Abstract
Alzheimer’s disease (AD), an intense neurological illness, severely impacts memory, behavior, and personality, posing a growing concern worldwide due to the aging population. Early and accurate detection is crucial as it enables preventive measures. However, current diagnostic methods are often inaccurate in identifying the disease in its early stages. Although deep learning-based bioimaging has shown promising results in medical image classification, challenges remain in achieving the highest accuracy for detecting AD. Existing approaches, such as ResNet50, VGG19, InceptionV3, and AlexNet have shown potential, but they often lack reliability and accuracy due to several issues. To address these gaps, this paper suggests a novel bioimaging technique by developing a custom Convolutional Neural Network (CNN) model for detecting AD. This model is designed with optimized layers to enhance feature extraction from medical images. The experiment’s first phase involved the construction of the custom CNN structure with three max-pooling layers, three convolutional layers, two dense layers, and one flattened layer. The Adam optimizer and categorical cross-entropy were adopted to compile the model. The model’s training was carried out on 100 epochs with the patience set to 10 epochs. The second phase involved augmentation of the dataset images and adding a dropout layer to the custom CNN model. Moreover, fine-tuned hyperparameters and advanced regularization methods were integrated to prevent overfitting. A comparative analysis of the proposed model with conventional models was performed on the dataset both before and after the data augmentation. The results validate that the proposed custom CNN model significantly overtakes pre-existing models, achieving the highest validation accuracy of 99.53% after data augmentation while maintaining the lowest validation loss of 0.0238. Its precision, recall, and F1 score remained consistently high across all classes, with perfect scores for the Moderate Demented and Non-Demented categories after augmentation, indicating superior classification capability.
1. Introduction
Alzheimer’s disease (AD) is a long-lasting neurodegenerative ailment that predominantly affects older adults, leading to memory loss, cognitive decline, and impaired reasoning. As the disease progresses, it severely affects a person’s ability to perform daily tasks, eventually leading to a complete loss of freedom. As stated by the World Health Organization (WHO), AD affects millions of people worldwide, with lots of new cases emerging each year [1]. The increasing aging population is expected to significantly increase this number, putting an immense burden on healthcare systems globally. Early detection of AD is vital, as it enables timely intervention strategies, personalized healthcare, and disease-modifying treatments that could slow disease progression or improve quality of life for patients. However, current diagnostic methods, which typically involve cognitive tests and procedures such as cerebrospinal fluid analysis, often fail to detect AD in its earliest stages. This highlights the urgent need for more reliable and accurate methods to detect AD at an early stage. Advances in Artificial Intelligence (AI), particularly in Machine Learning (ML), Federated Learning (FL), Deep Learning (DL), and Large Language Models (LLMs), have transformed various domains, including healthcare [2,3]. The recent rapid growth of AI has been fueled by increases in computational power, the availability of large datasets, and more sophisticated algorithms, all of which have significantly expanded AI’s potential applications. In healthcare, DL techniques are especially suited for medical image analysis, given their capability to learn difficult patterns and hierarchies from huge datasets. Models like Convolutional Neural Networks (CNNs) have been successfully applied in tasks, viz., image classification/segmentation and disease detection. The potential of AI/ML/DL in medical imaging lies in their capability to automatically take out and analyze features from imaging data, thereby assisting in the early diagnosis of diseases like AD. Various pre-trained DL models, such as ResNet50, VGG19, InceptionV3, and AlexNet, have demonstrated notable success in AD detection. Subjectivity in clinical diagnosis is a significant challenge, as human clinical evaluations are often prone to human bias. Another major issue is the high cost and time-consuming nature of diagnostic methods like PET scans and cerebrospinal fluid (CSF) analysis, which are expensive and widely inaccessible. This study proposes a custom CNN model designed specifically for AD detection using MRI scans to address these challenges. Unlike traditional clinical evaluations, the deep learning approach ensures automated detection, reducing human subjectivity and enhancing diagnostic consistency. The proposed method is also more cost-effective and accessible, as MRI scans are more widely available. Data augmentation techniques were employed to overcome the limitation of the dataset size and improve the model robustness. Data augmentation and dropout regularization were used to prevent overfitting, enhancing the model’s generalization ability.
The arrangement of the manuscript is as follows: Section 2 offers a broad literature review in the field of AD detection using AI techniques. Section 3 summarizes the methodology, including the details of the dataset and the experimental workflow. Section 4 talks about the results, highlighting the efficacy of the proposed model against the existing models (ResNet50, VGG19, InceptionV3, and AlexNet). Finally, Section 5 concludes by summarizing the main outcomes and proposing possible future research work.
2. Literature Review
Recently, a substantial body of research has been attentive to the growth of DL and ML models for the detection, classification, and prediction of AD. Non-invasive imaging techniques, particularly capillaroscopy, hold promise for diagnosing wide-ranging diseases, including neurodegenerative conditions like AD [4]. Studies have explored various data modalities, including MRI, PET, EEG, and genomic data, utilizing neural network architectures and ML approaches to enhance diagnostic accuracy and reliability. Table 1 summarizes key research works from recent literature, highlighting their objectives, methodologies, and relevance to the field of AD detection and classification. This comprehensive overview provides a clear understanding of the current advancements and evolving landscape of DL applications in AD diagnosis.

Table 1.
Summary of key works and reviews in the literature on AD detection.
The studies summarized in Table 1 demonstrate a range of approaches that leverage DL techniques for different aspects of AD detection and classification, from early diagnosis using non-invasive methods to predicting disease progression and identifying subtypes based on genetic data. While there is substantial progress in this research, issues with reliability and accuracy remain a challenge. In this view, this paper proposes a custom CNN model specifically designed for AD detection, aiming to enhance performance. This new approach leverages the strengths of DL and offers a more comprehensive solution for early and accurate AD detection.
3. Methodology
3.1. Dataset Description
The data used for the experiment comprise the Alzheimer’s MRI dataset, obtained from Kaggle [16]. It comprises 6400 MRI scanned images of the brain, divided into four classes: class 0 (Mild Demented), class 1 (Moderate Demented), class 2 (Non-Demented), and class 3 (Very Mild Demented) of 128 × 128 pixels each. Figure 1 shows sample images from each class of the dataset. As part of the preprocessing steps, pixel values were normalized to a range of [0, 1], a technique that aids in the faster convergence of DL models by scaling the inputs. Following normalization, the dataset was split into three subsets, allocating 80% for training, 10% for testing, and 10% for validation. The training dataset contains 5120 images; the testing and validation datasets contain 640 images each.
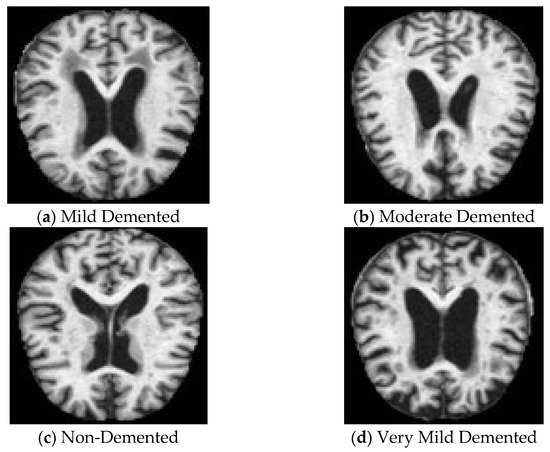
Figure 1.
Dataset images from each class.
3.2. Model Architecture
For the initial phase of the experiment, four pre-existing DL models: ResNet50, VGG19, InceptionV3, and AlexNet were chosen for transfer learning. ResNet50, with its deep residual connections [17], addresses the vanishing gradient problem and enables effective training of deeper networks. Despite its computational complexity, VGG19 is noted for its uniform architecture and ability to extract features. InceptionV3 was chosen due to its ability to capture multiscale spatial data through Inception modules. AlexNet was included to evaluate the performance of simpler architecture compared to more modern deep networks. To adapt these pre-trained models to the AD task, fine-tuning was performed by replacing their original classification layers with a custom fully connected head consisting of a global average pooling layer, a dense layer with 1024 neurons and ReLU activation, and followed by a softmax layer for multi-class classification. Accuracy, loss, precision, recall, F1 score, and AUROC were considered for the evaluation. In addition to pre-trained models, a custom CNN model was built.
The custom CNN architecture consisted of three convolutional layers with increasing filters of 16, 32, and 64 to capture low-level features in early layers and more complex patterns in deeper layers [18], and ReLU activation functions [19], each followed by max-pooling layers to reduce spatial dimensions and extract dominant features. ReLU activation was chosen based on its ability to mitigate the vanishing gradient problem. These were succeeded by a flattened layer, followed by two dense layers, containing 128 and 4 neurons each [8]. The final layer with four neurons employed a softmax activation function to yield probabilities for the four classes (Figure 2 and Figure 3).
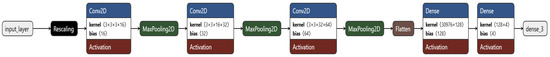
Figure 2.
Architecture of the custom CNN model in the first phase of the experiment.

Figure 3.
Architecture of the custom CNN model in the second phase of the experiment.
3.3. Data Augmentation
In the second phase of the experiment, data augmentation was employed. The augmentation techniques included horizontal flipping, random rotations, and zooming, which introduced variability and minimized overfitting by exposing the models to different perspectives of the MRI scans. First, a RandomFlip layer randomly flips the input images horizontally, which helps reduce overfitting by exposing the model to mirrored variations of the dataset images. Next, a RandomRotation layer introduces small random rotations to the images with a factor of 0.1, making the model invariant to rotational differences. Lastly, a RandomZoom layer randomly zooms into or out of the images with a factor of 0.1, enabling the model to handle varying scales and reinforcing its ability to detect features at different magnifications. In addition, to avoid overfitting and improve generalization, a 20% dropout layer was added in the second phase of the experiment, as shown in Figure 3. Dropout forces the network to learn redundant and robust feature representations by randomly deactivating neurons during training [20,21]. The input size of each model was changed based on its architectural needs.
3.4. Training Process
Figure 4 represents the flow of the experiment. The training was done over 100 epochs and a patience of 10 epochs. Patience is the number of epochs with no improvement in the validation performance, after which the training will be halted. It means an early stopping of 10 epochs. The list of hyperparameters used for the experiment is tabulated in Table 2.
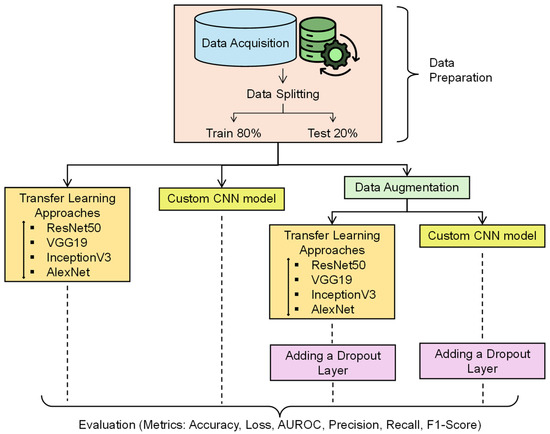
Figure 4.
Experimental workflow for AD detection.

Table 2.
Hyperparameters.
These models are trained again on the augmented dataset for performance assessment. A relative study was conducted across both phases to assess the impact of augmentation and dropout on accuracy, loss, precision, recall, F1 score, and AUROC.
4. Results and Discussion
In the first phase of the experiment, the classification performance of the models varied significantly. ResNet50, InceptionV3, and the custom CNN model demonstrated decent performance across all classes, whereas VGG19 and AlexNet struggled to distinguish between different dementia stages. ResNet50 achieved an F1 score of 98% or higher for all AD classes. It achieved 100% precision, recall, and F1 scores for the Moderate Demented case. ResNet50 also showed strong performance for the Very Mild Demented and Non-Demented cases, where precision and recall remained high. It can be observed from Table 3 that the validation accuracy for ResNet50 was 99.06%, with a validation loss of 0.0370, indicating that the model was able to generalize well to unseen data. The results are presented in Figure 5a–c. In contrast, Figure 5d–f depict that VGG19 exhibited poor classification performance. It failed to classify Mild Demented, Moderate Demented, and Very Mild Demented cases, achieving an F1 score of just 67% for Non-Demented cases. Its validation accuracy was 50.64%, with validation loss exceeding 1.0, reflecting its inability to perform on unseen data. InceptionV3 performed well by achieving an F1 score of 99% or higher for all AD categories. Its ability to generalize unseen data was reflected in a validation accuracy of 99.22%, with a validation loss of 0.0442, suggesting that InceptionV3 effectively captured essential patterns in the dataset and had better generalization than ResNet50, as shown in Table 3 and Figure 5g–i. Figure 5j–l depict that AlexNet struggled to differentiate Mild Demented, Moderate Demented, and Very Mild Demented cases, achieving an F1 score of 67% solely for Non-Demented cases. With a validation accuracy of 49.84% and a validation loss exceeding 1.0, the model demonstrated limited generalization ability on unseen data. The custom CNN model outperformed all existing architectures, achieving 100% F1 scores for Moderate Demented cases while maintaining precision and recall above 98% for all other classes. The model’s validation accuracy reached 99.28%, with a notably low validation loss of 0.0113, demonstrating its generalizability on unseen data and learning capability. The results are given in Figure 5m–o.

Table 3.
Accuracy and loss before augmentation.
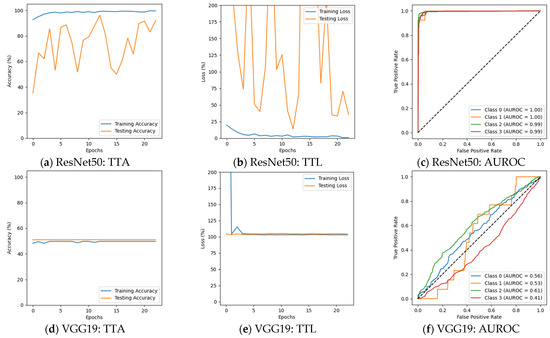
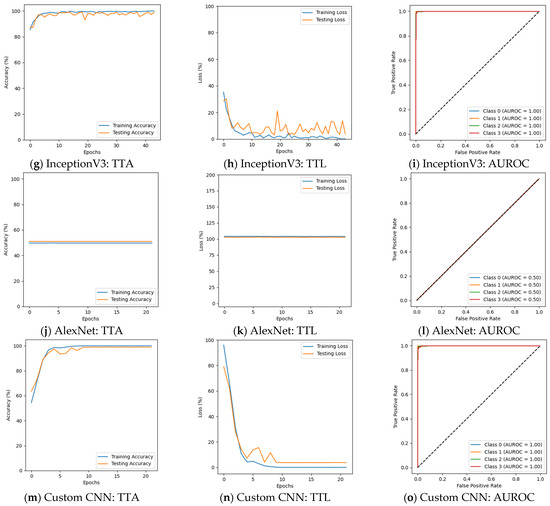
Figure 5.
Results before augmentation (training vs. testing accuracy: TTA; training vs. testing loss: TTL).
The results before data augmentation depict that the deeper architectures, such as ResNet50 and InceptionV3, were highly effective in learning discriminative features for AD detection. The validation accuracies of these models were among the highest, with low validation losses, indicating their strong generalization capabilities. The custom CNN model, despite being developed from scratch, outperformed all pre-trained architectures by achieving the highest validation accuracy and lowest validation loss. On the other hand, VGG19 and AlexNet exhibited poor performance, failing to classify Mild Demented, Moderate Demented, and Very Mild Demented cases. Their high validation losses and low validation accuracies suggest that these models struggled to capture the complex patterns present in MRI scans, making them unsuitable for AD classification.
Figure 6 depicts that the performance of several models changed, with some benefiting while others experienced a decline in performance in the second phase of the experiment. ResNet50 could not perform well, particularly for Moderate Demented cases, where its F1 score declined from 100% to 63%. The precision for this class fell from 100% to 55%, while recall dropped from 100% to 75%. Despite these declines, the model maintained a strong classification for Non-Demented and Very Mild Demented cases. Its validation accuracy was 69.64%, with a validation loss of 1.2924, suggesting that ResNet50 struggled to adapt to augmented data. The results are presented in Figure 6a–c and Table 4. VGG19 continued to show weak classification performance, failing to classify Mild Demented, Moderate Demented, and Very Mild Demented cases, as shown in Figure 6d–f. Its validation accuracy remained nearly unchanged at 50.94%, with validation losses above 1.0. InceptionV3 continued to generalize well, with F1 scores remaining above 91% for all classes. A decline in recall for Moderate Demented cases, dropping from 100% to 93%, slightly impacted its overall classification performance. The validation accuracy of InceptionV3 decreased slightly to 96.56%, with a validation loss of 0.1271. The results are given in Figure 6g–i. AlexNet faced challenges in distinguishing Mild Demented, Moderate Demented, and Very Mild Demented cases. However, it consistently identified Non-Demented cases with an F1 score of 67%, demonstrating its ability to recognize certain patterns in the data. It achieved a validation accuracy of 49.84% with a validation loss of 1.0244. The results are summarized in Figure 6j–l and Table 4. The custom CNN model showed superior performance, achieving 100% F1 scores for both Moderate Demented and Very Mild Demented cases, while maintaining precision and recall above 98% for all other categories. Its validation accuracy slightly improved to 99.53%, with a validation loss of 0.0238, and achieved an AUROC of 100% on Mild and Moderate Demented cases. The results are shown in Figure 6m–o and Table 4 and Table 5.
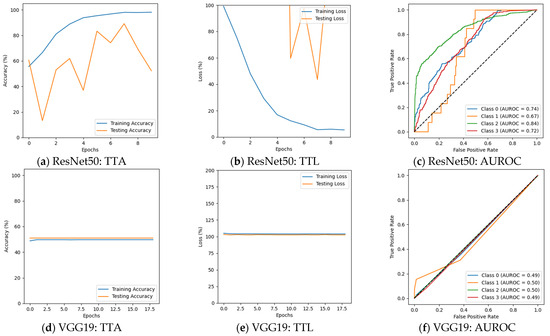
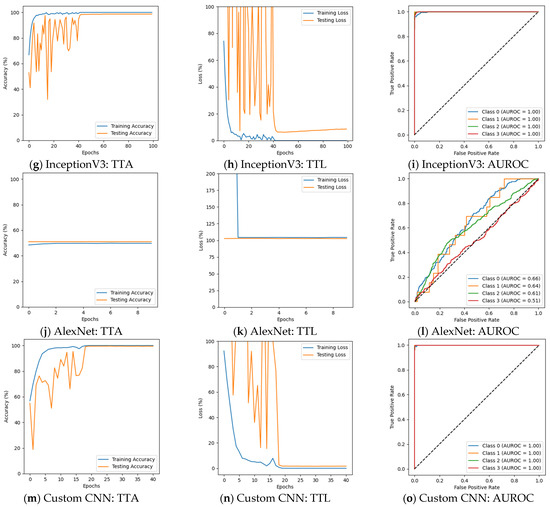
Figure 6.
Results after augmentation.

Table 4.
Accuracy and loss after augmentation.

Table 5.
Performance metrics for custom CNN model.
The introduction of data augmentation had a mixed impact on model performance. While it helped the custom CNN model and InceptionV3 maintain decent classification performance, others, like ResNet50, experienced significant declines. The sharp drop in validation accuracy and the increased validation loss for ResNet50 indicate that the model struggled to adapt to augmented data. VGG19 and AlexNet failed to show any improvement. Their inability to classify early-stage AD cases remained unchanged, showing their inability to learn complex spatial and texture-based features from MRI images.
Models such as ResNet50 (before augmentation), InceptionV3 (before and after augmentation), and the custom CNN model (before and after augmentation) demonstrated initial fluctuations in training and testing accuracy and loss, as shown in Figure 5 and Figure 6. Their eventual convergence, where training and testing curves align, indicates that the model successfully learned from the data without overfitting. The gradual stabilization of loss and the closing gap between training and testing accuracy suggest effective generalization. The presence of fluctuations before convergence can be due to learning rate adjustments and the complexity of feature extraction. On the other hand, ResNet50 (after augmentation) exhibited continuous fluctuations in both accuracy and loss curves without a clear convergence point. This suggests that while the model learned from the data, the optimization process was unstable. The ups and downs may be a result of the increased variability introduced by data augmentation. In contrast, VGG19 (before and after augmentation) and AlexNet (before and after augmentation) exhibited almost flat learning curves, with minimal fluctuations. Flat curves indicate that the model is struggling to capture complex patterns effectively. Before augmentation, ResNet50 exhibited strong performance across all classes, achieving more than 98% F1 score for each stage. After augmentation, ResNet50 achieved high precision and recall for Non-Demented and Very Mild Demented cases, suggesting improved generalization. After augmentation, the F1 score dropped for Moderate Demented cases from 100% to 63%. A decline in recall for Mild Demented cases from 100% to 83% suggests challenges in distinguishing early-stage AD patterns. For Non-Demented cases, VGG19’s recall was 100% in both the first and second phases of the experiment. VGG19 failed to classify Mild Demented and Moderate Demented cases. This suggests that VGG19 lacks discriminative power for early and moderate AD stages, making it unsuitable for clinical applications.
InceptionV3 performed well across all AD stages, with a precision and a recall of more than 95% before and after augmentation. The F1 score remained constant, indicating adaptability to augmented data. A slight recall reduction for Moderate Demented cases from 100% to 93% suggests a minor trade-off in classification performance after augmentation. AlexNet failed to classify Mild Demented, Moderate Demented, and Very Mild Demented cases. Even for Non-Demented cases, it only achieved 50% precision, indicating a high false-positive rate. The custom CNN model performed well across all AD stages before and after augmentation. The initial F1 scores of 97% and 98% for Mild Demented and Non-Demented cases were slightly lower than InceptionV3 but improved in the second phase of the experiment. F1 scores in post-augmentation results for Moderate Demented and Very Mild Demented cases highlight strong class separability.
High false-positive (FP) rates, as seen in VGG19 and AlexNet, which failed to classify Mild, Moderate, and Very Mild Demented cases, can lead to misdiagnosis, unnecessary anxiety, and increased medical costs. Patients wrongly diagnosed with AD may undergo unnecessary treatments, causing emotional and financial burdens. On the other hand, false negatives (FNs) delay early interventions, worsening disease progression. ResNet50′s post-augmentation recall for Moderate Demented cases dropped from 100% to 75%, while its F1 score declined from 100% to 63%, increasing the risk of missed diagnoses. This failure to identify affected individuals in time could reduce the effectiveness of treatment. Models like InceptionV3 and the custom CNN one demonstrated better generalization, maintaining high recall and precision even after augmentation. InceptionV3′s minor recall drop in Moderate Demented cases from 100% to 93% suggests a slight trade-off in classification performance. The custom CNN model’s post-augmentation 100% F1 score for Moderate and Very Mild Demented cases highlights its ability to minimize both FP and FN rates, making it highly reliable for detection and efficient in AD detection.
To enhance the interpretability of the custom CNN model, Local Interpretable Model-Agnostic Explanations (LIME) were applied to visualize the most influential regions in the MRI images contributing to classification decisions. Figure 7a–d present the LIME-generated heatmaps on sample images of each class of the validation set.
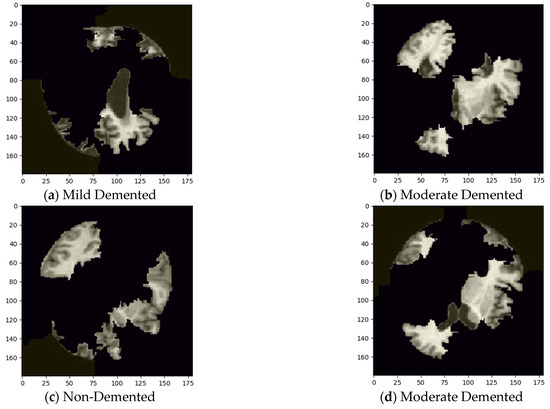
Figure 7.
Visualizing influential features using LIME.
5. Conclusions
This paper presents a new approach for detecting AD using advanced DL techniques, specifically by developing a custom CNN model. The proposed model was rigorously tested against several well-known DL models, including ResNet50, VGG19, InceptionV3, and AlexNet, using datasets both before and after data augmentation.
The custom CNN model demonstrated exceptional performance, achieving a validation accuracy of 99.28% before augmentation and 99.53% after augmentation, with minimal validation losses of 0.0113 and 0.0238, respectively. It also outperformed all other models in precision and recall across all classes, achieving an F1 score of 100% for Moderate Demented and Very Mild Demented cases post-augmentation. InceptionV3 also exhibited strong classification capabilities, with validation accuracies of 99.22% and 96.56% before and after augmentation, respectively, and an F1 score consistently above 90% for most classes. In contrast, ResNet50′s performance declined significantly post-augmentation, with validation accuracy dropping from 99.06% to 69.64% and its F1 score for Moderate Demented cases dropping from 100% to 63%, indicating its inability to adapt to augmented data. VGG19 and AlexNet consistently underperformed, both achieving a validation accuracy of around 50% across both phases and failing to classify Mild and Moderate Demented cases entirely, leading to an F1 score of 0% for those categories. These results highlight the superiority of the custom CNN model in classifying Alzheimer’s disease stages, particularly after augmentation. This model offers significant implications for clinical practice by enabling earlier diagnosis and more effective treatment strategies, thereby improving patient outcomes. This contributes to the growing literature on the application of DL in medical diagnostics, showing that customized models can provide superior performance compared to traditional ones.
5.1. Ethical Considerations
One of the primary concerns is the potential impact of false diagnoses. While false negatives could postpone important care and treatment, false positives could cause patients and their families unnecessary anxiety and result in unneeded medical diagnoses. The custom CNN model can be used as a decision support tool rather than a stand-alone diagnostic system. Additionally, ensuring patient privacy and informed consent is important when using medical imaging data. If clinical deployment of the model is pursued in the future, it will be accompanied by rigorous validation and real-world assessments to ensure both safety and effectiveness.
5.2. Limitations
Although the proposed model demonstrates strong performance metrics, certain limitations must be acknowledged to ensure a more comprehensive evaluation. One major limitation is its dependency on the dataset used for training. Since the dataset originates from a specific source, it may lack diversity in terms of demographic representation and scanner variations. This could limit the model’s ability to generalize when applied to MRI scans from different healthcare institutions and geographical regions. Additionally, the dataset used in this study is relatively small for deep learning applications, which may restrict the model’s ability to generalize across diverse populations effectively. The reliance on preprocessed MRI images may not fully capture the variability present in real-world AD diagnosis, leading to challenges when implementing the model in clinical settings. The absence of external validation on independent datasets limits the ability to assess its robustness across different populations. Additionally, the study does not incorporate clinical records and genetic information, which could provide a more comprehensive understanding of AD progression.
5.3. Future Scope
Future work will focus on expanding the dataset with larger, more diverse MRI scans from multiple sources to improve generalization and ensure that the model performs reliably across different demographic groups. Integrating with other diagnostic modalities, such as Positron Emission Tomography (PET), Electroencephalography (EEG), or cerebrospinal fluid biomarkers, could enhance the reliability and robustness of AD detection. Conducting external validation using independent datasets will help evaluate the model’s robustness and adaptability in real-world scenarios. Enhancing model explainability using techniques like Grad-CAM or SHAP will also be essential to ensure transparency and trust in AI-driven medical diagnoses.
Author Contributions
Conceptualization, G.P.R.; methodology, G.P.R., D.R. and S.M.A.K.; software, Y.V.P.K.; formal analysis, K.P.P. and M.J.; funding acquisition, Y.V.P.K.; investigation, G.P.R., D.R. and S.M.A.K.; resources, K.P.P. and M.J.; data curation, Y.V.P.K., K.P.P. and M.J.; supervision, Y.V.P.K.; validation, M.J.; visualization, K.P.P.; project administration, Y.V.P.K.; writing—original draft, G.P.R., D.R. and S.M.A.K.; writing—review and editing, Y.V.P.K., K.P.P. and M.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- World Health Organization. Available online: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed on 10 October 2024).
- Reddy, G.P.; Pavan, K.Y.V. A Beginner’s Guide to Federated Learning. In Proceedings of the 2023 Intelligent Methods, Systems, and Applications (IMSA), Giza, Egypt, 15 July 2023; pp. 557–562. [Google Scholar] [CrossRef]
- Reddy, G.P.; Pavan Kumar, Y.V.; Prakash, K.P. Hallucinations in Large Language Models (LLMs). In Proceedings of the 2024 IEEE Open Conference of Electrical, Electronic and Information Sciences (eStream), Vilnius, Lithuania, 25 April 2024; pp. 1–6. [Google Scholar] [CrossRef]
- Taormina, V.; Raso, G.; Gentile, V.; Abbene, L.; Buttacavoli, A.; Bonsignore, G.; Valenti, C.; Messina, P.; Scardina, G.A.; Cascio, D. Automated Stabilization, Enhancement and Capillaries Segmentation in Videocapillaroscopy. Sensors 2023, 23, 7674. [Google Scholar] [CrossRef] [PubMed]
- Rallabandi, S.V.P.; Seetharaman, K. Deep learning-based classification of healthy aging controls, mild cognitive impairment and alzheimer’s disease using fusion of MRI-PET imaging. Biomed. Signal Process. Control. 2023, 80, 104312. [Google Scholar] [CrossRef]
- Fouad, I.A.; El-Zahraa, M.L.F. Identification of alzheimer’s disease from central lobe EEG signals utilizing machine learning and residual neural network. Biomed. Signal Process. Control 2023, 86, 105266. [Google Scholar] [CrossRef]
- Qu, Z.; Yao, T.; Liu, X.; Wang, G. A graph convolutional network based on univariate neurodegeneration biomarker for alzheimer’s disease diagnosis. IEEE J. Transl. Eng. Health Med. 2023, 11, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Shamrat, F.J.M.; Akter, S.; Azam, S.; Karim, A.; Ghosh, P.; Tasnim, Z.; Hasib, K.M.; De Boer, F.; Ahmed, K. AlzheimerNet: An Effective Deep Learning Based Proposition for Alzheimer’s Disease Stages Classification from Functional Brain Changes in Magnetic Resonance Images. IEEE Access 2023, 11, 16376–16395. [Google Scholar] [CrossRef]
- Sharma, R.; Goel, T.; Tanveer, M.; Lin, C.T.; Murugan, R. Deep-Learning-Based Diagnosis and Prognosis of Alzheimer’s Disease: A Comprehensive Review. IEEE Trans. Cogn. Dev. Syst. 2023, 15, 1123–1138. [Google Scholar] [CrossRef]
- Klingenberg, M.; Stark, D.; Eitel, F.; Budding, C.; Habes, M.; Ritter, K. Higher Performance for Women than Men in MRI-Based Alzheimer’s Disease Detection. Alzheimer’s Res. Ther. 2023, 15, 84. [Google Scholar] [CrossRef] [PubMed]
- Shigemizu, D.; Akiyama, S.; Suganuma, M.; Furutani, M.; Yamakawa, A.; Nakano, Y.; Ozaki, K.; Niida, S. Classification and Deep-Learning–Based Prediction of Alzheimer Disease Subtypes by Using Genomic Data. Transl. Psychiatry 2023, 13, 232. [Google Scholar] [CrossRef] [PubMed]
- Arafa, D.A.; Moustafa, H.E.-D.; Ali-Eldin, A.M.T.; Ali, H.A. Early detection of alzheimer’s disease based on the state-of-the-art deep learning approach: A comprehensive survey. Multimed Tools Appl. 2022, 81, 23735–23776. [Google Scholar] [CrossRef]
- Diogo, V.S.; Ferreira, H.A.; Prata, D. Early Diagnosis of Alzheimer’s Disease Using Machine Learning: A Multi-Diagnostic, Generalizable Approach. Alzheimer’s Res. Ther. 2022, 14, 107. [Google Scholar] [CrossRef] [PubMed]
- Basher, A.; Kim, B.C.; Lee, K.H.; Jung, H.Y. Volumetric feature-based alzheimer’s disease diagnosis from sMRI data using a convolutional neural network and a deep neural network. IEEE Access 2021, 9, 29870–29882. [Google Scholar] [CrossRef]
- Lopez, M.M.; Nevado, A.; Carro, B. Detection of early stages of alzheimer’s disease based on meg activity with a randomized convolutional neural network. Artif. Intell. Med. 2020, 107, 101924. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.M.A.H.; Khan, M.Q.; Rizwan, A.; Jan, S.U.; Samee, N.A.; Jamjoom, M.M. Computer-aided diagnosis of Alzheimer’s disease and neurocognitive disorders with multimodal Bi-Vision Transformer (BiViT). Pattern Anal Applic 2024, 27, 76. [Google Scholar] [CrossRef]
- Hassan, N.; Musa Miah, A.S.; Shin, J. Residual-Based Multi-Stage Deep Learning Framework for Computer-Aided Alzheimer’s Disease Detection. J. Imaging 2024, 10, 141. [Google Scholar] [CrossRef] [PubMed]
- Alsubaie, M.G.; Luo, S.; Shaukat, K. Alzheimer’s Disease Detection Using Deep Learning on Neuroimaging: A Systematic Review. Mach. Learn. Knowl. Extr. 2024, 6, 464–505. [Google Scholar] [CrossRef]
- Pradhan, N.; Sagar, S.; Singh, A.S. Analysis of MRI Image Data for Alzheimer Disease Detection Using Deep Learning Techniques. Multimed Tools Appl. 2023, 83, 17729–17752. [Google Scholar] [CrossRef]
- Omar, A.; El-Hafeez, T.A. Optimizing Epileptic Seizure Recognition Performance with Feature Scaling and Dropout Layers. Neural Comput. Appl. 2024, 36, 2835–2852. [Google Scholar] [CrossRef]
- Menagadevi, M.; Devaraj, S.; Madian, N.; Thiyagarajan, D. Machine and Deep Learning Approaches for Alzheimer Disease Detection Using Magnetic Resonance Images: An Updated Review. Measurement 2024, 226, 114100. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).