A Comparative Assessment of Regular and Spatial Cross-Validation in Subfield Machine Learning Prediction of Maize Yield from Sentinel-2 Phenology
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Ground Truth Maize Yield Data
2.2. Sentinel-2 Time Series Data and Vegetation Indices
2.3. Phenological Modeling
2.4. Machine Learning Prediction and Accuracy Assessment Using Regular and Spatial Cross-Validation
3. Results
4. Discussion
5. Conclusions
- •
- Spatial cross-validation likely captured the effects of spatial autocorrelation, unlike regular cross-validation, thus providing a more realistic and conservative estimate of model generalizability in geospatial applications. Cross-validation results were sensitive to training/test split randomness, especially under low sample counts, reinforcing the importance of repeated validation with sufficient sample size. Therefore, ignoring spatial structure during model evaluation can lead to misleading conclusions, which may negatively affect precision agriculture decision-making.
- •
- EVI consistently outperformed WDRVI in terms of phenological model fitting accuracy and yield prediction reliability. Most notably, EVI produced notably more samples whose time series data enabled successful phenological modeling, which produced more robust yield prediction in comparison to WDRVI. Yield prediction models based on WDRVI were occasionally more accurate but were highly unstable due to low sample sizes after phenology modeling.
- •
- Among phenological fitting methods, AG, Beck, and Elmore demonstrated higher robustness due to their greater number of successfully fitted samples, making them more suitable for subfield crop yield prediction. Conversely, despite small differences in fitting accuracy, Gu, Klos, and Zhang’s methods had poor reliability due to a high failure rate in curve fitting, limiting their applicability in real-world scenarios.
- •
- Machine learning model performance varied across parcels, with RF and BGLM producing similar predictive accuracy depending on the parcel and index used.
- •
- Future studies should include longer-term and broader spatial datasets and consider integrating environmental covariates, including climate, soil, and topography data, to improve spatial model generalization.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Parcel ID | Vegetation Index | Fitting Method | Cross-Validation Method | RF Optimal Hyperparameters |
|---|---|---|---|---|
| Parcel 1 | EVI | AG | regular | mtry = 15, splitrule = “extratrees”, min.node.size = 5 |
| spatial | mtry = 19, splitrule = “extratrees”, min.node.size = 5 | |||
| Beck | regular | mtry = 13, splitrule = “extratrees”, min.node.size = 5 | ||
| spatial | mtry = 4, splitrule = “extratrees”, min.node.size = 5 | |||
| Elmore | regular | mtry = 19, splitrule = “extratrees”, min.node.size = 5 | ||
| spatial | mtry = 22, splitrule = “extratrees”, min.node.size = 5 | |||
| Gu | regular | mtry = 22, splitrule = “extratrees”, min.node.size = 5 | ||
| spatial | mtry = 13, splitrule = “extratrees”, min.node.size = 5 | |||
| Klos | regular | mtry = 22, splitrule = “extratrees”, min.node.size = 5 | ||
| spatial | mtry = 19, splitrule = “extratrees”, min.node.size = 5 | |||
| Zhang | regular | mtry = 4, splitrule = “extratrees”, min.node.size = 5 | ||
| spatial | mtry = 6, splitrule = “extratrees”, min.node.size = 5 | |||
| WDRVI | AG | regular | mtry = 6, splitrule = “extratrees”, min.node.size = 5 | |
| spatial | mtry = 10, splitrule = “extratrees”, min.node.size = 5 | |||
| Parcel 2 | EVI | AG | regular | mtry = 19, splitrule = “extratrees”, min.node.size = 5 |
| spatial | mtry = 13, splitrule = “extratrees”, min.node.size = 5 | |||
| Beck | regular | mtry = 6, splitrule = “extratrees”, min.node.size = 5 | ||
| spatial | mtry = 19, splitrule = “extratrees”, min.node.size = 5 | |||
| Elmore | regular | mtry = 22, splitrule = “extratrees”, min.node.size = 5 | ||
| spatial | mtry = 15, splitrule = “extratrees”, min.node.size = 5 | |||
| Gu | regular | mtry = 10, splitrule = “extratrees”, min.node.size = 5 | ||
| spatial | mtry = 15, splitrule = “extratrees”, min.node.size = 5 | |||
| Klos | regular | mtry = 13, splitrule = “extratrees”, min.node.size = 5 | ||
| spatial | mtry = 13, splitrule = “extratrees”, min.node.size = 5 | |||
| Zhang | regular | mtry = 13, splitrule = “extratrees”, min.node.size = 5 | ||
| spatial | mtry = 10, splitrule = “extratrees”, min.node.size = 5 | |||
| WDRVI | AG | regular | mtry = 4, splitrule = “extratrees”, min.node.size = 5 | |
| spatial | mtry = 6, splitrule = “extratrees”, min.node.size = 5 | |||
| Parcel 3 | EVI | AG | regular | mtry = 22, splitrule = “extratrees”, min.node.size = 5 |
| spatial | mtry = 8, splitrule = “extratrees”, min.node.size = 5 | |||
| Beck | regular | mtry = 15, splitrule = “extratrees”, min.node.size = 5 | ||
| spatial | mtry = 13, splitrule = “extratrees”, min.node.size = 5 | |||
| Elmore | regular | mtry = 2, splitrule = “extratrees”, min.node.size = 5 | ||
| spatial | mtry = 6, splitrule = “extratrees”, min.node.size = 5 | |||
| WDRVI | AG | regular | mtry = 2, splitrule = “extratrees”, min.node.size = 5 | |
| spatial | mtry = 2, splitrule = “extratrees”, min.node.size = 5 |
References
- Burdett, H.; Wellen, C. Statistical and Machine Learning Methods for Crop Yield Prediction in the Context of Precision Agriculture. Precis. Agric. 2022, 23, 1553–1574. [Google Scholar] [CrossRef]
- Kitchen, N.R.; Clay, S.A. Understanding and Identifying Variability. In Precision Agriculture Basics; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2018; pp. 13–24. ISBN 978-0-89118-367-9. [Google Scholar]
- Wang, N.; Wu, Q.; Gui, Y.; Hu, Q.; Li, W. Cross-Modal Segmentation Network for Winter Wheat Mapping in Complex Terrain Using Remote-Sensing Multi-Temporal Images and DEM Data. Remote Sens. 2024, 16, 1775. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, Y.; Yuan, F.; Liu, X.; Tian, Y.; Zhu, Y.; Cao, W.; Cao, Q. Optimizing Management Zone Delineation through Advanced Dimensionality Reduction Models and Clustering Algorithms. Precis. Agric. 2025, 26, 68. [Google Scholar] [CrossRef]
- McFadden, J.R.; Rosburg, A.; Njuki, E. Information Inputs and Technical Efficiency in Midwest Corn Production: Evidence from Farmers’ Use of Yield and Soil Maps. Am. J. Agric. Econ. 2022, 104, 589–612. [Google Scholar] [CrossRef]
- Radočaj, D.; Plaščak, I.; Jurišić, M. Phenology-Based Maize and Soybean Yield Potential Prediction Using Machine Learning and Sentinel-2 Imagery Time-Series. Appl. Sci. 2025, 15, 7216. [Google Scholar] [CrossRef]
- Getahun, S.; Kefale, H.; Gelaye, Y. Application of Precision Agriculture Technologies for Sustainable Crop Production and Environmental Sustainability: A Systematic Review. Sci. World J. 2024, 2024, 2126734. [Google Scholar] [CrossRef]
- Aarif KO, M.; Alam, A.; Hotak, Y. Smart Sensor Technologies Shaping the Future of Precision Agriculture: Recent Advances and Future Outlooks. J. Sens. 2025, 2025, 2460098. [Google Scholar] [CrossRef]
- Dhillon, R.; Moncur, Q. Small-Scale Farming: A Review of Challenges and Potential Opportunities Offered by Technological Advancements. Sustainability 2023, 15, 15478. [Google Scholar] [CrossRef]
- Longchamps, L.; Tisseyre, B.; Taylor, J.; Sagoo, L.; Momin, A.; Fountas, S.; Manfrini, L.; Ampatzidis, Y.; Schueller, J.K.; Khosla, R. Yield Sensing Technologies for Perennial and Annual Horticultural Crops: A Review. Precis. Agric. 2022, 23, 2407–2448. [Google Scholar] [CrossRef]
- Phiri, D.; Simwanda, M.; Salekin, S.; Nyirenda, V.R.; Murayama, Y.; Ranagalage, M. Sentinel-2 Data for Land Cover/Use Mapping: A Review. Remote Sens. 2020, 12, 2291. [Google Scholar] [CrossRef]
- Sun, H.; Ma, X.; Liu, Y.; Zhou, G.; Ding, J.; Lu, L.; Wang, T.; Yang, Q.; Shu, Q.; Zhang, F. A New Multiangle Method for Estimating Fractional Biocrust Coverage From Sentinel-2 Data in Arid Areas. IEEE Trans. Geosci. Remote Sens. 2024, 62, 4404015. [Google Scholar] [CrossRef]
- Sentinel-2 L2A—Documentation. Available online: https://documentation.dataspace.copernicus.eu/APIs/SentinelHub/Data/S2L2A.html (accessed on 3 June 2025).
- Zou, X.; Zhu, S.; Mõttus, M. Estimation of Canopy Structure of Field Crops Using Sentinel-2 Bands with Vegetation Indices and Machine Learning Algorithms. Remote Sens. 2022, 14, 2849. [Google Scholar] [CrossRef]
- Swoish, M.; Da Cunha Leme Filho, J.F.; Reiter, M.S.; Campbell, J.B.; Thomason, W.E. Comparing Satellites and Vegetation Indices for Cover Crop Biomass Estimation. Comput. Electron. Agric. 2022, 196, 106900. [Google Scholar] [CrossRef]
- Arshad, A.; Raza, M.A.; Zhang, Y.; Zhang, L.; Wang, X.; Ahmed, M.; Habib-ur-Rehman, M. Impact of Climate Warming on Cotton Growth and Yields in China and Pakistan: A Regional Perspective. Agriculture 2021, 11, 97. [Google Scholar] [CrossRef]
- Arshad, A.; Zhang, Y.; Zhang, P.; Wang, X.; Chen, Y.; Ahmed, M.; Zhang, L. APSIM-Cotton Model Calibration for Phenology-Driven Sowing and Yield Optimization in Drip Irrigated Arid Climate. Smart Agric. Technol. 2025, 12, 101325. [Google Scholar] [CrossRef]
- Diao, C.; Li, G. Near-Surface and High-Resolution Satellite Time Series for Detecting Crop Phenology. Remote Sens. 2022, 14, 1957. [Google Scholar] [CrossRef]
- Ma, Y.; Liang, S.-Z.; Myers, D.B.; Swatantran, A.; Lobell, D.B. Subfield-Level Crop Yield Mapping without Ground Truth Data: A Scale Transfer Framework. Remote Sens. Environ. 2024, 315, 114427. [Google Scholar] [CrossRef]
- Dossa, K.F.; Bissonnette, J.-F.; Barrette, N.; Bah, I.; Miassi, Y.E. Projecting Climate Change Impacts on Benin’s Cereal Production by 2050: A SARIMA and PLS-SEM Analysis of FAO Data. Climate 2025, 13, 19. [Google Scholar] [CrossRef]
- Radočaj, D.; Plaščak, I.; Jurišić, M. A Machine-Learning Approach for the Assessment of Quantitative Changes in the Tractor Diesel-Engine Oil During Exploitation. Poljoprivreda 2024, 30, 108–114. [Google Scholar] [CrossRef]
- Yates, L.A.; Aandahl, Z.; Richards, S.A.; Brook, B.W. Cross Validation for Model Selection: A Review with Examples from Ecology. Ecol. Monogr. 2023, 93, e1557. [Google Scholar] [CrossRef]
- Tziachris, P.; Nikou, M.; Aschonitis, V.; Kallioras, A.; Sachsamanoglou, K.; Fidelibus, M.D.; Tziritis, E. Spatial or Random Cross-Validation? The Effect of Resampling Methods in Predicting Groundwater Salinity with Machine Learning in Mediterranean Region. Water 2023, 15, 2278. [Google Scholar] [CrossRef]
- Koldasbayeva, D.; Tregubova, P.; Gasanov, M.; Zaytsev, A.; Petrovskaia, A.; Burnaev, E. Challenges in Data-Driven Geospatial Modeling for Environmental Research and Practice. Nat. Commun. 2024, 15, 10700. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Li, G.; Qi, Z. Integration of Remote Sensing and Machine Learning for Precision Agriculture: A Comprehensive Perspective on Applications. Agronomy 2024, 14, 1975. [Google Scholar] [CrossRef]
- Beigaitė, R.; Mechenich, M.; Žliobaitė, I. Spatial Cross-Validation for Globally Distributed Data. In Proceedings of the Discovery Science; Pascal, P., Ienco, D., Eds.; Springer Nature Switzerland: Cham, Switzerland, 2022; pp. 127–140. [Google Scholar]
- John, K.; Saurette, D.D.; Heung, B. The Problematic Case of Data Leakage: A Case for Leave-Profile-out Cross-Validation in 3-Dimensional Digital Soil Mapping. Geoderma 2025, 455, 117223. [Google Scholar] [CrossRef]
- Kattenborn, T.; Schiefer, F.; Frey, J.; Feilhauer, H.; Mahecha, M.D.; Dormann, C.F. Spatially Autocorrelated Training and Validation Samples Inflate Performance Assessment of Convolutional Neural Networks. ISPRS Open J. Photogramm. Remote Sens. 2022, 5, 100018. [Google Scholar] [CrossRef]
- Roberts, D.R.; Bahn, V.; Ciuti, S.; Boyce, M.S.; Elith, J.; Guillera-Arroita, G.; Hauenstein, S.; Lahoz-Monfort, J.J.; Schröder, B.; Thuiller, W.; et al. Cross-Validation Strategies for Data with Temporal, Spatial, Hierarchical, or Phylogenetic Structure. Ecography 2017, 40, 913–929. [Google Scholar] [CrossRef]
- Wang, Y.; Khodadadzadeh, M.; Zurita-Milla, R. Spatial+: A New Cross-Validation Method to Evaluate Geospatial Machine Learning Models. Int. J. Appl. Earth Obs. Geoinf. 2023, 121, 103364. [Google Scholar] [CrossRef]
- Meyer, H.; Pebesma, E. Predicting into Unknown Space? Estimating the Area of Applicability of Spatial Prediction Models. Methods Ecol. Evol. 2021, 12, 1620–1633. [Google Scholar] [CrossRef]
- Radočaj, D.; Jurišić, M. A Phenology-Based Evaluation of the Optimal Proxy for Cropland Suitability Based on Crop Yield Correlations from Sentinel-2 Image Time-Series. Agriculture 2025, 15, 859. [Google Scholar] [CrossRef]
- Banaj, A.; Banaj, Đ.; Stipešević, B.; Horvat, D. The Impact of Planting Technology on the Maize Yield. Poljoprivreda 2024, 30, 100–107. [Google Scholar] [CrossRef]
- CLAAS Connect|CLAAS. Available online: https://www.claas.com/en-us/smart-farming/claas-connect (accessed on 24 July 2025).
- Paccioretti, P.; Córdoba, M.; Giannini-Kurina, F.; Balzarini, M. Paar: Precision Agriculture Data Analysis. Available online: https://cran.r-project.org/web/packages/paar/index.html (accessed on 24 July 2025).
- Paccioretti, P.; Córdoba, M.; Balzarini, M. FastMapping: Software to Create Field Maps and Identify Management Zones in Precision Agriculture. Comput. Electron. Agric. 2020, 175, 105556. [Google Scholar] [CrossRef]
- Radočaj, D.; Plaščak, I.; Jurišić, M. Fusion of Sentinel-2 Phenology Metrics and Saturation-Resistant Vegetation Indices for Improved Correlation with Maize Yield Maps. Agronomy 2025, 15, 1329. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the Radiometric and Biophysical Performance of the MODIS Vegetation Indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Gitelson, A.A. Wide Dynamic Range Vegetation Index for Remote Quantification of Biophysical Characteristics of Vegetation. J. Plant Physiol. 2004, 161, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Xiao, M.; Zhang, Y.; Gu, X.; Cui, J. Phenofit: Extract Remote Sensing Vegetation Phenology. Available online: https://cran.r-project.org/web/packages/phenofit/index.html (accessed on 4 June 2025).
- Kong, D.; Zhang, Y.; Wang, D.; Chen, J.; Gu, X. Photoperiod Explains the Asynchronization Between Vegetation Carbon Phenology and Vegetation Greenness Phenology. J. Geophys. Res. Biogeosciences 2020, 125, e2020JG005636. [Google Scholar] [CrossRef]
- Rapčan, I.; Radočaj, D.; Jurišić, M. A Length-of-Season Analysis for Maize Cultivation from the Land- Surface Phenology Metrics Using the Sentinel-2 Images. Poljoprivreda 2025, 31, 92–98. [Google Scholar] [CrossRef]
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; R Core Team; et al. Caret: Classification and Regression Training. Available online: https://cran.r-project.org/web/packages/caret/index.html (accessed on 17 July 2025).
- Wright, M.N.; Wager, S.; Probst, P. Ranger: A Fast Implementation of Random Forests. Available online: https://cran.r-project.org/web/packages/ranger/index.html (accessed on 4 June 2025).
- Gelman, A.; Su, Y.-S.; Yajima, M.; Hill, J.; Pittau, M.G.; Kerman, J.; Zheng, T.; Dorie, V. Arm: Data Analysis Using Regression and Multilevel/Hierarchical Models. Available online: https://cran.r-project.org/web/packages/arm/index.html (accessed on 28 July 2025).
- Meyer, H.; Ludwig, M.; Milà, C.; Linnenbrink, J.; Schumacher, F. The CAST Package for Training and Assessment of Spatial Prediction Models in R. arXiv 2024, arXiv:2404.06978. [Google Scholar] [CrossRef]
- Sáenz, C.; Cicuéndez, V.; García, G.; Madruga, D.; Recuero, L.; Bermejo-Saiz, A.; Litago, J.; de la Calle, I.; Palacios-Orueta, A. New Insights on the Information Content of the Normalized Difference Vegetation Index Sentinel-2 Time Series for Assessing Vegetation Dynamics. Remote Sens. 2024, 16, 2980. [Google Scholar] [CrossRef]
- POLJ-2023-2-6 Area and Production of Cereals and Other Crops in 2023—Provisional Data|State Bureau of Statistics. Available online: https://podaci.dzs.hr/2023/hr/58457 (accessed on 25 July 2025).
- Mo, Y.; Zhang, X.; Liu, Z.; Zhang, J.; Hao, F.; Fu, Y. Effects of Climate Extremes on Spring Phenology of Temperate Vegetation in China. Remote Sens. 2023, 15, 686. [Google Scholar] [CrossRef]
- Zhang, D.; Hou, L.; Lv, L.; Qi, H.; Sun, H.; Zhang, X.; Li, S.; Min, J.; Liu, Y.; Tang, Y.; et al. Precision Agriculture: Temporal and Spatial Modeling of Wheat Canopy Spectral Characteristics. Agriculture 2025, 15, 326. [Google Scholar] [CrossRef]
- Croci, M.; Ragazzi, M.; Grassi, A.; Impollonia, G.; Amaducci, S. Assessing the Temporal Transferability of Machine Learning Models for Predicting Processing Pea Yield and Quality Using Sentinel-2 and ERA5-Land Data. Smart Agric. Technol. 2025, 12, 101207. [Google Scholar] [CrossRef]
- Perich, G.; Turkoglu, M.O.; Graf, L.V.; Wegner, J.D.; Aasen, H.; Walter, A.; Liebisch, F. Pixel-Based Yield Mapping and Prediction from Sentinel-2 Using Spectral Indices and Neural Networks. Field Crops Res. 2023, 292, 108824. [Google Scholar] [CrossRef]
- Fernando, H.; Ha, T.; Nketia, K.A.; Attanayake, A.; Shirtliffe, S. Machine Learning Approach for Satellite-Based Subfield Canola Yield Prediction Using Floral Phenology Metrics and Soil Parameters. Precis. Agric. 2024, 25, 1386–1403. [Google Scholar] [CrossRef]
- Crusiol, L.G.T.; Sun, L.; Sibaldelli, R.N.R.; Junior, V.F.; Furlaneti, W.X.; Chen, R.; Sun, Z.; Wuyun, D.; Chen, Z.; Nanni, M.R.; et al. Strategies for Monitoring Within-Field Soybean Yield Using Sentinel-2 Vis-NIR-SWIR Spectral Bands and Machine Learning Regression Methods. Precis. Agric. 2022, 23, 1093–1123. [Google Scholar] [CrossRef]
- An, C.; Park, Y.W.; Ahn, S.S.; Han, K.; Kim, H.; Lee, S.-K. Radiomics Machine Learning Study with a Small Sample Size: Single Random Training-Test Set Split May Lead to Unreliable Results. PLoS ONE 2021, 16, e0256152. [Google Scholar] [CrossRef] [PubMed]
- Darra, N.; Anastasiou, E.; Kriezi, O.; Lazarou, E.; Kalivas, D.; Fountas, S. Can Yield Prediction Be Fully Digitilized? A Systematic Review. Agronomy 2023, 13, 2441. [Google Scholar] [CrossRef]
- Mancini, A.; Solfanelli, F.; Coviello, L.; Martini, F.M.; Mandolesi, S.; Zanoli, R. Time Series from Sentinel-2 for Organic Durum Wheat Yield Prediction Using Functional Data Analysis and Deep Learning. Agronomy 2024, 14, 109. [Google Scholar] [CrossRef]
- Shi, Q.; Dai, W.; Santerre, R.; Liu, N. A Modified Spatiotemporal Mixed-Effects Model for Interpolating Missing Values in Spatiotemporal Observation Data Series. Math. Probl. Eng. 2020, 2020, 1070831. [Google Scholar] [CrossRef]
- Hounkpatin, K.O.L.; Stendahl, J.; Lundblad, M.; Karltun, E. Predicting the Spatial Distribution of Soil Organic Carbon Stock in Swedish Forests Using a Group of Covariates and Site-Specific Data. Soil 2021, 7, 377–398. [Google Scholar] [CrossRef]
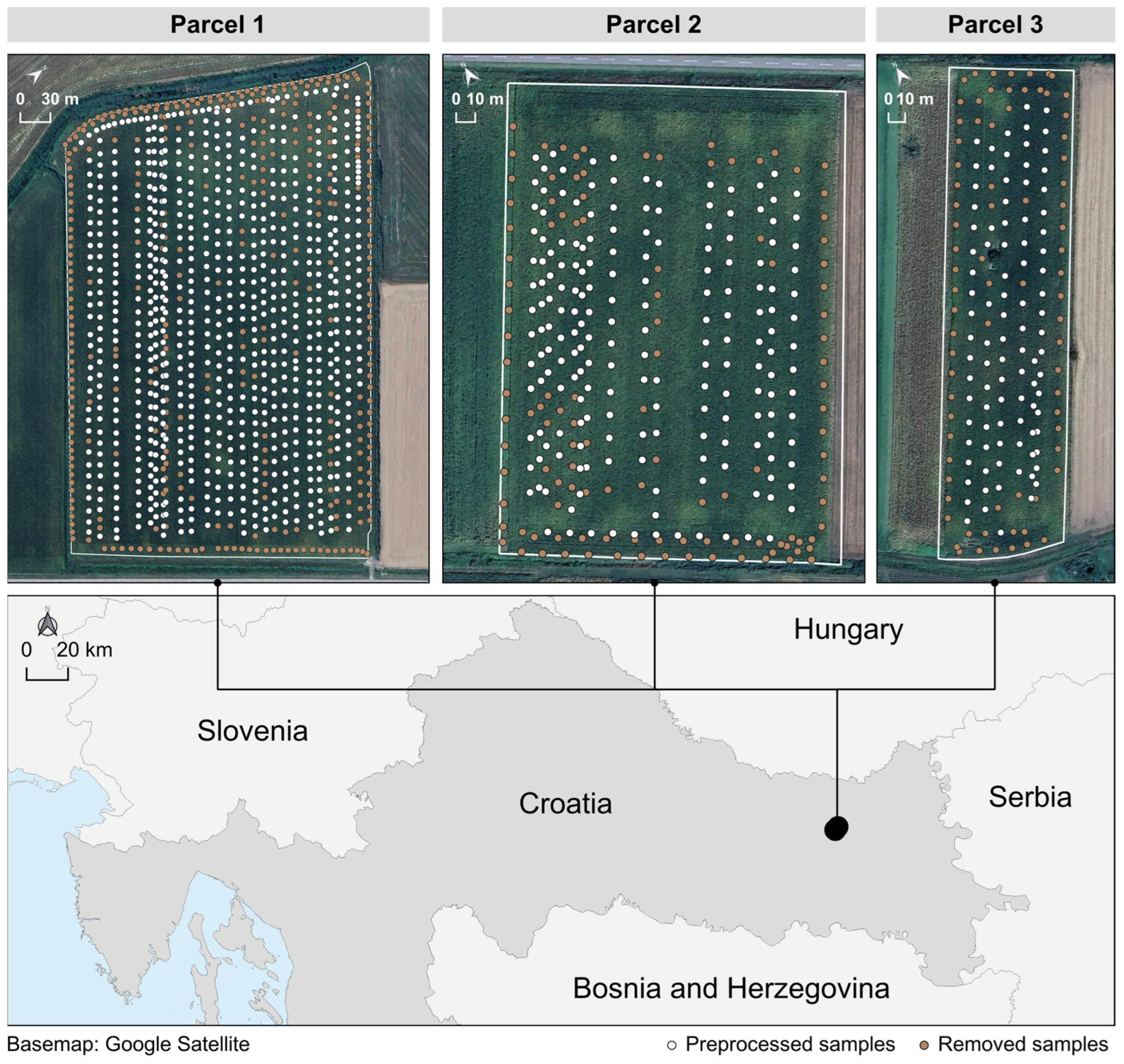

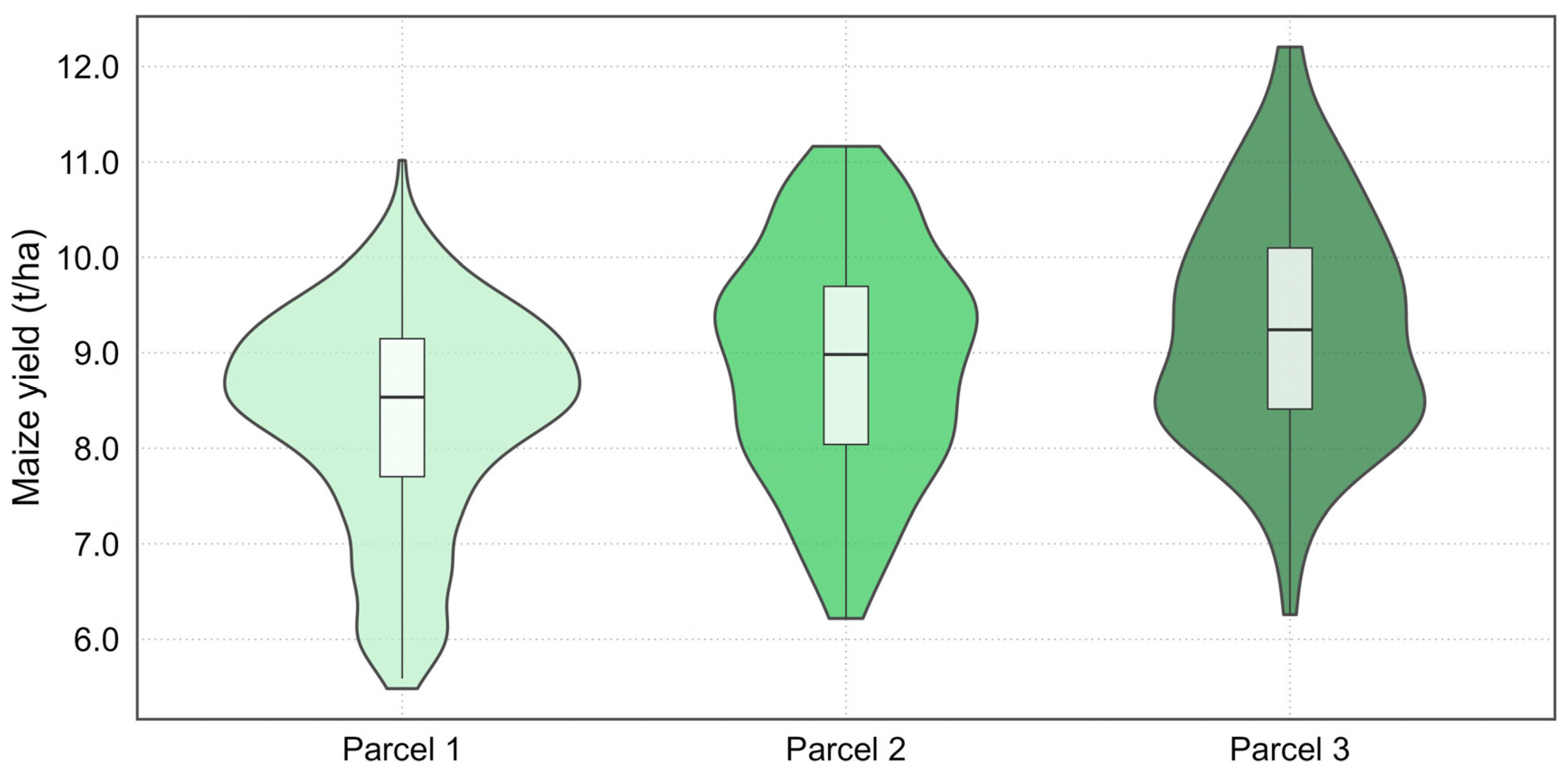

| Parcel ID | Area | Preprocessing Stage | Sample Count | Mean | Median | CV | Shapiro–Wilk Test p-Value |
|---|---|---|---|---|---|---|---|
| Parcel 1 | 11.4 ha | Before preprocessing | 1094 | 7.69 | 8.16 | 0.242 | <0.0001 |
| After preprocessing | 749 | 8.34 | 8.54 | 0.135 | <0.0001 | ||
| Parcel 2 | 4.4 ha | Before preprocessing | 302 | 8.25 | 8.44 | 0.229 | <0.0001 |
| After preprocessing | 188 | 8.91 | 8.98 | 0.134 | 0.0217 | ||
| Parcel 3 | 2.2 ha | Before preprocessing | 170 | 8.20 | 8.46 | 0.275 | <0.0001 |
| After preprocessing | 94 | 9.27 | 9.24 | 0.129 | 0.4941 |
| Parcel ID | Fitting Method | EVI | WDRVI | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSE | NSE | nfit | R2 | RMSE | NSE | nfit | ||
| Parcel 1 | AG | 0.924 | 0.066 | 0.917 | 610 | 0.251 | 0.500 | −0.941 | 35 |
| Beck | 0.922 | 0.067 | 0.916 | 610 | 0.266 | 0.500 | −0.939 | 35 | |
| Elmore | 0.921 | 0.067 | 0.915 | 610 | 0.265 | 0.500 | −0.941 | 35 | |
| Gu | 0.928 | 0.064 | 0.922 | 259 | 0.160 | 0.488 | −1.071 | 3 | |
| Klos | 0.928 | 0.064 | 0.923 | 259 | 0.160 | 0.490 | −1.087 | 3 | |
| Zhang | 0.928 | 0.064 | 0.922 | 259 | 0.212 | 0.489 | −1.078 | 3 | |
| Parcel 2 | AG | 0.603 | 0.284 | 0.582 | 146 | 0.258 | 0.491 | −0.755 | 65 |
| Beck | 0.599 | 0.284 | 0.582 | 146 | 0.284 | 0.491 | −0.749 | 65 | |
| Elmore | 0.600 | 0.284 | 0.583 | 146 | 0.285 | 0.491 | −0.755 | 65 | |
| Gu | 0.577 | 0.300 | 0.558 | 55 | 0.252 | 0.476 | −0.700 | 10 | |
| Klos | 0.583 | 0.299 | 0.564 | 55 | 0.245 | 0.476 | −0.702 | 10 | |
| Zhang | 0.577 | 0.300 | 0.559 | 55 | 0.265 | 0.477 | −0.706 | 10 | |
| Parcel 3 | AG | 0.941 | 0.064 | 0.927 | 71 | 0.221 | 0.474 | −0.927 | 28 |
| Beck | 0.940 | 0.062 | 0.935 | 71 | 0.230 | 0.474 | −0.928 | 28 | |
| Elmore | 0.939 | 0.062 | 0.935 | 71 | 0.237 | 0.473 | −0.921 | 28 | |
| Gu | 0.938 | 0.064 | 0.934 | 14 | 0.203 | 0.473 | −0.824 | 11 | |
| Klos | 0.941 | 0.062 | 0.937 | 14 | 0.275 | 0.472 | −0.817 | 11 | |
| Zhang | 0.933 | 0.068 | 0.925 | 14 | 0.237 | 0.474 | −0.836 | 11 | |
| Parcel ID | Vegetation Index | Fitting Method | Cross-Validation Method | RF | BGLM | n | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSE | MAE | R2 | RMSE | MAE | |||||
| Parcel 1 | EVI | AG | regular | 0.569 | 0.754 | 0.578 | 0.449 | 0.863 | 0.654 | 610 |
| spatial | 0.134 | 0.902 | 0.720 | 0.149 | 0.916 | 0.706 | 610 | |||
| Beck | regular | 0.571 | 0.749 | 0.564 | 0.457 | 0.841 | 0.651 | 608 | ||
| spatial | 0.162 | 0.875 | 0.698 | 0.148 | 0.872 | 0.696 | 608 | |||
| Elmore | regular | 0.505 | 0.801 | 0.611 | 0.329 | 0.932 | 0.740 | 606 | ||
| spatial | 0.141 | 0.950 | 0.764 | 0.117 | 0.997 | 0.817 | 606 | |||
| Gu | regular | 0.568 | 0.776 | 0.588 | 0.471 | 0.862 | 0.671 | 259 | ||
| spatial | 0.172 | 0.920 | 0.762 | 0.250 | 0.943 | 0.776 | 259 | |||
| Klos | regular | 0.498 | 0.839 | 0.642 | 0.360 | 0.964 | 0.724 | 258 | ||
| spatial | 0.131 | 1.042 | 0.851 | 0.197 | 1.062 | 0.844 | 258 | |||
| Zhang | regular | 0.576 | 0.772 | 0.582 | 0.450 | 0.884 | 0.683 | 259 | ||
| spatial | 0.195 | 0.923 | 0.747 | 0.198 | 0.954 | 0.772 | 259 | |||
| WDRVI | AG | regular | 0.461 | 0.433 | 0.387 | 0.551 | 0.596 | 0.528 | 34 | |
| spatial | 0.588 | 0.449 | 0.393 | 0.624 | 0.736 | 0.643 | 34 | |||
| Parcel 2 | EVI | AG | regular | 0.309 | 1.023 | 0.823 | 0.362 | 1.008 | 0.800 | 146 |
| spatial | 0.175 | 1.069 | 0.885 | 0.179 | 1.025 | 0.825 | 146 | |||
| Beck | regular | 0.354 | 0.987 | 0.781 | 0.360 | 1.009 | 0.814 | 144 | ||
| spatial | 0.160 | 1.035 | 0.844 | 0.173 | 1.013 | 0.833 | 144 | |||
| Elmore | regular | 0.357 | 0.992 | 0.796 | 0.238 | 1.111 | 0.894 | 146 | ||
| spatial | 0.251 | 1.025 | 0.845 | 0.200 | 1.197 | 0.988 | 146 | |||
| Gu | regular | 0.677 | 0.897 | 0.767 | 0.700 | 0.837 | 0.724 | 55 | ||
| spatial | 0.216 | 0.977 | 0.854 | 0.332 | 0.894 | 0.783 | 55 | |||
| Klos | regular | 0.751 | 0.754 | 0.643 | 0.640 | 0.915 | 0.791 | 55 | ||
| spatial | 0.356 | 0.833 | 0.737 | 0.329 | 1.041 | 0.878 | 55 | |||
| Zhang | regular | 0.656 | 0.904 | 0.750 | 0.679 | 0.869 | 0.741 | 55 | ||
| spatial | 0.180 | 1.087 | 0.937 | 0.319 | 1.078 | 0.912 | 55 | |||
| WDRVI | AG | regular | 0.277 | 0.727 | 0.604 | 0.235 | 1.234 | 0.882 | 64 | |
| spatial | 0.240 | 0.740 | 0.647 | 0.203 | 1.267 | 0.900 | 64 | |||
| Parcel 3 | EVI | AG | regular | 0.336 | 0.953 | 0.760 | 0.436 | 0.899 | 0.708 | 71 |
| spatial | 0.150 | 1.135 | 0.937 | 0.282 | 1.000 | 0.810 | 71 | |||
| Beck | regular | 0.397 | 0.925 | 0.749 | 0.395 | 1.004 | 0.806 | 71 | ||
| spatial | 0.195 | 1.062 | 0.888 | 0.285 | 1.081 | 0.890 | 71 | |||
| Elmore | regular | 0.298 | 1.043 | 0.842 | 0.237 | 1.135 | 0.896 | 71 | ||
| spatial | 0.203 | 1.194 | 1.017 | 0.194 | 1.262 | 1.047 | 71 | |||
| WDRVI | AG | regular | 0.594 | 1.044 | 0.865 | 0.598 | 1.650 | 1.357 | 28 | |
| spatial | 0.725 | 1.139 | 1.009 | 0.720 | 1.922 | 1.714 | 28 | |||
| Test Statistics | RF | BGLM | ||||
|---|---|---|---|---|---|---|
| R2 | RMSE | MAE | R2 | RMSE | MAE | |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radočaj, D.; Plaščak, I.; Jurišić, M. A Comparative Assessment of Regular and Spatial Cross-Validation in Subfield Machine Learning Prediction of Maize Yield from Sentinel-2 Phenology. Eng 2025, 6, 270. https://doi.org/10.3390/eng6100270
Radočaj D, Plaščak I, Jurišić M. A Comparative Assessment of Regular and Spatial Cross-Validation in Subfield Machine Learning Prediction of Maize Yield from Sentinel-2 Phenology. Eng. 2025; 6(10):270. https://doi.org/10.3390/eng6100270
Chicago/Turabian StyleRadočaj, Dorijan, Ivan Plaščak, and Mladen Jurišić. 2025. "A Comparative Assessment of Regular and Spatial Cross-Validation in Subfield Machine Learning Prediction of Maize Yield from Sentinel-2 Phenology" Eng 6, no. 10: 270. https://doi.org/10.3390/eng6100270
APA StyleRadočaj, D., Plaščak, I., & Jurišić, M. (2025). A Comparative Assessment of Regular and Spatial Cross-Validation in Subfield Machine Learning Prediction of Maize Yield from Sentinel-2 Phenology. Eng, 6(10), 270. https://doi.org/10.3390/eng6100270








