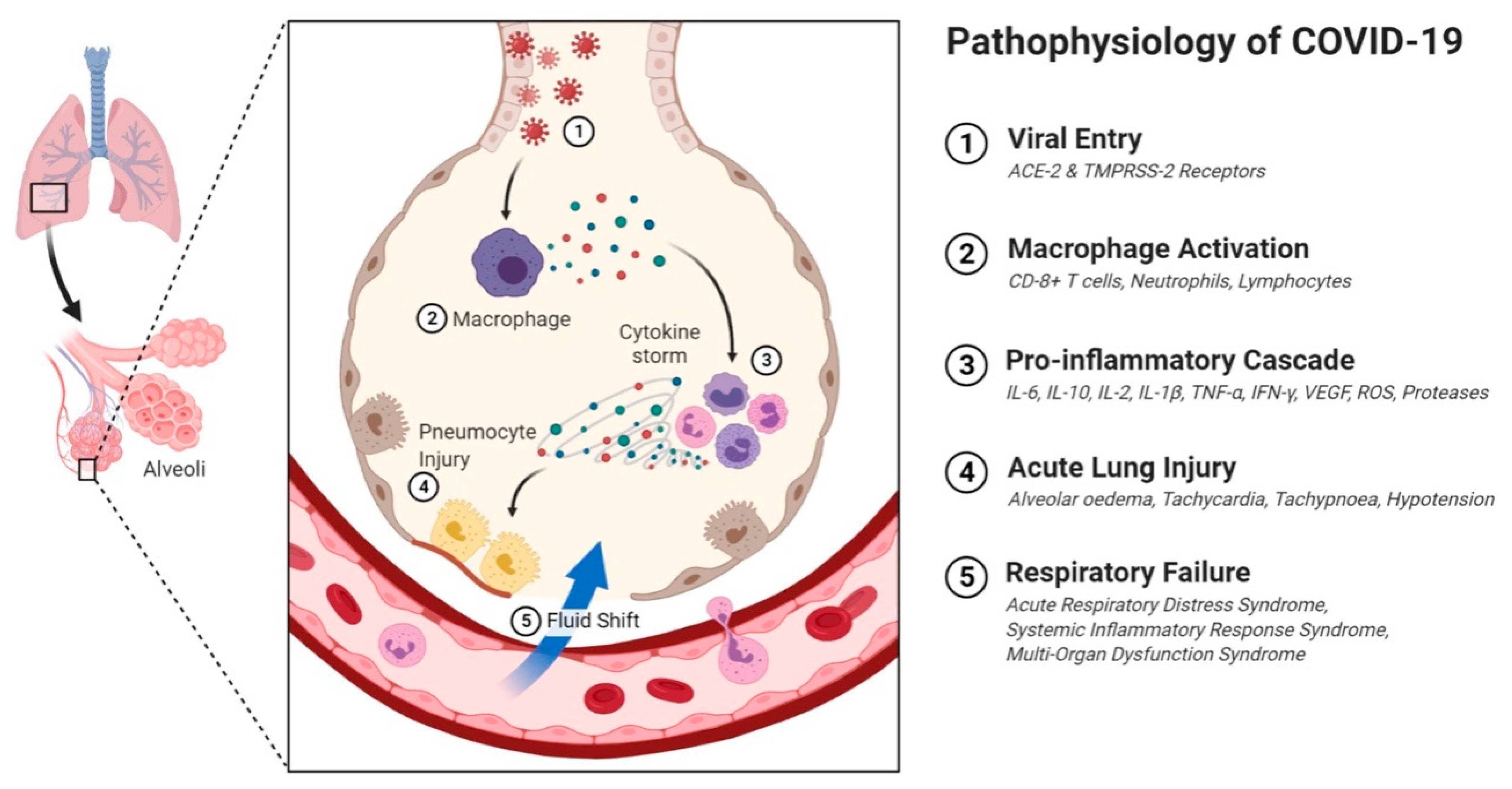
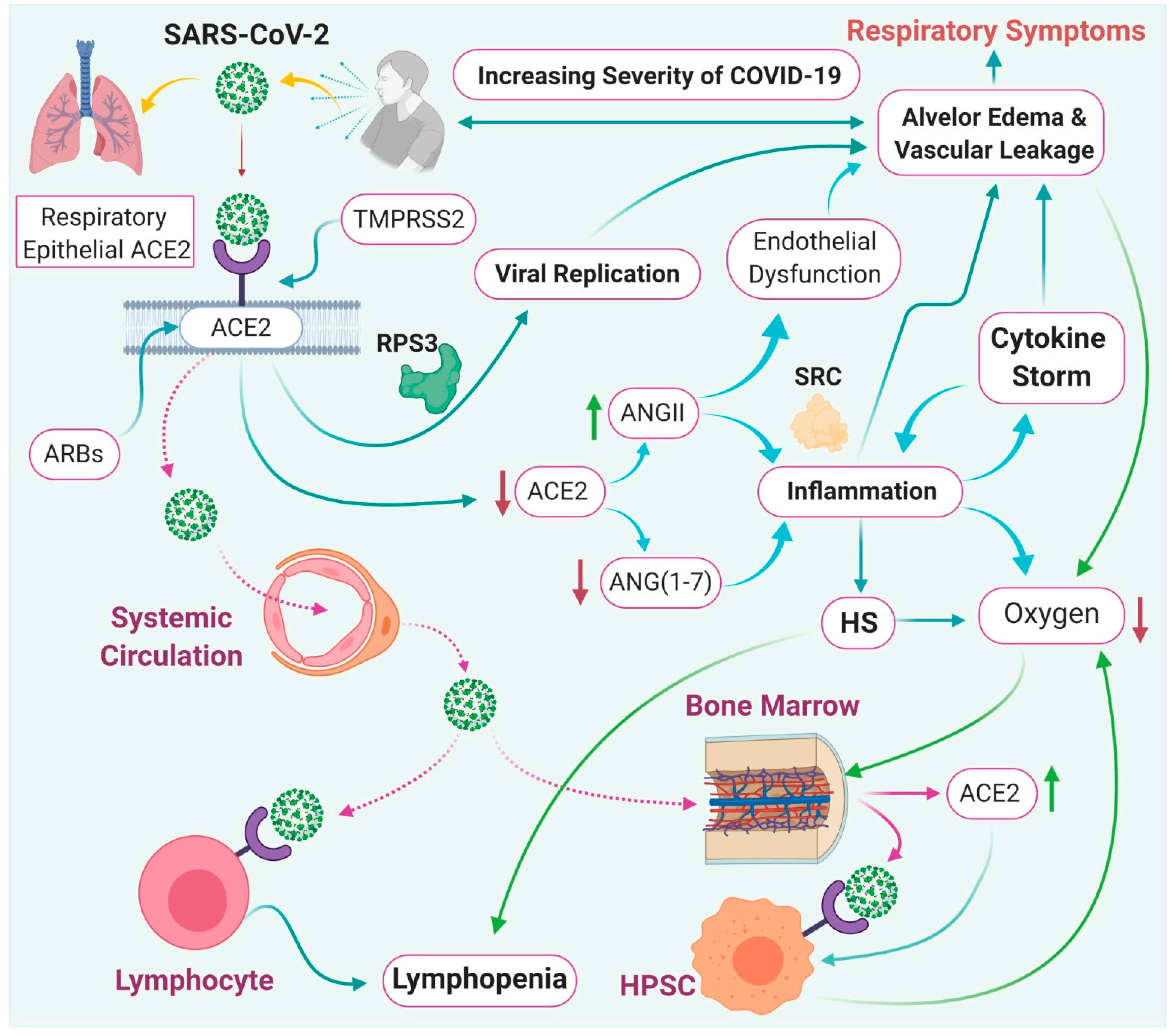
| S.No. | Name | Host Organism | Genera Name | Clinical Manifestations |
|---|---|---|---|---|
| 1 | Feline infectious peritonitis virus | Cat | Alpha | Vasculitis, fever, serositis, with or without effusions |
| 2 | Camel alphacoronavirus isolate camel/Riyadh | Camel | Alpha | Asymptomatic |
| 3 | Canine CoV/TU336/F/2008 | Dog | Alpha | Diarrhea and mild clinical signs |
| 4 | SeACoV-CH/GD-01 | Pig | Alpha | Acute and severe diarrhea and vomiting |
| 5 | TGEV/PUR46-MAD | Pig | Alpha | Diarrhea |
| 6 | PRCV/ISU-1 | Pig | Alpha | Mild respiratory tract infections (RTIs) |
| 7 | PEDV/ZJU-G1-2013 | Pig | Alpha | Severe watery diarrhea |
| 8 | Human CoV-NL63 | Human | Alpha | Mild RTIs |
| 9 | Human CoV-229E | Human | Alpha | Mild RTIs |
| 10 | MHV-A59 | Mouse | Beta | Severe lung injuries and acute pneumonia |
| 11 | Equine CoV/Obihiro12-1 | Horse | Beta | Leucopenia, fever, and anorexia |
| 12 | Bovine CoV/ENT | Cow | Beta | Diarrhea |
| 13 | MERS-CoV | Human | Beta | Severe acute respiratory syndrome |
| 14 | SARS-CoV | Human | Beta | Severe acute respiratory syndrome |
| 15 | Human CoV-OC43 | Human | Beta | Mild RTIs |
| 16 | Human CoV-HKU1 | Human | Beta | Pneumonia |
| 17 | IBV | Chicken | Gamma | Severe respiratory disease |
| 18 | Beluga Whale CoV/SW1 | Whale | Gamma | Terminal acute liver failure and pulmonary disease |
| 19 | Sparrow coronavirus HKU17 | Sparrow | Delta | Respiratory disease |
| 20 | Bulbul coronavirus HKU11 | Bulbul | Delta | Respiratory disease |
| S.No. | Name | Associated Functions | References |
|---|---|---|---|
| 1 | nsp1 | Inhibits IFNsignaling and involves in cellular mRNA degradation | [67,68] |
| 2 | nsp2 | Unclear | [69,70] |
| 3 | nsp3 | Promotescytokine expression, PLP, polypeptides cleaving and blocks hostinnate immune response | [71,72] |
| 4 | nsp4 | Involves in double-membrane vesicle (DMV) formation | [73,74] |
| 5 | nsp5 | Inhibits IFNsignaling, acts as a chymotrypsin-like protease (3CLpro), main protease (Mpro), and cleaves polypeptides | [75,76,77] |
| 6 | nsp6 | Restricts DMV formation and autophagosome expansion | [78,79] |
| 7 | nsp7 | Acts as a cofactor with nsp8 and nsp12 | [64,80] |
| 8 | nsp8 | Primase activity and also acts as a cofactor with nsp7 and nsp12 | [66,80,81] |
| 9 | nsp9 | Involves in dimerization and RNA binding | [82,83] |
| 10 | nsp10 | acts as a scaffold protein for nsp14 and nsp16 | [84,85,86,87] |
| 11 | nsp11 | Unclear | [88] |
| 12 | nsp12 | Primer dependent RdRp | [66,89,90] |
| 13 | nsp13 | 5′ triphosphatase and RNA helicase | [91,92,93] |
| 14 | nsp14 | N7-Mtase and exoribonuclease | [94,95,96,97] |
| 15 | nsp15 | Acts as an endoribonuclease and evasion of double-stranded RNA viruses (dsRNA) sensors | [98,99,100] |
| 16 | nsp16 | 2′-O-Mtase avoids MDA5 recognition and negatively regulates innate immunity | [85,86,101] |
| Categories | Compound Name | Proposed Mode of Actions | Involved Viruses | References |
|---|---|---|---|---|
| Antiviral drugs | Remdesivir (GS-5734, Nucleoside analogue of Remdesivir triphosphate) (RDV-TP) | Inhibitor of RdRp | SARS-CoV-2 | [156] |
| Lopinavir/Ritonavir | HIV protease inhibitor | HIV infection, SARS-CoV-1, and MERS-CoV | [157,158] | |
| Darunavir/Cobicistat | Protease inhibitor | SARS-CoV-2 | [159] | |
| Favipiravir (T-705) Purine nucleotide | RNA polymerase inhibitor | RNA viruses and SARS-CoV-2 | [160] | |
| Ribavirin (Guanine analogue) | Inhibits viral RdRp | SARS-CoV-1 and SARS-CoV-2 | [161] | |
| Umifenovir (Arbidol) | Targeting the S protein/ACE2 and inhibits the membrane fusion of the envelope of the virus | Influenza and SARS-CoV-2 | [162,163] | |
| Antimalarial drugs | Chloroquine (Synthetic version of quinine and is found in the bark of cinchona trees) | Reduces the rate of replication | Malaria, systemic inflammatory diseases, and SARS-CoV-2 | [164] |
| Hydroxychloroquine | Inhibition of glycosylation of host receptors, proteolytic processing, and acidification of endosomes | SARS-CoV-2 and autoimmune diseases | [165] | |
| Antiparasitic drugs | Ivermectin | Inhibits nuclear transport | Parasitic Infections and SARS-CoV-2 | [166] |
| Nitazoxanide (Anti-helminthic drug) | Unclear | MERS and SARS-CoV-2 | [167] | |
| Adjunctive drugs | Corticosteroids/quinolone (n combination) | Prevents ARDS | SARS-CoV and SARS-CoV-2 | [168] |
| Monoclonal Antibodies (Tocilizumab, Sarilumab, Eculizumab, Fingolimod, Bevacizumab) | Immunomodulatory effect, inhibition of terminal complement, and anti–VEGF medication | SARS-CoV-2 and Chronic Inflammatory disorders | [169,170] | |
| ACE-Inhibitors and ARBs (enzyme) | Activates RAAS mechanism | SARS-CoV-2 | [171] | |
| Interferon-(α and β) | Unclear | MERS-CoV and SARS-CoV-2 | [161] | |
| Vitamin-D (Adjunct with vitamin C and zinc) | Inhibits inflammatory response and attenuates cytokine storm | SARS-CoV-2 | [172,173] |
| S.No | Name | Vaccine Type | Primary Developer | Country of Origin | List of Countries Approved for Use |
|---|---|---|---|---|---|
| 1 | Comirnaty (BNT162b2) | mRNA-based vaccine | Pfizer, BioNTech; Fosun Pharma | Multinational | Albania, Andorra, Argentina, Aruba, Australia, Bahrain, Brazil, Canada, Caribbean, Chile, Colombia, Costa Rica, Ecuador, EU, Faroe Islands, Greenland, Hong Kong, Iceland, Iraq, Israel, Japan, Jordan, Kuwait, Liechtenstein, Malaysia, Mexico, Monaco, New Zealand, North Macedonia, Norway, Oman, Panama, Philippines, Qatar, Rwanda, Saint Vincent and the Grenadines, Saudi Arabia, Serbia, Singapore, South Korea, Suriname, Switzerland, UAE, UK, US, Vatican City, WHO |
| 2 | Moderna COVID-19 Vaccine (mRNA-1273) | mRNA-based vaccine | Moderna, BARDA, NIAID | US | Canada, EU, Faroe Islands, Greenland, Iceland, Israel, Liechtenstein, Norway, Qatar, Saint Vincent and the Grenadines, Singapore, Switzerland, United Kingdom, United States, Vietnam |
| 3 | COVID-19 Vaccine AstraZeneca (AZD1222); also known as Covishield | Adenovirus vaccine | BARDA, OWS | UK | Australia, Argentina, Bahrain, Bangladesh, Barbados, Brazil, Canada, Chile, Dominican Republic, Ecuador, El Salvador, Egypt, EU, Ghana, Guyana, Hungary, India, Indonesia, Iraq, Ivory Coast, Malaysia, Maldives, Mauritius, Mexico, Morocco, Myanmar, Nepal, Nigeria, Pakistan, Philippines, Saint Vincent and the Grenadines, South Africa, South Korea, Sri Lanka, Taiwan, Thailand, UK, Vietnam |
| 4 | Sputnik V | Recombinant adenovirus vaccine (rAd26 and rAd5) | Gamaleya Research Institute, Acellena Contract Drug Research and Development | Russia | Algeria, Angola, Argentina, Armenia, Bahrain, Belarus, Bolivia, Congo, Djibouti, Egypt, Gabon, Ghana, Guatemala, Guinea, Guyana, Honduras, Hungary, Iran, Iraq, Jordan, Kazakhstan, Kenya, Kyrgyzstan, Laos, Lebanon, Mexico, Moldova, Mongolia, Montenegro, Morocco, Myanmar, Nicaragua, North Macedonia, Pakistan, Palestine, Paraguay, Republika Srpska, Russia, Saint Vincent and the Grenadines, San Marino, Serbia, Slovakia, Sri Lanka, Syria, Tunisia, Turkmenistan, United Arab Emirates, Uzbekistan, Venezuela, Zimbabwe |
| 5 | COVID-19 Vaccine Janssen (JNJ-78436735; Ad26.COV2.S) | Non-replicating viral vector | Janssen Vaccines (Johnson & Johnson) | The Netherlands, US | Bahrain, Canada, EU, Saint Vincent and the Grenadines, US, WHO |
| 6 | CoronaVac | Inactivated vaccine (formalin with alum adjuvant) | Sinovac | China | Azerbaijan, Bolivia, Brazil, Cambodia, China, Chile, Colombia, Ecuador, Hong Kong, Indonesia, Laos, Malaysia, Mexico, Thailand, Tunisia, Turkey, Philippines, Ukraine, Uruguay, Zimbabwe |
| 7 | BBIBP-CorV | Inactivated vaccine | Beijing Institute of Biological Products; China National Pharmaceutical Group (Sinopharm) | China | Argentina, Bahrain, Cambodia, China, Egypt, Hungary, Iraq, Jordan, Laos, Macau, Morocco, Nepal, Pakistan, Peru, Senegal, Serbia, Seychelles, UAE, Venezuela, Zimbabwe |
| 8 | EpiVacCorona | Peptide vaccine | Federal Budgetary Research Institution State Research Center of Virology and Biotechnology | Russia | Russia, Turkmenistan |
| 9 | Convidicea (Ad5-nCoV) | Recombinant vaccine (adenovirus type 5 vector) | CanSino Biologics | China | Mexico, China, Pakistan |
| 10 | Covaxin | Inactivated vaccine | Bharat Biotech, ICMR | India | India, Zimbabwe |
| 11 | No name announced | Inactivated vaccine | Wuhan Institute of Biological Products; China National Pharmaceutical Group (Sinopharm) | China | China |
| 12 | CoviVac | Inactivated vaccine | Chumakov Federal Scientific Center for Research and Development of Immune and Biological Products | Russia | Russia |
| 13 | ZF2001 | Recombinant vaccine | Anhui ZhifeiLongcom Biopharmaceutical, Institute of Microbiology of the Chinese Academy of Sciences | China, Uzbekistan | Uzbekistan |
| S.No | Candidate Name | Vaccine Type | Sponsor/Developer | Clinical Trial Stage | Companies/Universities |
|---|---|---|---|---|---|
| 1 | NVX-CoV2373 | Nanoparticle vaccine | Novavax | Phase 3 | Novavax |
| 2 | ZyCoV-D | DNA vaccine (plasmid) | Zydus Cadila | Phase 3 | Zydus Cadila |
| 3 | Abdala (CIGB 66) | Protein subunit vaccine | Center for Genetic Engineering and Biotechnology | Phase 3 | Center for Genetic Engineering and Biotechnology |
| 4 | CVnCoV | mRNA-based vaccine | CureVac; GSK | Phase 2b/3 | CureVac |
| 5 | Bacillus Calmette-Guerin (BCG) vaccine | Live-attenuated vaccine | University of Melbourne and Murdoch Children’s Research Institute; Radboud University Medical Center; Faustman Lab at Massachusetts General Hospital | Phase 2/3 | University of Melbourne and Murdoch Children’s Research Institute; Radboud University Medical Center; Faustman Lab at Massachusetts General Hospital |
| 6 | INO-4800 | DNA vaccine (plasmid) | Inovio Pharmaceuticals | Phase 2/3 | Center for Pharmaceutical Research, Kansas City. Mo.; University of Pennsylvania, Philadelphia |
| 7 | VIR-7831 | Plant-based adjuvant vaccine | Medicago; GSK; Dynavax | Phase 2/3 | Medicago |
| 8 | No name announced | Adenovirus-based vaccine | ImmunityBio; NantKwest | Phase 2/3 | NA |
| 9 | UB-612 | Multitope peptide-based vaccine | COVAXX | Phase 2/3 | United Biomedical Inc. (UBI) |
| 10 | No name announced | Recombinant protein vaccine | Sanofi; GlaxoSmithKline | Phase 2 | Various |
| 11 | BNT162 | mRNA-based vaccine | Pfizer, BioNTech | Phase 1/2/3 | Multiple study sites in Europe, North America and China |
| 12 | Soberana 1 and 2 | Monovalent/conjugate vaccine | Finlay Institute of Vaccines | Phase 1/2/3 | Finlay Institute of Vaccines |
| 13 | AdCLD-CoV19 | Adenovirus-based vaccine | Cellid; LG Chem | Phase 1/2a | Korea University Guro Hospital |
| 14 | Nanocovax | Recombinant vaccine (Spike protein) | Nanogen Biopharmaceutical | Phase 1/2 | Military Medical Academy (Vietnam) |
| 15 | EuCorVac-19 | nanoparticle vaccine | EuBiologics | Phase 1/2 | Eunpyeong St. Mary’s Hospital |
| 16 | Mambisa (CIGB 669) | Protein subunit vaccine | Center for Genetic Engineering and Biotechnology | Phase 1/2 | Center for Genetic Engineering and Biotechnology |
| 17 | IIBR-100 | Recombinant vesicular stomatitis virus (rVSV) vaccine | Israel Institute for Biological Research | Phase 1/2 | Hadassah Medical Center; Sheba Medical Center Hospital |
| 18 | No name announced | SF9 cell vaccine candidate | West China Hospital, Sichuan University | Phase 1/2 | West China Hospital, Sichuan University |
| 19 | VLA2001 | Inactivated vaccine | Valneva; National Institute for Health Research (NIHR) | Phase 1/2 | Multiple NIHR testing sites |
| 20 | No name announced | Adjuvanted protein subunit vaccine | CEPI | Phase 1/2 | NA |
| 21 | AG0301-COVID19 | DNA vaccine | AnGes, Inc. | Phase 1/2 | AnGes, Inc.; Japan Agency for Medical Research and Development |
| 22 | GX-19N | DNA vaccine | Genexine | Phase 1/2 | |
| 23 | ARCT-021 (LUNAR-COV19) | Self-replicating RNA vaccine | Arcturus Therapeutics and Duke-NUS Medical School | Phase 1/2 | Duke-NUS Medical School, Singapore |
| 24 | No name announced | Inactivated vaccine | Chinese Academy of Medical Sciences, Institute of Medical Biology | Phase 1/2 | West China Second University Hospital, Yunnan Center for Disease Control and Prevention |
| 25 | HDT-301 (HGCO19) | RNA vaccine | University of Washington; National Institutes of Health Rocky Mountain Laboratories; HDT Bio Corp; Gennova Biopharmaceuticals | Phase 1/2 | NA |
| 26 | AV-COVID-19 | Dendritic cell vaccine | Aivita Biomedical, Inc. | Phase 1b/2 | Rumah Sakit Umum Pusat DrKariadi |
| 27 | PTX-COVID19-B | mRNA-based vaccine | Providence Therapeutics; Canadian government | Phase 1 | NA |
| 28 | COVI-VAC | Intranasal vaccine | Codagenix; Serum Institute of India | Phase 1 | NA |
| 29 | CORVax12 | DNA vaccine (plasmid) | OncoSec; Providence Cancer Institute | Phase 1 | Providence Portland Medical Center |
| 30 | MVA-SARS-2-S | Modified vaccinia virus ankara (MVA) vector vaccine candidate | Universitätsklinikum Hamburg-Eppendorf; German Center for Infection Research; Philipps University Marburg Medical Center; Ludwig-Maximilians-University of Munich | Phase 1 | University Medical Center Hamburg-Eppendorf |
| 31 | COH04S1 | Modified vaccinia virus ankara (MVA) vector vaccine candidate | City of Hope Medical Center; National Cancer Institute | Phase 1 | City of Hope Medical Center |
| 32 | pVAC | Multi-peptide vaccine candidate | University Hospital Tuebingen | Phase 1 | University Hospital Tuebingen |
| 33 | AdimrSC-2f | Protein subunit vaccine | Adimmune | Phase 1 | Adimmune |
| 34 | bacTRL-Spike | Monovalent oral vaccine (bifidobacteria) | Symvivo | Phase 1 | Symvivo Corporation |
| 35 | COVAX-19 | Monovalent recombinant protein vaccine | Vaxine Pty Ltd. | Phase 1 | Royal Adelaide Hospital |
| 36 | DelNS1-2019-nCoV-RBD-OPT1 | Replicating viral vector | Xiamen University, Beijing Wantai Biological Pharmacy | Phase 1 | Jiangsu Provincial Centre For Disease Control and Prevention |
| 37 | GRAd-COV2 | Adenovirus-based vaccine | ReiThera; Leukocare; Univercells | Phase 1 | Lazzaro Spallanzani National Institute for Infectious Diseases |
| 38 | UQ-CSL V451 | Protein subunit vaccine | CSL; The University of Queensland | Phase 1 | NA |
| 39 | SCB-2019 | Protein subunit vaccine | GlaxoSmithKline, Sanofi, Clover Biopharmaceuticals, Dynavax and Xiamen Innovax; CEPI | Phase 1 | Linear Clinical Research (Australia) |
| 40 | VXA-CoV2-1 | Recombinant vaccine (adenovirus type 5 vector) | Vaxart | Phase 1 | Vaxart |
| 41 | AdCOVID | Intranasal vaccine | Altimmune | Phase 1 | University of Alabama at Birmingham |
| 42 | AAVCOVID | Gene-based vaccine | Massachusetts Eye and Ear; Massachusetts General Hospital; University of Pennsylvania | Pre-clinical | NA |
| 43 | ChAd-SARS-CoV-2-S | Adenovirus-based vaccine | Washington University School of Medicine in St. Louis | Pre-clinical | Washington University School of Medicine in St. Louis |
| 44 | HaloVax | Self-assembling vaccine | Voltron Therapeutics, Inc.; Hoth Therapeutics, Inc. | Pre-clinical | MGH Vaccine and Immunotherapy Center |
| 45 | LineaDNA | DNA vaccine | Takis Biotech | Pre-clinical | Takis Biotech |
| 46 | MRT5500 | mRNA-based vaccine | Sanofi, Translate Bio | Pre-clinical | NA |
| 47 | No name announced | Ii-Key peptide COVID-19 vaccine | Generex Biotechnology | Pre-clinical | Generex |
| 48 | No name announced | Protein subunit vaccine | University of Saskatchewan Vaccine and Infectious Disease Organization-International Vaccine Centre | Pre-clinical | University of Saskatchewan Vaccine and Infectious Disease Organization-International Vaccine Centre |
| 49 | No name announced | mRNA-based vaccine | Chulalongkorn University’s Center of Excellence in Vaccine Research and Development | Pre-clinical | NA |
| 50 | No name announced | gp96-based vaccine | Heat Biologics | Pre-clinical | University of Miami Miller School of Medicine |
| 51 | No name announced | Inactivated vaccine | Shenzhen Kangtai Biological Products | Pre-clinical | NA |
| 52 | PittCoVacc | Recombinant protein subunit vaccine (delivered through microneedle array) | UPMC/University of Pittsburgh School of Medicine | Pre-clinical | University of Pittsburgh |
| 53 | T-COVIDTM | Intranasal vaccine | Altimmune | Pre-clinical | NA |
| 54 | LNP-nCoVsaRNA | Self-amplifying RNA vaccine | Imperial College London | No longer being studied | Imperial College London |
| 55 | V590 | Recombinant vaccine (vesicular stomatitis virus) | Merck; IAVI | No longer being studied | NA |
| 56 | V591 | Measles vector vaccine | University of Pittsburgh’s Center for Vaccine Research | No longer being studied | University of Pittsburgh; Themis Biosciences; Institut Pasteur |
| Target/Binding Site | Natural Products/Metabolites | Binding Energy (kcal mol−1) | Reference |
|---|---|---|---|
| ACE2 | Zhebeininoside | −6.8 | [189] |
| Verdine | −6.6 | ||
| Pseudojervine | −6.8 | ||
| Imperialine-3-b-D-glucoside | −7.1 | ||
| Hupehemonside | −7.1 | ||
| Nobiletin | −5.42 | [190] | |
| Neohesperidin | −3.78 | ||
| Hesperetin | −6.09 | ||
| Hesperidin | −4.21 | ||
| Naringenin | −6.05 | ||
| Narigin | −6.85 | ||
| Chloroquine | −8.019 | [191] | |
| Philligenin | −7.807 | ||
| Hinokinin | −7.11 | ||
| Withaferin A | −9.631 | ||
| Quercetin | −8.664 | ||
| Isoaloresin | −7.835 | ||
| Aloin | −8.383 | ||
| Corydine | −6.041 | ||
| Tetrahydrocurcumin | −8.009 | ||
| Silybin | −10.572 | ||
| Isoquercitrin | −7.8 | [192] | |
| Afzelin | −7.1 | ||
| Oriciacridone F | −6.7 | ||
| Remdesivir | −7.8 | ||
| Cassameridin | −8.1 | ||
| (-)-Asperlicin C | −9.5 | ||
| Kaempferol | −7.2 | ||
| Apigenin | −7.1 | ||
| Myricitrin | −7.1 | ||
| Vitetrifolin D | −7.3 | ||
| Lactucopicrin | −8.3 | ||
| Lactucopicrin 15-oxalate | −8.3 | ||
| Taiwanhomoflavone A | −7.6 | ||
| Epicatechin-(4b,8)-epicatechin-(4b,6)-catechin | −8.2 | ||
| Epicatechin-4-epigallocatechin | −7.2 | ||
| Quercetin 3-glucosyl-(1,4)-rhamnoside | −6.5 | ||
| 3CLpro | Epicatechin-gallate | −6.27 | [193] |
| α-Copaene | −20.08 | [194] | |
| (E)-β-Farnesene | −27.56 | [194] | |
| Gingerol | −5.38 | [193] | |
| Zingerol | −5.4 | ||
| Apigenin-7-glucoside | −7.83 | ||
| Quercetin | −8.47 | ||
| Kaempferol | −8.58 | ||
| Lopinavir | −9.41 | ||
| Nelfinavir | −10.74 | ||
| Sugiol | −6.04 | [195] | |
| N-cis-feruloyltyramine | −6.25 | ||
| Cryptotanshinone | −6.23 | ||
| Betulinic acid | −4.23 | ||
| Amaranthin | −18.14 | [196] | |
| Methyl rosmarinate | −20.62 | ||
| 5,7,3',4'-tetrahydroxy-2'-(3,3-dimethylallyl) isoflavone | −29.57 | ||
| Mirycitrin | 22.13 | ||
| Zeylanone | −9.12 | [197] | |
| Glabrolide | −9.16 | ||
| Amentoflavone | −9.28 | ||
| Isoquercitrin | −8.2 | [192] | |
| Afzelin | −8.8 | ||
| Oriciacridone F | −9.1 | ||
| Remdesivir | −8.2 | ||
| Cassameridin | −9.3 | ||
| Kaempferol | −7.8 | ||
| (-)-Asperlicin C | −9.7 | ||
| Apigenin | −7.8 | ||
| Myricitrin | −8.9 | ||
| Vitetrifolin D | −7.6 | ||
| Lactucopicrin 15-oxalate | −8.2 | ||
| Lactucopicrin | −7.8 | ||
| Quercetin 3-glucosyl-(1,4)-rhamnoside | −9.9 | ||
| Epicatechin-(4',8)-epigallocatechin | −10 | ||
| Epicatechin-(4b,8)-epicatechin-(4b,6)-catechin | -10.6 | ||
| Taiwanhomoflavone A | −9.6 | ||
| TMPRSS2 | Isogemichalcone B | −13.07 | [184] |
| Microcarpin | −13.31 | ||
| Durumolide K | −13.92 | ||
| Dictyosphaeric acid A | −14.02 | ||
| Geniposide | −14.69 | ||
| Baicalin | −8.46 | [198] | |
| Silybin | −11.928 | [190] | |
| Tetrahydrocurcumin | −8.793 | ||
| Corydine | −7.91 | ||
| Aloin | −9.18 | ||
| HSPA5 | Caffeic acid | −6.2 | [199] |
| Chlorogenic acid | −6.5 | ||
| Palmitic acid | −5.5 | ||
| Biochanin A | −6.9 | ||
| Formotein | −7.5 | ||
| Genistein | −7.5 | ||
| Diadiazin | −8.6 | ||
| NSP1 | Shogaol | −2.64 | [200] |
| Gingerenone | −4.39 | ||
| Remdesivir | −5.8 | ||
| Lactose | −11.66 | ||
| Esculin | −6.88 | ||
| (-)-Epicatechin 3-O-(3'-O-methyl) gallate | −13.1 | [184] | |
| Curtisian L | −13.38 | ||
| 5-Methoxyhydnocarpin | −13.92 | ||
| Citicoline | −13.96 | ||
| Isoaloresin | −9.759 | [190] | |
| Withaferin A | −11.242 | ||
| Hinokinin | −7.67 | ||
| Philligenin | −9.503 | ||
| Excavatolide M | −14.38 | [184] | |
| Schisphenin A | −14.27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jha, N.K.; Jeyaraman, M.; Rachamalla, M.; Ojha, S.; Dua, K.; Chellappan, D.K.; Muthu, S.; Sharma, A.; Jha, S.K.; Jain, R.; et al. Current Understanding of Novel Coronavirus: Molecular Pathogenesis, Diagnosis, and Treatment Approaches. Immuno 2021, 1, 30-66. https://doi.org/10.3390/immuno1010004
Jha NK, Jeyaraman M, Rachamalla M, Ojha S, Dua K, Chellappan DK, Muthu S, Sharma A, Jha SK, Jain R, et al. Current Understanding of Novel Coronavirus: Molecular Pathogenesis, Diagnosis, and Treatment Approaches. Immuno. 2021; 1(1):30-66. https://doi.org/10.3390/immuno1010004
Chicago/Turabian StyleJha, Niraj Kumar, Madhan Jeyaraman, Mahesh Rachamalla, Shreesh Ojha, Kamal Dua, Dinesh Kumar Chellappan, Sathish Muthu, Ankur Sharma, Saurabh Kumar Jha, Rashmi Jain, and et al. 2021. "Current Understanding of Novel Coronavirus: Molecular Pathogenesis, Diagnosis, and Treatment Approaches" Immuno 1, no. 1: 30-66. https://doi.org/10.3390/immuno1010004
APA StyleJha, N. K., Jeyaraman, M., Rachamalla, M., Ojha, S., Dua, K., Chellappan, D. K., Muthu, S., Sharma, A., Jha, S. K., Jain, R., Jeyaraman, N., GS, P., Satyam, R., Khan, F., Pandey, P., Verma, N., Singh, S. K., Roychoudhury, S., Dholpuria, S., ... Kesari, K. K. (2021). Current Understanding of Novel Coronavirus: Molecular Pathogenesis, Diagnosis, and Treatment Approaches. Immuno, 1(1), 30-66. https://doi.org/10.3390/immuno1010004


















