Molecular Modelling of Withanolides Against EP2 Receptor for Treatment of Endometrial Cancer: A Pharmacokinetic and Molecular Docking Study †
Abstract
1. Introduction
2. Materials and Methods
2.1. Software, Hardware, and Databases
2.2. In Silico Anticancer Studies
2.2.1. Evaluation of Theoretical Oral Bioavailability
2.2.2. Protein Structure Preparation
2.2.3. Ligand Structure Preparation
2.2.4. Molecular Docking Analysis
3. Results
3.1. Creation of Library
3.2. Analysis of Theoretical Oral Bioavailability
ADMET Profile
3.3. Molecular Docking Studies
3.3.1. Grid Box
3.3.2. Validation of Docking Procedures
3.3.3. Binding Affinity Ligands to EP2 Enzyme
3.3.4. Binding Poses and Binding Interaction Analysis of Isolated Compounds Against EP2 Enzyme
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EP2 | Prostaglandin E2 Receptor Subtype EP2 |
| ADMET | Absorption, Distribution, Metabolism, Excretion, Toxicity. |
| PDB ID | Protein Data Bank Identifier |
| EC | Endometrial Cancer |
| PTEN | Phosphatase and TENsin Homolog |
| PIK3CA | Phosphatidylinositol-4,5-Biphosphate 3-Kinase Catalytic Subunit Alpha |
| ARID1A | AT-rich Interaction Domain-Containing Protein 1A |
| TP53 | Tumor Protein 53 |
| PI3K/AKT/Mtor | Phosphatidylinositol-3-Kinase/Protein Kinase B/Mammalian Target of Rapamycin |
| NF-Κb | Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B Cells |
| UCSF | University of California, San Franscisco |
| PDBQT | Protein Data Bank, Quaternion, Torsion |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Anakwenze, C.P.; Ewongwo, A.; Onyewadume, L.; Oyekan, A.; Chigbo, C.O.; Valle, L.; Geng, Y.; Olapade, P.; Okwunze, K.; Lasebikan, N.; et al. A systematic review of endometrial cancer clinical research in Africa. Infect. Agent. Cancer 2024, 19, 2. [Google Scholar] [CrossRef]
- Nzeribe, E.A.; Ododo, N.A.; Eteike, P.O. Profile of gynecological cancers in a tertiary hospital, Eastern Nigeria. Pan Afr. Med. J. 2023, 44, 139. [Google Scholar]
- Olatunde, O.A.; Samaila, M.O.; Imam, M.I.; Uchime, K.E.; Dauda, S.E. Histopathological patterns of endometrial carcinoma in a tertiary hospital in North-West Nigeria. Ecancermedicalscience 2024, 18, 1651. [Google Scholar] [CrossRef]
- Jabbour, H.N.; Sales, K.J. Prostaglandin receptor signalling and function in human endometrial pathology. Trends Endocrinol. Metab. 2004, 15, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Dongol, S.; Qiu, C.; Xu, Y.; Sun, C.; Zhang, Z.; Yang, X.; Zhang, Q.; Kong, B. MiR-652 promotes tumor proliferation and metastasis by targeting RORA in endometrial cancer. Mol. Cancer Res. 2018, 16, 1927–1939. [Google Scholar] [CrossRef] [PubMed]
- Finetti, F.; Travelli, C.; Ercoli, J.; Colombo, G.; Buoso, E.; Trabalzini, L. Prostaglandin E2 and Cancer: Insight into Tumor Progression and Immunity. Biology 2020, 9, 434. [Google Scholar] [CrossRef]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef]
- Bestvina, C.M.; Fleming, G.F. Chemotherapy for Endometrial Cancer in Adjuvant and Advanced Disease Settings. Oncologist 2016, 21, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Oaknin, A.; Bosse, T.J.; Creutzberg, C.L.; Giornelli, G.; Harter, P.; Joly, F.; Lorusso, D.; Marth, C.; Makker, V.; Mirza, M.R.; et al. Endometrial cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2022, 33, 860–877. [Google Scholar] [CrossRef] [PubMed]
- Eskander, R.N.; Sill, M.W.; Beffa, L.; Moore, R.G.; Hope, J.M.; Musa, F.B.; Mannel, R.; Shahin, M.S.; Cantuaria, G.H.; Girda, E.; et al. Pembrolizumab plus Chemotherapy in Advanced Endometrial Cancer. N. Engl. J. Med. 2023, 388, 2159–2170. [Google Scholar] [CrossRef]
- Afewerky, H.K.; Ayodeji, A.E.; Tiamiyu, B.B.; Orege, J.I.; Okeke, E.S.; Oyejobi, A.O.; Bate, P.N.N.; Adeyemi, S.B. Critical review of the Withania somnifera (L.) Dunal: Ethnobotany, pharmacological efficacy, and commercialization significance in Africa. Bull. Natl. Res. Cent. 2021, 45, 176. [Google Scholar] [CrossRef]
- Palliyaguru, D.L.; Singh, S.V.; Kensler, T.W. Withania somnifera: From prevention to treatment of cancer. Mol. Nutr. Food Res. 2016, 60, 1342–1353. [Google Scholar] [CrossRef]
- Choudhary, M.I.; Hussain, S.; Yousuf, S.; Dar, A.; Mudassar; Atta-Ur-Rahman. Chlorinated and diepoxy withanolides from Withania somnifera and their cytotoxic effects against human lung cancer cell line. Phytochemistry 2010, 71, 2205–2209. [Google Scholar] [CrossRef]
- Atteeq, M. Evaluating anticancer properties of Withaferin A—A potent phytochemical. Front. Pharmacol. 2022, 13, 975320. [Google Scholar] [CrossRef]
- Silva, G.W.d.S.e.; Marques, A.M.; Sampaio, A.L.F. Anticancer Effects of Withanolides: In Silico Prediction of Pharmacological Properties. Molecules 2025, 30, 2457. [Google Scholar] [CrossRef]
- Saleem, S.; Muhammad, G.; Hussain, M.A.; Altaf, M.; Abbas Bukhari, S.N. Withania somnifera L.: Insights into the phytochemical profile, therapeutic potential, clinical trials, and future prospective. Iran. J. Basic Med. Sci. 2020, 23, 1501–1526. [Google Scholar] [CrossRef] [PubMed]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W. Computational methods in drug discovery. Pharmacol. Rev. 2013, 66, 334–395. [Google Scholar] [CrossRef]
- Sanches, R.C.O.; Tiwari, S.; Ferreira, L.C.G.; Oliveira, F.M.; Lopes, M.D.; Passos, M.J.F.; Maia, E.H.B.; Taranto, A.G.; Kato, R.; Azevedo, V.A.C.; et al. Immunoinformatics Design of Multi-Epitope Peptide-Based Vaccine Against Schistosoma mansoni Using Transmembrane Proteins as a Target. Front. Immunol. 2021, 12, 621706. [Google Scholar] [CrossRef] [PubMed]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Baroroh, U.; Biotek, M.; Muscifa, Z.S.; Destiarani, W.; Rohmatullah, F.G.; Yusuf, M. Molecular interaction analysis and visualization of protein-ligand docking using Biovia Discovery Studio Visualizer. Indones. J. Comput. Biol. 2023, 2, 22–30. [Google Scholar] [CrossRef]
- ChemSpider: Search and Share Chemistry—Homepage. Available online: https://www.chemspider.com/ (accessed on 25 September 2025).
- SwissADME. Available online: http://www.swissadme.ch/index.php (accessed on 25 September 2025).
- ProTox-3.0—Prediction of TOXicity of Chemicals. Available online: https://tox.charite.de/protox3/ (accessed on 25 September 2025).
- Qu, C.; Mao, C.; Xiao, P.; Shen, Q.; Zhong, Y.; Yang, F.; Shen, D.; Tao, X.; Zhang, H.; Yan, X.; et al. Cryo-EM Structure of the PGE2-Bound EP2-Gs Complex. In Full wwPDB EM Validation Report; wwPDB: Cambridgeshire, UK, 2021. [Google Scholar] [CrossRef]
- El-Hachem, N.; Haibe-Kains, B.; Khalil, A.; Kobeissy, F.H.; Nemer, G. Chapter 20 AutoDock and AutoDockTools for Protein-Ligand Docking: Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) as a Case Study. In Neuroproteomics. Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1598. [Google Scholar] [CrossRef]
- Lipinski, C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 2000, 44, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Yano, A.; Takahashi, Y.; Moriguchi, H.; Inazumi, T.; Koga, T.; Otaka, A.; Sugimoto, Y. An aromatic amino acid within intracellular loop 2 of the prostaglandin EP2 receptor is a prerequisite for selective association and activation of Gαs. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2017, 1862, 615–622. [Google Scholar] [CrossRef] [PubMed]
 Lig 1 (withanolide D) | 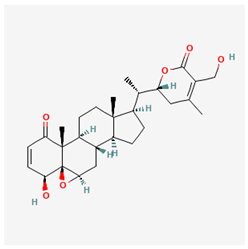 Lig 2 (withaferin A) |
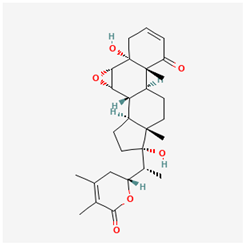 Lig 3 (withanone) | 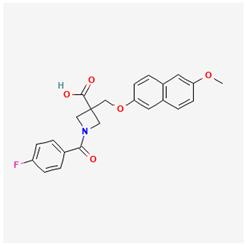 PF-04418948 |
| Properties | Lig1 | Lig2 | Lig3 |
|---|---|---|---|
| CID | 161671 | 265237 | 21679027 |
| Molecular Formula | C28H38O6 | C28H38O7 | C28H38O6 |
| Mol. Wt (g/mol) | 470.60 | 486.60 | 470.60 |
| Heavy atoms | 34 | 35 | 34 |
| Aromatic Heavy atoms | 0 | 0 | 0 |
| Fraction Csp3 | 0.79 | 0.79 | 0.79 |
| HbA | 6 | 7 | 6 |
| HbD | 2 | 3 | 2 |
| Nrb | 2 | 3 | 2 |
| MR | 127.53 | 128.69 | 127.53 |
| TPSA (Å2) | 96.36 | 116.59 | 96.36 |
| MlogP | 2.75 | 1.95 | 2.75 |
| Lipinski violation | No | No | No |
| Inference | Pass | Pass | Pass |
| Ghose violations | 1 | 2 | 1 |
| Veber violations | 0 | 0 | 0 |
| Egan violations | 0 | 0 | 0 |
| Muegge violations | 0 | 0 | 0 |
| Bioavailability score | 0.55 | 0.55 | 0.55 |
| Synthetic accessibility | 6.85 | 6.88 | 6.38 |
| Properties | Lig1 | Lig2 | Lig3 |
|---|---|---|---|
| LogS (Silicos-IT) | −3.78 | −3.20 | −4.00 |
| Silicos-class | Moderately soluble | Moderately soluble | Moderately soluble |
| Consensus Log Po/w | 3.39 | 2.64 | 3.36 |
| Log Kp (skin permeation) (cm/s) | −6.96 | −7.55 | −7.01 |
| GI Absorption | High | High | High |
| BBB Permeant | No | No | No |
| Pgp substrate | Yes | Yes | Yes |
| CYP1A2 inhibitor | No | No | No |
| CYP2C19 inhibitor | No | No | No |
| CYP2C9 inhibitor | No | No | No |
| CYP2D6 inhibitor | No | No | No |
| CYP3A4 inhibitor | No | No | No |
| Properties | Lig1 | Lig2 | Lig3 |
|---|---|---|---|
| Oral acute toxicity | Class III | Class III | Class II |
| Ames Mutagenesis | - | - | - |
| Carcinogenicity | - | - | - |
| Hepatotoxicity | - | - | - |
| Androgen receptor binding | - | - | - |
| Thyroid receptor binding | - | - | - |
| Estrogen receptor binding | - | - | - |
| Aromatase binding | - | - | - |
| Enzyme | Grid Box Size | Center | ||||
|---|---|---|---|---|---|---|
| X | Y | Z | X | Y | Z | |
| 7CX2 | 22 | 16 | 22 | 3.719 | 114.893 | 119.227 |
| Enzyme Code and Name | Crystal Structure Complex (Enzyme and Native Ligand) | Crystal Structure Complex (Enzyme, Native Ligand, and Re-Docked Ligand) (Validation) |
|---|---|---|
| 7CX2 PGE2-bound EP2-Gs complex |  | 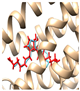 |
| Enzyme | Affinity (kcal/mol) | |||
|---|---|---|---|---|
| PF-04418948 | Lig1 | Lig2 | Lig3 | |
| 7CX2 | −9.6 | −12.6 | −12.0 | −11.3 |
| Compounds | Interaction Type | Interacting Residues |
|---|---|---|
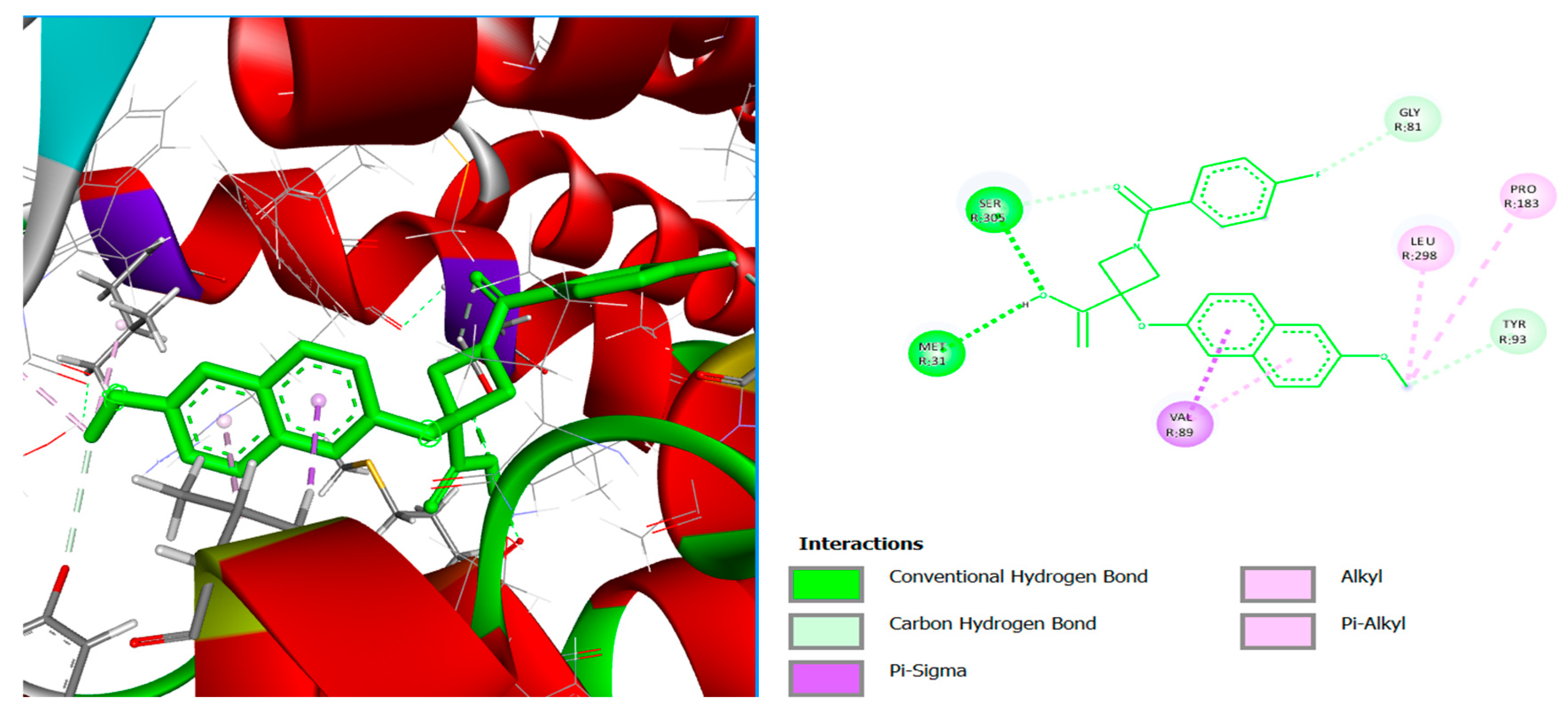
| PF-04418948 | Conventional Hydrogen Bond | SER305, MET31 |
| Alkyl | LEU298, PRO183 | |
| Pi-Alkyl | VAL89 | |
| Carbon Hydrogen Bond | TYR93, GLY81 | |
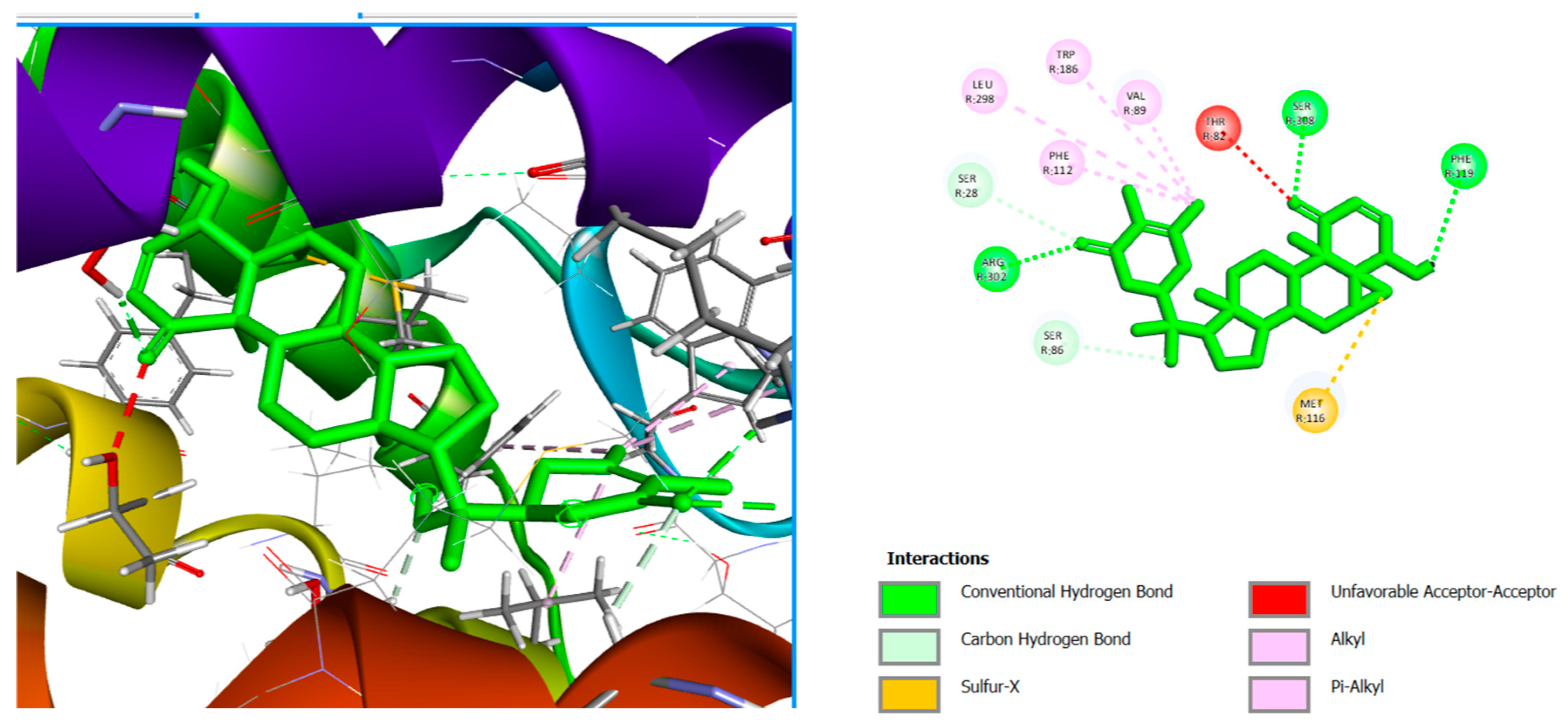
| Lig 1 | Pi-Alkyl | PHE112, TRP186 |
| Alkyl | VAL89, LEU298 | |
| Conventional Hydrogen Bond | ARG302, SER308, PHE119 | |
| Sulfur-X | MET116 | |
| Carbon Hydrogen Bond | SER28, SER86 | |
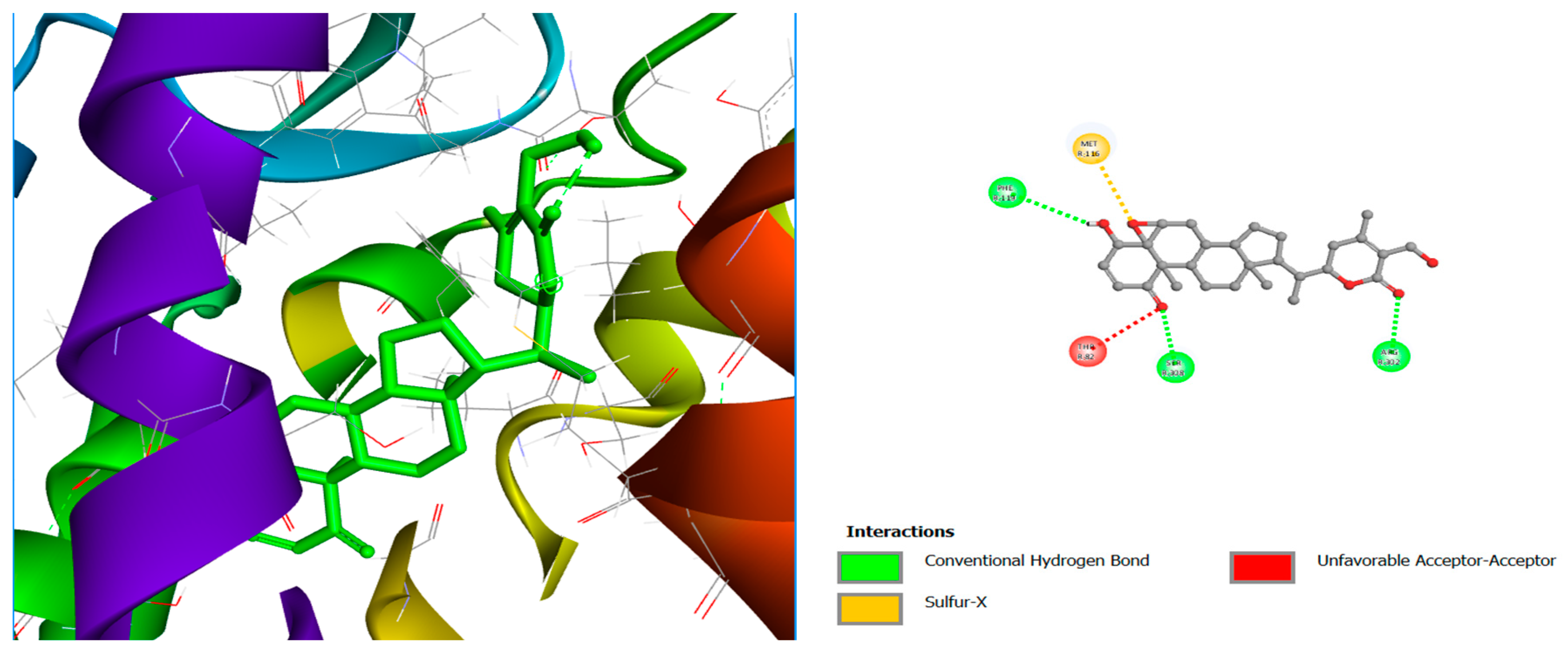
| Lig 2 | Conventional Hydrogen bond | SER305, ARG302 |
| Sulfur-X | MET31 | |
| Carbon Hydrogen Bond | SER308 | |
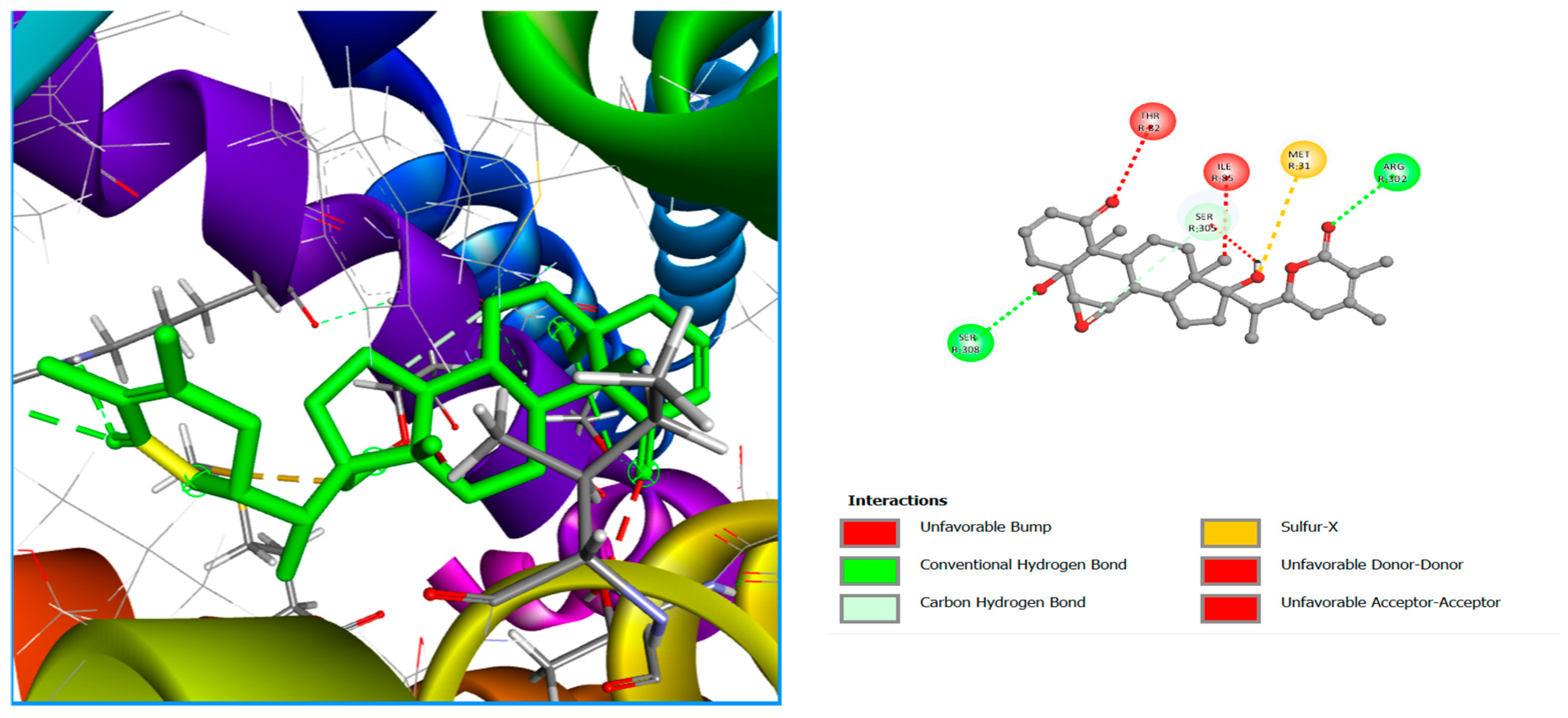
| Lig3 | Conventional hydrogen bond | ARG302, SER302 |
| Carbon Hydrogen Bond | SER305, SER308 | |
| Sulfur-X | Met31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adaji, G.; Hamza, A.N. Molecular Modelling of Withanolides Against EP2 Receptor for Treatment of Endometrial Cancer: A Pharmacokinetic and Molecular Docking Study. Chem. Proc. 2025, 18, 7. https://doi.org/10.3390/ecsoc-29-26724
Adaji G, Hamza AN. Molecular Modelling of Withanolides Against EP2 Receptor for Treatment of Endometrial Cancer: A Pharmacokinetic and Molecular Docking Study. Chemistry Proceedings. 2025; 18(1):7. https://doi.org/10.3390/ecsoc-29-26724
Chicago/Turabian StyleAdaji, Grace, and Asmau Nasir Hamza. 2025. "Molecular Modelling of Withanolides Against EP2 Receptor for Treatment of Endometrial Cancer: A Pharmacokinetic and Molecular Docking Study" Chemistry Proceedings 18, no. 1: 7. https://doi.org/10.3390/ecsoc-29-26724
APA StyleAdaji, G., & Hamza, A. N. (2025). Molecular Modelling of Withanolides Against EP2 Receptor for Treatment of Endometrial Cancer: A Pharmacokinetic and Molecular Docking Study. Chemistry Proceedings, 18(1), 7. https://doi.org/10.3390/ecsoc-29-26724






