Performance Analysis of YOLOv11: Nano, Small, and Medium Models for Herbal Leaf Classification †
Abstract
1. Introduction
2. Related Works
2.1. Previous Related Research
2.2. Herbal Leaf
- Annona muricata contains bioactive compounds such as acetogenins, alkaloids, flavonoids, and sterols that play an important role in health benefits, including anticancer, antioxidant, antimicrobial, and anti-inflammatory activities [15].
- Anredera cordifolia contains active compounds such as flavonoids, alkaloids, terpenoids, and saponins. Flavonoids function as antibiotics by disrupting the function of microorganisms such as bacteria and viruses. In addition, this plant also has antimicrobial properties that can prevent bacterial growth. Its antibacterial activity is proven to help heal wounds infected with Staphylococcus aureus, while its antifungal activity is effective against Candida albicans [16,17].
- Psidium guajava contains important compounds such as tannins, triterpenes, flavonoids (quercetin), saponins, and ursolic acid, which have various benefits, including antioxidant, hepatoprotective, antimicrobial, antidiabetic, anti-inflammatory, and antiallergic activities. The plant also supports the treatment of rotavirus enteritis in children, diarrhea, and diabetes [24].
- Isotoma longiflora contains compounds such as flavonoids, alkaloids, saponins, and polyteros, which are used in traditional medicine to relieve headaches and respiratory problems [4].
- Coleus scutellarioides contains compounds such as alkaloids, steroids, tannins, saponins, and flavonoids. This plant is used in traditional medicine as an antibacterial, anthelmintic, antidiabetic, and appetite enhancer, as well as to treat coughs, hemorrhoids, stomach ulcers, fever, and diarrhea and facilitate the menstrual cycle [25,26].
- Ageratum conyzoides is a widespread weed, especially in tropical and subtropical areas [27]. The active compounds contained in Ageratum conyzoides can be used as antibacterials to treat skin infections caused by Staphylococcus epidermidis and Propionibacterium acnes bacteria.
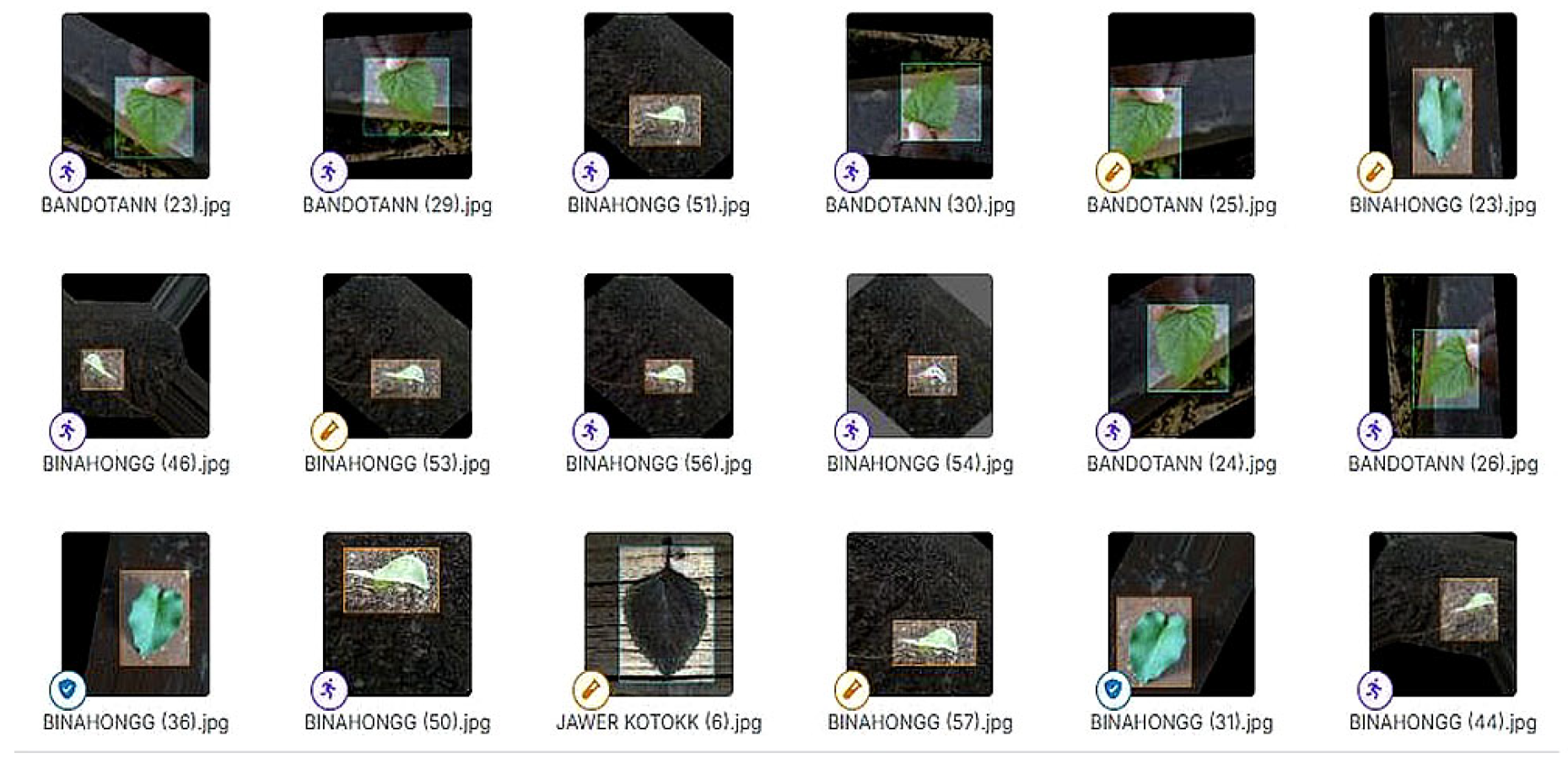
- Syzygium polyanthum contains secondary compounds such as flavonoids, alkaloids, tannins, phenols, saponins, and essential oils. The leaves of this plant have antihypertensive, anti-inflammatory, antioxidant, cholesterol-lowering, gout medication, analgesic, and antimicrobial activities [28]. This makes herbal leaves an attractive alternative in natural medicine and a worthy subject for further research. Ten types of herbal leaves as shown in Figure 1 below.
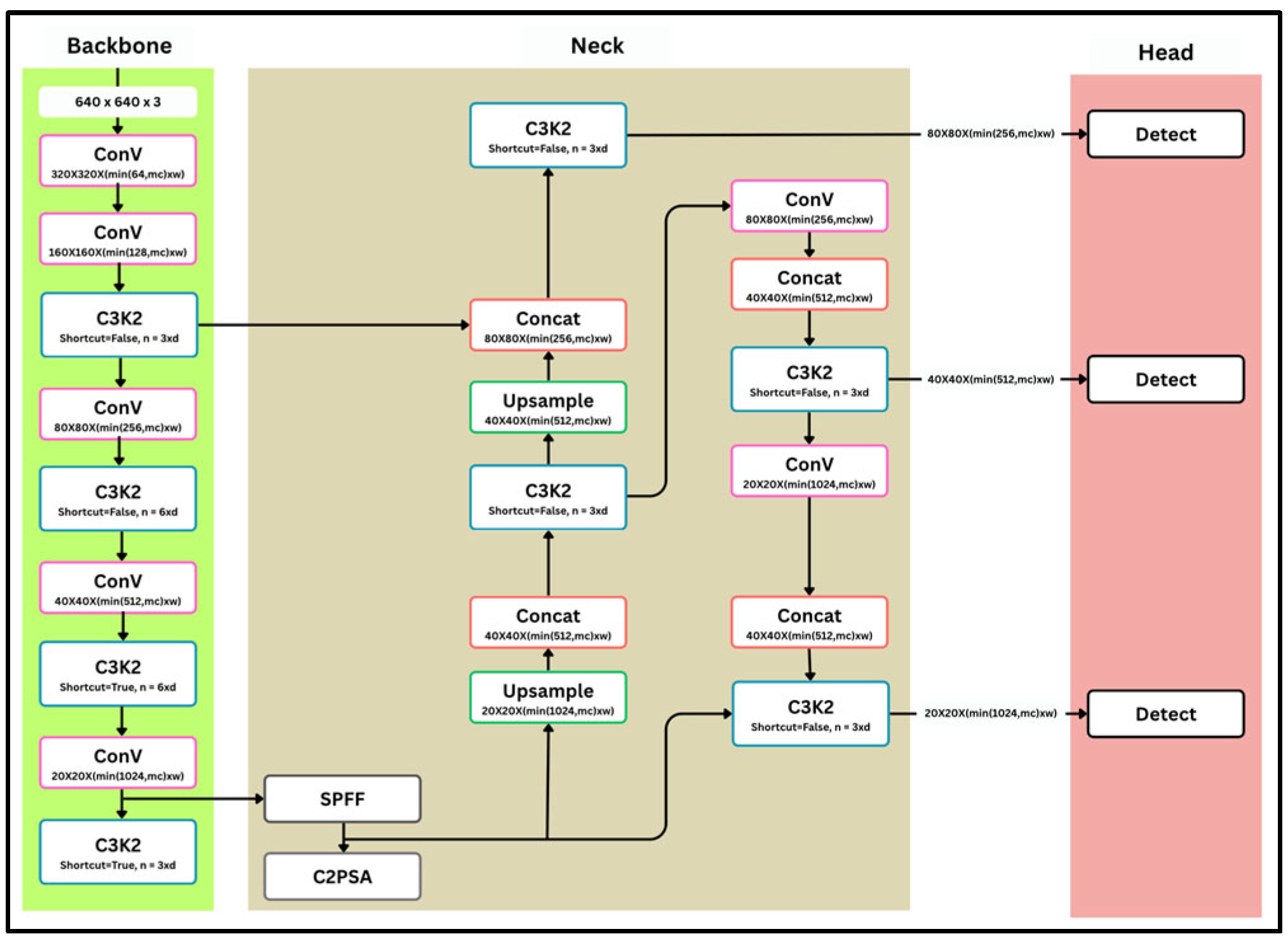
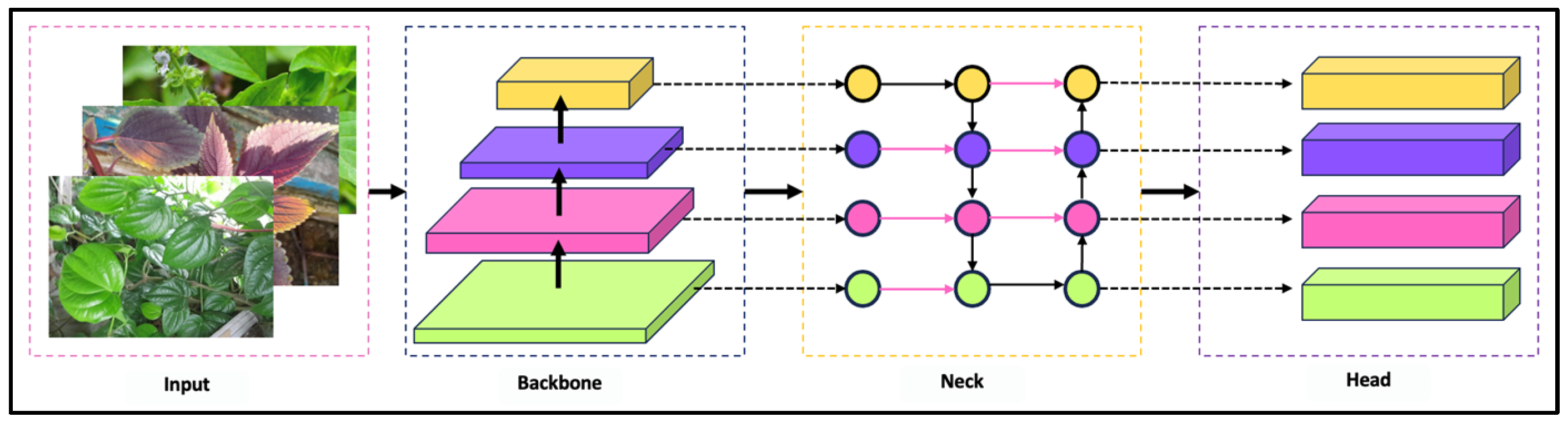
2.3. YOLOv11
3. Materials and Methods
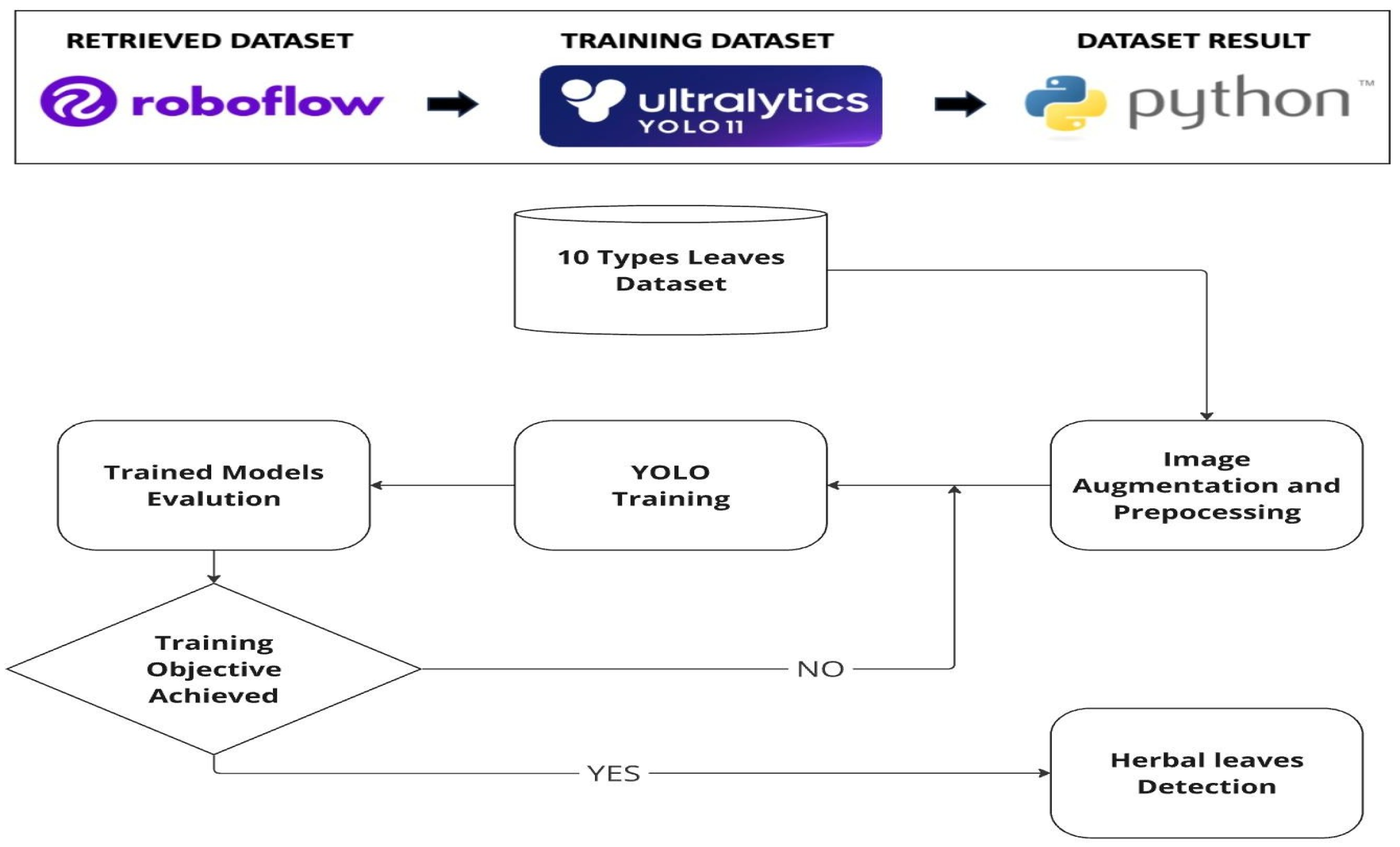
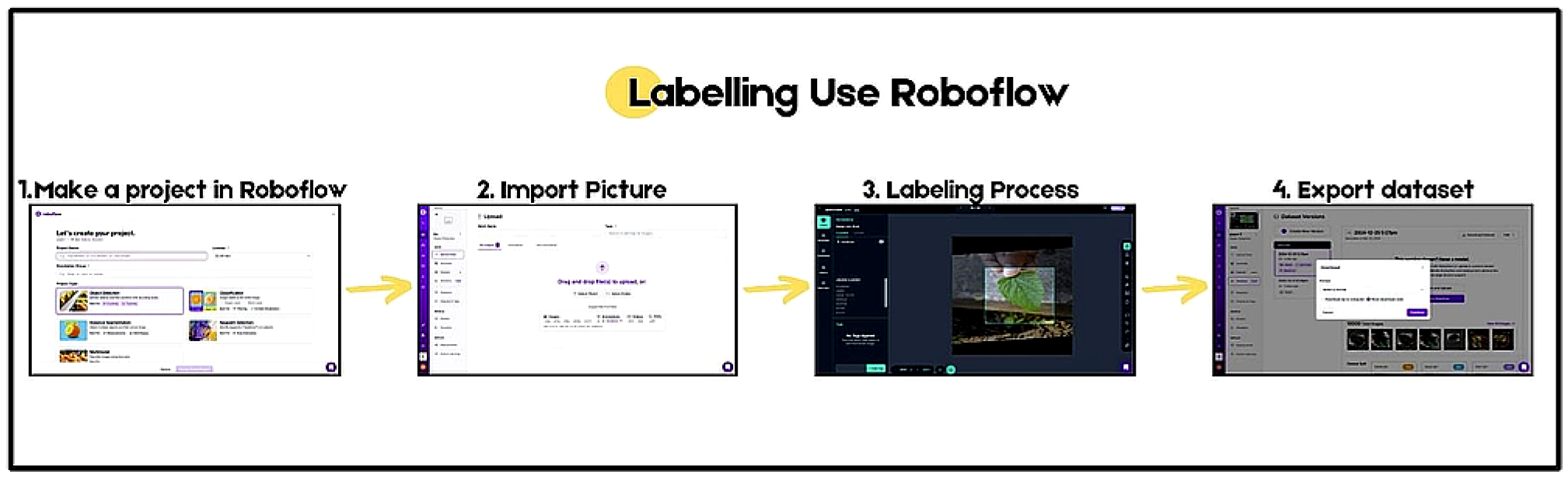
3.1. Dataset Retrieval
3.2. Training Data
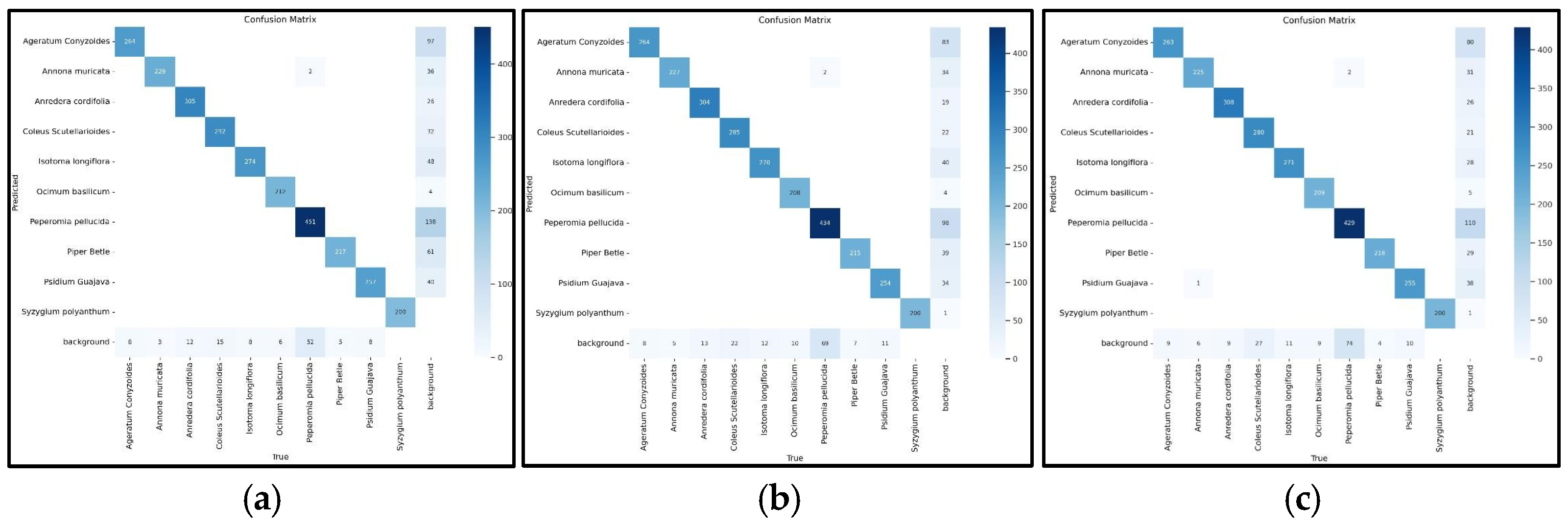
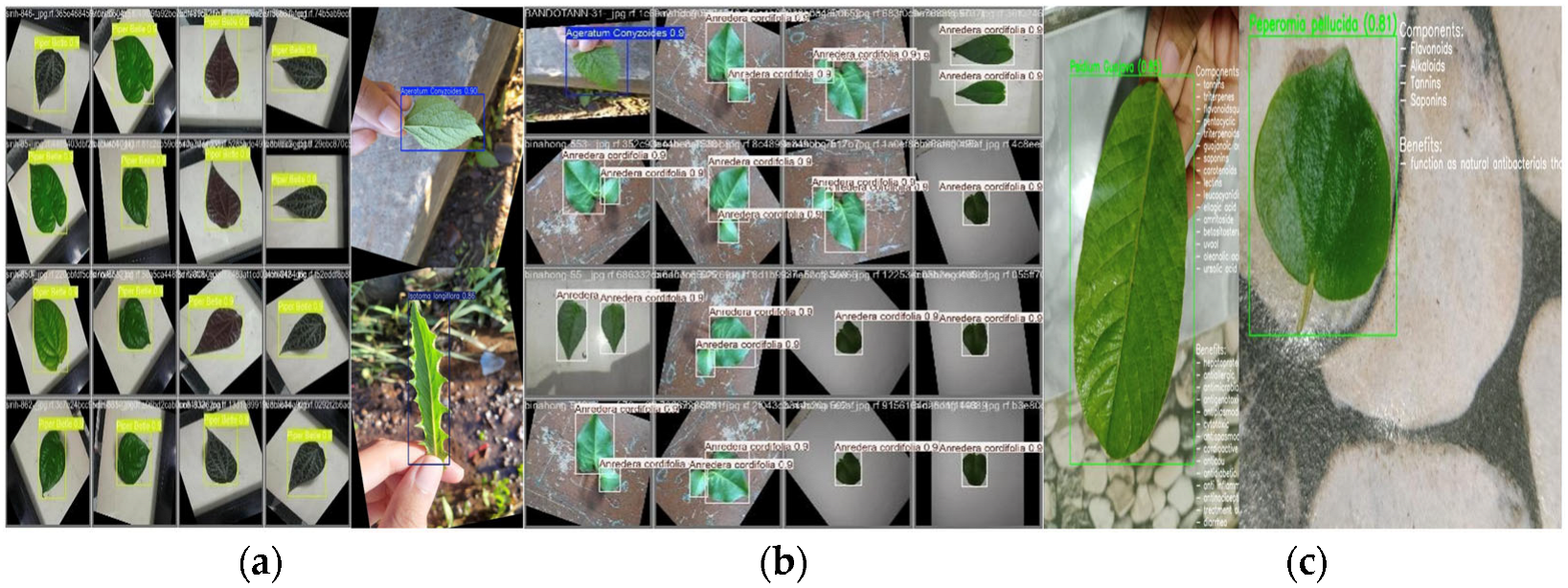
4. Result and Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rani, D.M.; Wongso, H.; Purwoko, R.Y.; Winarto, N.B.; Shalas, A.F.; Triatmoko, B.; Pratama, A.N.W.; Keller, P.A.; Nugraha, A.S. Anti-cancer bioprospecting on medicinal plants from Indonesia: A review. Phytochemistry 2023, 216, 113881. [Google Scholar] [CrossRef]
- Kaur, A.; Kaur, S.; Singh, H.P.; Datta, A.; Chauhan, B.S.; Ullah, H.; Kohli, R.K.; Batish, D.R. Ecology, Biology, Environmental Impacts, and Management of an Agro-Environmental Weed Ageratum conyzoides. Plants 2023, 12, 2329. [Google Scholar] [CrossRef]
- Sura, S.P. The Phytochemical Landscape of Herbal Plants: Insights and Implications. Int. J. Adv. Res. Sci. Commun. Technol. 2023, 916–918. [Google Scholar] [CrossRef]
- Basir, H.; Purnamasari, I.; Thalib, M.; Daeng Pine, A.T.; Saldi; Nurka Sari, N.P. Formulasi Dan Uji Stabilitas Fisik Facial Wash Ekstrak Etanol Daun Kitolod (Isotoma longiflora L.) Serta Aktivitasnya Terhadap Staphylococcus epidermidis. J. Kesehat. Yamasi Makassar 2024, 8, 33–45. [Google Scholar] [CrossRef]
- Nwafor, C.; Manduna, I.T. Local processing methods for commonly used medicinal plants in South Africa. Med. Plants-Int. J. Phytomed. Relat. Ind. 2021, 13, 289–301. [Google Scholar] [CrossRef]
- Javid, A.; Fatima, M.; Hamad, M.; Ahmed, M. From roots to codes: Applications of computer-aided drug discovery from medicinal plants. S. Afr. J. Bot. 2024, 173, 159–174. [Google Scholar] [CrossRef]
- Purwono, S.; Nisa, U.; Astana, P.; Wijayaningsih, R.; Wicaksono, A.; Wahyuningsih, M.; Kertia, N.; Mustofa, M.; Wahyuono, S.; Fakhrudin, N. Factors Affecting the Perception of Indonesian Medical Doctors on Herbal Medicine Prescription in Healthcare Facilities: Qualitative and Quantitative Studies. J. Herb. Med. 2023, 42, 100747. [Google Scholar] [CrossRef]
- Bajpai, M. YOLO Models for Security and Surveillance Applications. Int. J. Res. Appl. Sci. Eng. Technol. 2024, 12, 2513–2518. [Google Scholar] [CrossRef]
- Musyaffa, M.S.I.; Yudistira, N.; Rahman, M.A.; Basori, A.H.; Mansur, A.B.F.; Batoro, J. IndoHerb: Indonesia medicinal plants recognition using transfer learning and deep learning. Heliyon 2024, 10, e40606. [Google Scholar] [CrossRef]
- Sharma, V.; Kumar, V.; Longchamps, L. Comparative performance of YOLOv8, YOLOv9, YOLOv10, YOLOv11 and Faster R-CNN models for detection of multiple weed species. Smart Agric. Technol. 2024, 9, 100648. [Google Scholar] [CrossRef]
- Siahaan, R.D.; Simbolon, I.N. Enhancing Real-Time Herbal Plant Detection in Agricultural Environments with YOLOv8. J. ISI 2024, 6, 2491–2507. [Google Scholar] [CrossRef]
- Mia, J.; Bijoy, H.I.; Uddin, S.; Raza, D.M. Real-Time Herb Leaves Localization and Classification Using YOLO. In Proceedings of the 2021 12th International Conference on Computing Communication and Networking Technologies (ICCCNT), Kharagpur, India, 6–8 July 2021; pp. 1–7. [Google Scholar] [CrossRef]
- Susa, J.A.B.; Dellosa, R.M.; Doculan, J.A.D.; Jallorina, R.D.; Evangelista, R.S.; Zapanta, G.S.; Mindoro, J.N. Identification of Philippine Therapeutic Leave Using Deep Learning. In Proceedings of the 2022 International Conference on Emerging Technologies in Electronics, Computing and Communication (ICETECC), Jamshoro, Pakistan, 7–9 December 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Meiriyama, M.; Devella, S.; Adelfi, S.M. Klasifikasi Daun Herbal Berdasarkan Fitur Bentuk dan Tekstur Menggunakan KNN. JATISI 2022, 9, 2573–2584. [Google Scholar] [CrossRef]
- Errayes, A.O.; Abdussalam-Mohammed, W.; Darwish, M.O. Review of Phytochemical and Medical Applications of Annona Muricata Fruits. J. Chem. Rev. 2020, 2, 70–79. [Google Scholar] [CrossRef]
- Aruperes, G.Y.; Pangemanan, D.H.C.; Mintjelungan, C.N. Daya Hambat Ekstrak Daun Binahong (Anredera cordifolia Steenis) Terhadap Pertumbuhan Bakteri Streptococcus mutans. e-GiGi 2021, 9, 250. [Google Scholar] [CrossRef]
- Alba, T.M.; de Pelegrin, C.M.G.; Sobottka, A.M. Ethnobotany, ecology, pharmacology, and chemistry of Anredera cordifolia (Basellaceae): A review. Rodriguesia 2020, 71, 01042019. [Google Scholar] [CrossRef]
- Sadiah, H.H.; Cahyadi, A.I.; Windria, S. Kajian Daun Sirih Hijau (Piper betle L) Sebagai Antibakteri. J. Sain Vet. 2022, 40, 128. [Google Scholar] [CrossRef]
- Metasari, D.; Sianipar, B.K.; Arfianti, M. Pemanfaatan pekarangan rumah untuk tanaman daun sirih (piper betle) bahan pembuatan handsanitizer di Panti Asuhan Bumi Raflesia. J. Indones. RAYA (Pengabdi. Pada Masy. Bid. Sos. Hum. Kesehat. Ekon. Dan Umum) 2021, 2, 69–73. [Google Scholar] [CrossRef]
- Kumalasari, M.L.F.; Andiarna, F. Uji Fitokimia Ekstrak Etanol Daun Kemangi (Ocimum basilicum L). Indones. J. Health Sci. 2020, 4, 39. [Google Scholar] [CrossRef]
- Tondolambung, A.H.; Edy, H.J.; Lebang, J.S. Uji Efektivitas Antibakteri Sediaan Krim Ekstrak Etanol Daun Kemangi (Ocimum basilicum L.) Terhadap Staphylococcus aureus. Pharmacon 2021, 10, 661. [Google Scholar] [CrossRef]
- Permadani, A.; Nikmah, H.; Halimatussakdiah, H.; Mastura, M.; Amna, U. Skrining Fitokimia Daun Sirih Cina (Peperomia pellucida L.) dari Kecamatan Bireun Bayeun, Aceh Timur. Quim. J. Kim. Sains Dan Terap. 2024, 6, 6–12. [Google Scholar] [CrossRef]
- Rahmawati, R.P.; Anggun, L.; Primandana, A.Z.; Dwiyanti, U. Pengaruh Pemberian Ekstrak Etanol 96% Daun Suruhan (Peperonia pellucida (L.) Kunth) terhadap Pertumbuhan Bakteri Propionibacterium acnes dengan Metode Difusi Cakram. Indones. J. Farm. 2021, 6, 22. [Google Scholar] [CrossRef]
- Sravani, S.; Charitha, C.S.; Nadendla, R.R. A Complete Review on Psidium guajava Linn (Medicinal Plant). Int. J. Pharm. Sci. Rev. Res. 2021, 67, 13–17. [Google Scholar] [CrossRef]
- Aji, N.P.; Noviyanty, Y.; Fahlevi, R. Skrining Fitokimia dan Profil KLT Metabolit Sekunder dari Ekstrak Etanol Daun Miana (Coleus scutellarioides Benth). J. Farm. Malahayati 2023, 6, 149–157. [Google Scholar] [CrossRef]
- Rante, D.; Asmuruf, F.A.; Jannah, L.R.; Nadeak, E.S.M.; Simaremare, E.S. Anticoagulant Activity Test of Ethanol Extract Coleus scutellarioides (L.) Benth. Int. J. Pharm. Bio-Med. Sci. 2024, 4, 351–357. [Google Scholar] [CrossRef]
- Baral, D.; Chaudhary, M.; Lamichhane, G.; Pokhrel, B. Ageratum conyzoides: A Potential Source for Medicinal and Agricultural Products. Turkish J. Agric.-Food Sci. Technol. 2022, 10, 2307–2313. [Google Scholar] [CrossRef]
- Haryanto, F.K.; Jesica, I.A.; Arafi, A.R.; Pranasti, E.A.; Rosa, D. Review Jurnal: Pemanfaatan Daun Salam (Syzygium polyanthum (Wight) Walp.) Sebagai Pengobatan Tradisional di Indonesia. Pharm. J. Pharmacy, Med. Health Sci. 2023, 4, 20–33. [Google Scholar] [CrossRef]
- Sulake, N.R. A Comprehensive Guide to YOLOv11 Object Detection. Data Science Blogathon. Available online: https://www.analyticsvidhya.com/blog/2024/10/yolov11-object-detection/ (accessed on 26 December 2024).
- Jegham, N.; Koh, C.Y.; Abdelatti, M.; Hendawi, A. Evaluating the Evolution of YOLO (You Only Look Once) Models: A Comprehensive Benchmark Study of YOLO11 and Its Predecessors. 2024. Available online: http://arxiv.org/abs/2411.00201 (accessed on 26 December 2024).
- Wu, M.; Yun, L.; Wang, Y.; Chen, Z.; Cheng, F. Detection algorithm for dense small objects in high altitude image. Digit. Signal Process. 2024, 146, 104390. [Google Scholar] [CrossRef]


| Model | Training Time (Hours) | GPU |
|---|---|---|
| Yolov11 Nano | 2.2 | 2.56 GB (Tesla T4) |
| Yolov11 Small | 2.6 | 4.35 GB (Tesla T4) |
| Yolov11 Medium | 4 | 8.75 GB (Tesla T4) |
| Class | img | Instance | Precision | Recall | mAP 50 | mAP 50–59 |
|---|---|---|---|---|---|---|
| All | 2000 | 2820 | 0.933 | 0.921 | 0.972 | 0.734 |
| Ageratum conyzoides | 199 | 272 | 0.856 | 0.923 | 0.947 | 0.778 |
| Annona muricata | 208 | 232 | 0.921 | 0.957 | 0.983 | 0.647 |
| Anredera cordifolia | 220 | 317 | 0.963 | 0.934 | 0.989 | 0.869 |
| Coleus scutellarioides | 199 | 307 | 0.942 | 0.899 | 0.972 | 0.831 |
| Isotoma longiflora | 198 | 282 | 0.927 | 0.947 | 0.964 | 0.754 |
| Ocimum basilicum | 198 | 218 | 0.995 | 0.928 | 0.993 | 0.606 |
| Peperomia pellucida | 203 | 505 | 0.917 | 0.722 | 0.914 | 0.554 |
| Piper betle | 179 | 222 | 0.893 | 0.955 | 0.977 | 0.88 |
| Psidium guajava | 195 | 265 | 0.922 | 0.942 | 0.982 | 0.708 |
| Syzygium polyanthum | 200 | 200 | 0.998 | 1 | 0.995 | 0.708 |
| (a) | ||||||
| Class | img | Instance | Precision | Recall | mAP 50 | mAP 50–59 |
| All | 2000 | 2820 | 0.947 | 0.906 | 0.973 | 0.739 |
| Ageratum conyzoides | 199 | 272 | 0.858 | 0.912 | 0.962 | 0.795 |
| Annona muricata | 208 | 232 | 0.945 | 0.957 | 0.986 | 0.653 |
| Anredera cordifolia | 220 | 317 | 0.965 | 0.927 | 0.989 | 0.872 |
| Coleus scutellarioides | 199 | 307 | 0.964 | 0.876 | 0.968 | 0.845 |
| Isotoma longiflora | 198 | 282 | 0.942 | 0.927 | 0.973 | 0.754 |
| Ocimum basilicum | 198 | 218 | 0.988 | 0.917 | 0.987 | 0.6 |
| Peperomia pellucida | 203 | 505 | 0.928 | 0.693 | 0.913 | 0.556 |
| Piper betle | 179 | 222 | 0.93 | 0.95 | 0.976 | 0.884 |
| Psidium guajava | 195 | 265 | 0.952 | 0.902 | 0.982 | 0.711 |
| Syzygium polyanthum | 200 | 200 | 0.994 | 1 | 0.995 | 0.715 |
| (b) | ||||||
| Class | img | Instance | Precision | Recall | mAP 50 | mAP 50–59 |
| All | 2000 | 2820 | 0.932 | 0.928 | 0.974 | 0.743 |
| Ageratum conyzoides | 199 | 272 | 0.86 | 0.929 | 0.964 | 0.797 |
| Annona muricata | 208 | 232 | 0.906 | 0.961 | 0.98 | 0.656 |
| Anredera cordifolia | 220 | 317 | 0.955 | 0.933 | 0.989 | 0.872 |
| Coleus scutellarioides | 199 | 307 | 0.941 | 0.883 | 0.971 | 0.841 |
| Isotoma longiflora | 198 | 282 | 0.94 | 0.95 | 0.973 | 0.767 |
| Ocimum basilicum | 198 | 218 | 0.994 | 0.94 | 0.991 | 0.612 |
| Peperomia pellucida | 203 | 505 | 0.886 | 0.787 | 0.91 | 0.564 |
| Piper betle | 179 | 222 | 0.926 | 0.96 | 0.981 | 0.885 |
| Psidium guajava | 195 | 265 | 0.913 | 0.936 | 0.982 | 0.721 |
| Syzygium polyanthum | 200 | 200 | 0.997 | 1 | 0.995 | 0.71 |
| (c) | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Insany, G.P.; Indriyani, R.; Ma’wa, N.J.; Safitri, S. Performance Analysis of YOLOv11: Nano, Small, and Medium Models for Herbal Leaf Classification. Eng. Proc. 2025, 107, 102. https://doi.org/10.3390/engproc2025107102
Insany GP, Indriyani R, Ma’wa NJ, Safitri S. Performance Analysis of YOLOv11: Nano, Small, and Medium Models for Herbal Leaf Classification. Engineering Proceedings. 2025; 107(1):102. https://doi.org/10.3390/engproc2025107102
Chicago/Turabian StyleInsany, Gina Purnama, Ranti Indriyani, Nadila Jannatul Ma’wa, and Sherly Safitri. 2025. "Performance Analysis of YOLOv11: Nano, Small, and Medium Models for Herbal Leaf Classification" Engineering Proceedings 107, no. 1: 102. https://doi.org/10.3390/engproc2025107102
APA StyleInsany, G. P., Indriyani, R., Ma’wa, N. J., & Safitri, S. (2025). Performance Analysis of YOLOv11: Nano, Small, and Medium Models for Herbal Leaf Classification. Engineering Proceedings, 107(1), 102. https://doi.org/10.3390/engproc2025107102






