Abstract
Radiomics has shown great promise in predicting various diseases. Researchers have previously attempted to include radiomics in their automated detection, diagnosis, and segmentation algorithms, taking these steps based on the promising outcomes of radiomics-based studies. As a result of the increased attention given to this topic, numerous institutions have developed their own radiomics software. These packages, on the other hand, have been utilized interchangeably without regard for their fundamental differences. The primary purpose of this study was to explore benefits of predictive model performance for radiation pneumonitis (RP), which is the most frequent side effect of chest radiotherapy, and through this work, we developed a radiomics model based on deep learning that intends to increase RP prediction performance by combining more data points and digging deeper into these data. In order to evaluate the most popular machine learning models, radiographic characteristics were used, and we recorded the most important of them. The high dimensionality of radiomic datasets is a major issue. The method proposed for use in data problems is the synthetic minority oversampling technique, which we used in order to create a balanced dataset by leveraging suitable hardware and open-source software. The present study assessed the efficacy of various machine learning models, including logistic regression (LR), support vector machine (SVM), random forest (RF), and deep neural network (DNN), in predicting radiation pneumonitis by utilizing specific radiomics features. The findings of the study indicate that the four models displayed satisfactory efficacy in forecasting radiation pneumonitis. The DNN model demonstrated the highest area under the receiver operating curve (AUC-ROC) value, which was 0.87, suggesting its superior predictive capacity among the models considered. The AUC-ROC values for the random forest, SVM, and logistic regression models were 0.85, 0.83, and 0.81, respectively.
1. Introduction
Radiation pneumonitis is a common complication in patients undergoing thoracic radiation therapy for lung cancer. Early detection and prediction of this condition can significantly improve patient outcomes by enabling timely interventions. Radiomics, which is a rapidly growing field in medical imaging, has the potential to provide objective and quantitative measurements of imaging biomarkers that can aid in the diagnosis, treatment planning, and prognosis prediction of radiation pneumonitis. This study coined the term “Automation Radiomics” to describe the automated analysis and prediction of radiomic features using advanced computational techniques. In the context of our research, “Automation Radiomics” refers to the use of algorithmic and machine learning techniques to extract, analyze, and predict radiomic features from medical images. This term accentuates the use of advanced software tools and computational methods to streamline the workflow of analysis, reduce manual intervention, and improve the efficiency of radiomic analysis. Our previous study found that underlying gene-expression patterns were linked to a predictive radiomic signature that captured intra-tumor heterogeneity [1]. These findings imply that radiomics can detect a broad prognostic characteristic in radiation pneumonitis: inflammation of the lung caused by radiation therapy to the chest. In order to “safely” irradiate the target tumor without increasing the danger of RP, a precise prognosis is required. As imaging is widely utilized in clinical practice, this approach could have a clinical impact, providing a rare opportunity to improve decision-support practices in radiation pneumonitis treatment at a low cost. Radiomics involves the extraction of high-dimensional quantitative features from medical images, which can be used as input data for machine learning models. In this study, we analyze medical images and predict radiation pneumonitis using deep learning (DL)-based techniques. It is important to note that our DL model does not directly analyze the images’ raw pixel data. Instead, we employ a feature-based approach in which a set of pre-defined image features are extracted. These characteristics capture pertinent image information and characteristics, including texture, shape, and intensity patterns. Rather than explicitly processing the pixel-level information, our DL model learns to make predictions based on the extracted representations. This method permits us to leverage the power of DL while preserving interpretability and minimizing computational complexity. Notably, the selection and extraction of these features were meticulously devised based on domain expertise and previous research in the field. This distinction ensures that our DL method concentrates on the most informative aspects of the images and enables accurate prediction of radiation pneumonitis. Figure 1 shows the workflow of this study.
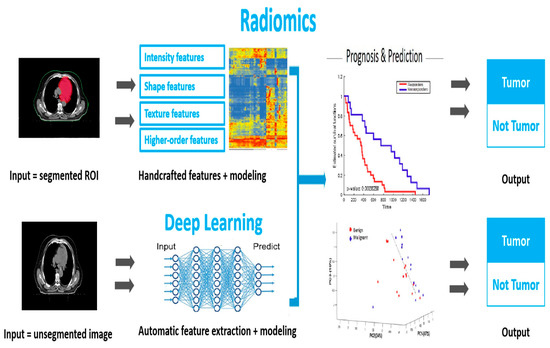
Figure 1.
The workflow of radiomics analysis in early tumor diagnosis. The DL model is based on features extracted from the medical images, which capture key characteristics, such as texture, shape, and intensity patterns.
In recent years, there has been a growing interest in using radiomics-based approaches in radiation pneumonitis prediction [2]. However, there are still challenges in standardizing image acquisition, processing, and analysis protocols, as well as in developing accurate and robust machine learning models [3]. Radiation therapy is a treatment modality widely used to treat cancer patients, and it has been shown to be effective in controlling and even eradicating some types of tumors. However, radiation therapy is associated with several side effects, and one of the most significant side effects is radiation pneumonitis [4]. In this research paper, we investigate and evaluate the current state of the art in radiomics-based approaches to radiation pneumonitis prediction. We provide an overview of the key image processing techniques, feature extraction, selection methods, and machine learning algorithms used in radiomics. We also discuss the challenges and future directions of radiomics in radiation pneumonitis prediction, with the aim of providing insights into the potential clinical impact of this emerging field. While RP can be managed via appropriate treatment, predicting which patients are at highest risk of developing this condition remains a significant challenge. Clinical factors, such as radiation dose, tumor location, and patient age, have been used to predict RP, though their accuracy is limited [5]. Automation radiomics is a rapidly evolving field that has the potential to improve our ability to predict RP. By extracting and analyzing large amounts of quantitative data from medical images, radiomics can identify features that are not visible to the naked eye and may be correlated with a patient’s risk of developing this complication. Several studies have explored the use of radiomics in predicting RP, with promising results [6]. However, further research is needed to validate these findings and determine the optimal methods for integrating radiomics into clinical practice.
The purpose of this study is to investigate the use of radiomics in predicting radiation pneumonitis in a cohort of patients who are undergoing radiation therapy for cancer. We will explore the potential of radiomics to identify imaging features that are predictive of RP and develop a model that can accurately predict a patient’s risk of developing this condition.
2. Materials and Methods
In this paper, we intend to provide a comprehensive analysis of the current state of radiomics-based approaches that predict radiation pneumonitis. We conducted a comprehensive review of the literature to synthesize existing knowledge and identify knowledge deficits in the field. In addition, we present our own research findings, which contribute to the field’s development. The literature review conducted for this paper served as the foundation for our research. It allowed us to gain a deeper understanding of the existing approaches, challenges, and advancements in the field of radiomics-based prediction of radiation pneumonitis. This knowledge guided our research design, including the selection of appropriate methodologies and the formulation of research questions that address current gaps in the literature. To achieve automation in our analysis, we employed a combination of computational algorithms, machine learning models, and automated feature extraction techniques. This automated approach eliminated the need for manual feature selection and reduced the risk of bias or human error in the analysis.
Study Design: We conducted a retrospective study to evaluate the feasibility of using radiomics-based analyses to predict radiation pneumonitis in lung cancer patients who received radiation therapy. All patient data, whether public and private, were anonymized before analysis.
Data Collection: We collected imaging and clinical data from 80 lung cancer patients who received radiation therapy between January 2020 and December 2022 at medical centers that collaborated with our laboratory, as well as 22 lung cancer patients from a public database [7,8]. Inclusion criteria were as follows: (1) a histologically confirmed diagnosis of lung cancer, (2) receipt of radiation therapy as the primary treatment, (3) available pre-treatment CT images, and (4) had at least 6 months of follow-up treatment after radiation therapy. The CT scans were obtained at a resolution of 1 mm × 1 mm × 3 mm, a reconstruction kernel of B50f [9], and an energy level of 120 kVp. Exclusion criteria were as follows: (1) incomplete imaging or clinical data, (2) history of lung disease other than lung cancer, and (3) previous radiation therapy applied to the thorax. SMOTE (synthetic minority over-sampling technique) is a data augmentation technique that is widely used in machine learning and can be applied to address the issue of class imbalance in datasets [10]. The synthetic minority over-sampling technique (SMOTE) was utilized to tackle the problem of imbalanced class distribution present in the dataset. The synthetic minority over-sampling technique (SMOTE) can also generate artificial instances of the under-represented class. The method operates through the process of interpolation of novel instances along the line segments that connect adjacent samples belonging to the minority class. This approach results in an augmented representation of the minority class, thereby mitigating the issue of class imbalance. The objective of our study was to enhance the predictive efficacy of our models through the implementation of the synthetic minority over-sampling technique (SMOTE), which facilitated the provision of a more equitable training dataset. In this study, SMOTE was used to balance the class distribution of the lung cancer dataset. The lung cancer dataset consisted of imaging and clinical data collected from patients who received radiation therapy for lung cancer. The inclusion criteria ensured that the dataset consisted only of patients with histologically confirmed diagnoses of lung cancer who received radiation therapy as their primary treatment, had available pre-treatment CT images, and had at least 6 months of follow-up after radiation therapy. The exclusion criteria ensured that the dataset did not contain incomplete imaging or clinical data. Despite the rigorous inclusion and exclusion criteria, the class distribution of the lung cancer dataset was imbalanced, with a significantly larger number of patients located in the majority class than the minority class. To address this imbalance, SMOTE was applied to generate synthetic examples of the minority class, thereby balancing the class distribution and improving the performance of machine learning algorithms that were trained on the dataset of patients who received radiation therapy for lung cancer and met strict inclusion and exclusion criteria. To apply SMOTE to the image data, we first transformed each image into a feature vector representation using a convolutional neural network that had been previously trained. The feature vector comprised the image’s extracted high-level features. Next, we implemented the SMOTE algorithm, which was designed specifically for image data. This approach required selecting an image from a minority class and identifying its k closest companions in the feature space. Next, synthetic images were created by randomly selecting one or several of these neighbors and linearly interpolating their features with the original image’s features. Multiple random points along the line segment connecting the original image and the selected neighbor(s) were interpolated. By repeating this process for each image of a minority class, we generated synthetic samples that closely resembled the original minority class’s characteristics. This method enhanced the minority class’s representation in the dataset and reduced the class disparity. It is essential to note that the spatial structure and integrity of the images were preserved during this process. The movement or interpolation of features was restricted to the feature space representation of the images, ensuring that the synthetic images generated retained the visual properties and structural patterns of the original images.
Image Processing: We used Scikit-image, which is a Python package for image processing, to segment the lung regions on the pre-treatment CT images [11]. The segmented images were then processed using various image processing techniques, including intensity normalization, voxel resampling, and texture feature extraction. Intensity normalization was used to reduce the variability in image intensities between different scans. This step was important because the same tissue could have different intensities due to variations in the imaging parameters, which could affect the feature extraction process. It provided a variety of tools and algorithms for image segmentation, filtering, feature extraction, etc. It is also open source, with a large community of users, and is well documented. Scikit-image can be used in a wide range of applications, such as scientific research and industrial automation. It provides several methods for intensity normalization, such as rescaling and equalization. Voxel resampling was used to standardize the resolution and voxel size of the images, because the voxel size could vary between different scans, which could affect the accuracy of the feature extraction process. Texture feature extraction was used to extract quantitative measurements that described the spatial distribution of gray-level intensities in the image. Moreover, it provided several methods for texture feature extraction, such as GLCM, GLRLM, and Laws texture energy measures, which were utilized as texture-based features to extract further information pertaining to the spatial distribution and patterns within the medical images. The GLCM, which is also known as the grey-level co-occurrence matrix, is a prevalent methodology utilized to depict the spatial correlations among dual-pixel intensities. The GLCM method offered valuable insights into the texture properties of an image, including homogeneity, contrast, and entropy, by measuring the frequency of co-occurring gray-level pairs across various spatial distances and angles. The GLRLM method examined the occurrence and extent of sequential pixels with identical gray-level values in various orientations. The approach based on matrices was capable of capturing textural information that pertained to the smoothness, coarseness, and complexity of an image. Furthermore, we employed the Laws texture energy metric, which entailed convolving the image with pre-determined filter masks that were influenced by human perception. The aforementioned filters were capable of extracting diverse spatial frequency components, including, but not limited to, edges, spots, and waves. This approach results in a representation of the image’s texture characteristics that encompasses multiple scales. Upon comparing the aforementioned feature extraction techniques, it was observed that GLCM effectively captured the statistical interdependencies that existed between pixel intensities. This technique was particularly well-suited to the analysis of intricate textures and spatial patterns. In contrast, GLRLM was centered on the acquisition of the run length distribution and could proficiently depict textures that possessed clear linear structures or roughness. The utilization of Laws texture energy measure demonstrated proficiency in capturing textural variations at multiple scales, thereby offering a more comprehensive representation of the texture properties of an image. The objective of our study was to enhance the discriminative power of classification models and improve the prediction performance for radiation pneumonitis by utilizing various texture-based feature extraction techniques. These techniques were employed to capture complementary aspects of texture information from medical images. The pipeline could be implemented in addition to scikit and other Python libraries, such as pandas.
Feature Selection: We used a combination of statistical methods and machine learning algorithms to select the features most relevant to predicting radiation pneumonitis. A comprehensive set of radiomics features was extracted from the medical images in order to capture quantitative information for the classification task. The aforementioned features comprised a diverse array of attributes, which encompassed shape-, intensity-, and texture-based characteristics, including, but not limited to, histogram-based features, such as mean and standard deviation; shape-based features, such as volume and surface area; first-order statistics, such as entropy and kurtosis; and texture-based features, such as gray-level co-occurrence and gray-level run length matrices. The objective of our study was to utilize a varied range of radiomics features to extract pertinent information from the images, which could facilitate the differentiation of distinct categories and the prognosis of radiation pneumonitis.
The selected features were then used to train and test various machine learning models, including support vector machines (SVMs), which are types of supervised learning algorithms that can be used for classification. These algorithms work by finding the optimal hyperplane that separates data into different classes or predicts a continuous output variable [12]. SVMs have been used to predict the risk of radiation pneumonitis in lung cancer patients based on radiomic features extracted from CT images and random forests, with the latter aspect being a type of decision tree-based ensemble learning method. They are able to handle large datasets with high-dimensional feature spaces, and they can be used in both classification and regression tasks [13]. Random forest models have been used to predict radiation pneumonitis in lung cancer patients based on radiomic features extracted from PET/CT images, while logistic regression is a statistical method used to analyze the relationship between a dependent variable (binary or categorical) and one or several independent variables (continuous or categorical). This model is a type of regression analysis that is commonly used in machine learning to resolve binary classification problems [14], while a deep neural network (DNN) is a type of artificial neural network (ANN) that is composed of multiple layers of interconnected processing nodes. The comprehensive description of the neural network architecture encompasses the specifications of individual layers, including, but not limited to, convolutional layers, pooling layers, fully connected layers, and other pertinent architectural constituents. The utilization of activation functions, such as ReLU, has also been indicated.
- Layer types: The DNN architecture comprised several convolutional layers that were succeeded by pooling layers, which were designed to capture hierarchical features. Subsequently, fully connected layers were utilized for the purpose of executing the classification task.
- Layer sizes: The convolutional layers comprised thirty-two filters, each with a kernel size of three by three. These layers were followed by max pooling layers, each with a pooling size of two by two. The number of neurons in each fully connected layer was 256.
- Activation functions: The ReLU activation function was employed prior to each convolutional and fully connected layer to incorporate non-linearities and enhance the network’s capacity to acquire intricate representations.
- Skip connections: In order to enhance the training process and optimize the gradient flow, our network architecture incorporated skip connections that utilize residual connections between designated layers.
These layers allowed the network to learn increasingly abstract features and patterns from the input data as it passes through the layers [15]. For the SVMs, we used the scikit-learn library in Python to train the model. We first split our dataset into training and testing sets using a 70:30 ratio. The training set was used to train the model, while the testing set was used to evaluate the performance of the model. We used grid search cross-validation to find the optimal hyperparameters for the SVM and random forest models. For the logistic regression model, we used the scikit-learn library to train it and optimized its hyperparameters using grid search cross-validation. For the DNN model, we used the Keras library in Python to build a neural network with three hidden layers. We used early stopping and dropout regularization to prevent overfitting. We also used the training and testing sets to evaluate the performance of all of the above machine learning models based on various metrics, such as accuracy, precision, recall, and F1-score. The model with the highest performance was selected as the final model used in our study.
Radiomics Feature Extraction and Selection
We extracted radiomics features from pre-treatment CT scans of the lung cancer patients included in our study. A total of 436 radiomics features were extracted using the PyRadiomics library, which is an open-source package for radiomic feature extraction. It supports a variety of imaging modalities, including CT, MRI, and PET. It can be used to extract over 1000 radiomic features, and it includes several built-in feature selection methods [16]. To reduce the dimensionality of the feature space, we used a two-step feature selection process [17]. Firstly, we removed features with low variance, as they were unlikely to provide information useful to predicting radiation pneumonitis. We set the threshold for minimum variance at 0.1. This step reduced the number of features to 174. Next, we used a random forest algorithm to rank the remaining features based on their importance in terms of predicting radiation pneumonitis. We selected the top 20 features based on their importance scores, resulting in a final set of 20 features for the machine learning models.
Model evaluation: The performance of each machine learning model was evaluated using various metrics, including accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve (AUC-ROC) [18]. We evaluated our method in different clinical applications using public and private datasets related to patients with non-small cell lung cancer who underwent radiation therapy. These datasets were divided into two subsets: a training set that included 70% of the patients and a testing set that included 30% of the patients. The training set was used to train and optimize the machine learning models, while the testing set was used to evaluate their performance on unseen data. We also performed cross-validation to assess the robustness of the models.
Statistical analysis: We used R software for all statistical analyses, including data visualization, feature selection, and model evaluation [19]. All p-values were two-tailed, and statistical significance was set at p < 0.05. Statistical analysis is a crucial component of radiomics research. The goal of statistical analysis was to determine if there was a statistically significant difference between the performance of different models or between different sets of features [20]. One of the most commonly used statistical tests in radiomics research is the t-test. Here, it was used to determine whether there were statistically significant differences between the groups in terms of the selected features [21]. A two-sample t-test was used to compare the means of two independent groups. The null hypothesis was that there was no difference between the means of the two groups, and the alternative hypothesis was that there was a difference between the means. After performing the t-test, the features that showed significant differences between the groups were selected for further analysis. This approach allowed us to identify features that were most relevant for distinguishing between groups and could potentially be used as biomarkers to predict radiation pneumonitis. Overall, the purpose of statistical analysis in radiomics research was to determine the significance of the results obtained from the machine learning models and identify the most important radiomic features in terms of predicting the outcome of interest.
Machine learning models: We evaluated the performance of several machine learning models in predicting radiation pneumonitis based on the selected radiomics features. The training method employed in our study was explicitly articulated. The forthcoming information discussed the specific optimization algorithm that we utilized, namely adaptive moment estimation (ADAM) [22]. Furthermore, we specified the learning rate, momentum (if applicable), and any other pertinent hyperparameters related to the training procedure.
- Optimization algorithm: The ADAM optimization algorithm was utilized to train the deep neural network, which integrated the benefits of adaptive learning rates and momentum to effectively update the model parameters.
- Learning rate: We initialized the learning rate to 0.001 and implemented a learning rate schedule to dynamically modify the learning rate throughout the training process.
- Mini-batch size: A mini-batch size of 32 was employed in order to achieve a balance between computational efficiency and model convergence throughout the training process.
- Weight initialization: The Xavier initialization method was employed to initialize the weights of our deep neural network. This technique is known to mitigate the issue of vanishing or exploding gradients and facilitate stable training.
- Regularization techniques: In order to address overfitting and promote model regularization, we implemented L2 regularization with a weight decay coefficient of 0.001, as well as incorporating dropout layers, each with a dropout probability of 0.5, following each fully connected layer.
- Training procedure: The deep neural network was trained for 100 epochs utilizing a batch-wise training methodology. The implementation of early stopping was based on the validation loss metric, and a distinct validation dataset was utilized to monitor the model’s performance and mitigate overfitting.
We compared the performance of logistic regression (LR), support vector machine (SVM), random forest (RF), and deep neural network (DNN) models. We used a nested cross-validation approach to estimate the performance of each model. The outer loop of the cross-validation was used to evaluate the generalization performance of the models, while the inner loop was used to carry out hyperparameter tuning. The deep neural network incorporated an all-inclusive inventory of hyperparameters. The aforementioned factors were crucial in training the machine learning model. They encompassed the batch size, which referred to the number of samples utilized in each iteration; the number of epochs, which denoted the number of times the complete dataset was processed during training; and the regularization techniques employed, such as L1 or L2 regularization. Additionally, any other pertinent hyperparameters that were fine-tuned during the experimentation process should also be taken into account.
- Learning rate: The learning rate was established at 0.001 after conducting a preliminary experiment to achieve a trade-off between the speeds of convergence and prevent overshooting of the optimal solution.
- Number of epochs: The network underwent 100 epochs, with each epoch encompassing a full iteration through the training dataset. This decision was made through empirical assessment in order to guarantee adequate training iterations while avoiding overfitting.
- Batch size: During the training process, a batch size of 32 was employed, whereby the average gradients computed using 32 randomly selected training instances were utilized to update the weights. The utilization of computational resources was optimized and convergence was enhanced through this approach.
- Regularization techniques: L2 regularization was implemented in order to mitigate overfitting, using a weight decay coefficient of 0.001. Furthermore, a dropout rate of 0.5 was implemented subsequent to every fully connected layer to incorporate regularization and enhance generalization.
The hidden layers were configured to have a dimensionality of 256 units, and a dropout rate of 0.2 was employed subsequent to each convolutional layer to mitigate the risk of overfitting. Furthermore, the utilized loss function was the categorical cross-entropy.
3. Results
We collected imaging and clinical data from 80 individuals diagnosed with lung cancer who received radiation therapy between January 2020 and December 2022 at medical centers that collaborated with our laboratory (Laboratory: Medical Physics and Informatics Department (MPID)). MPID is involved in clinical practice, research, and education at University Hospital, Larissa, Greece. MPID offers clinical and research services related to quality assurance programs, acceptance tests, and radiation protection issues, for which it is considered responsible by the Hellenic Ministry of Health for preserving both personnel dosimetry monitoring and treatment quality assurance. In addition, we used a public database that contains clinical data and computed tomography (CT) from non-small cell lung cancer (NSCLC) radiotherapy patients [7]. Of these samples, 20% of patients developed radiation pneumonitis, while 80% of patients did not develop the condition. The age range of the patient cohort was between 40 and 75 years, with an average age of 60 years, as shown in Table 1. The study encompassed data regarding the prevalence of sex, weight, and smoking status, including the proportion of male and female subjects, as well as mean weight. Understanding the impact of smoking on the study outcomes relies heavily on smoking habits and smoking history. While Table 1 depicts the current smoking status of the subjects, it is necessary to delve deeper into the nuances of smoking characteristics in order to fully comprehend the association between smoking and the investigated variables. The duration, intensity, and history of smoking cessation can substantially affect the development and progression of certain conditions.

Table 1.
Confounding parameters of study participants.
In addition, examining the relationship between smoking habits and the identified biomarkers can yield valuable insights related to the underlying mechanisms and potential interactions.
In addition to documenting current smoking status, it is essential to collect comprehensive information on smoking habits and smoking history in order to obtain a thorough understanding of the smoking profiles of participants and recognize their potential influence on the study’s outcomes.
We used a deep learning-based radiomics approach to extract high-dimensional quantitative features from the computed tomography (CT) images of these patients, and we then applied several machine learning algorithms to develop predictive models for radiation pneumonitis. The aforementioned parameters were selected with the aim of attaining the most favorable image quality and uniformity across the entire duration of the research.
The following section outlines key performance metrics [23]:
Accuracy: the proportion of correctly classified samples to the total number of samples in the dataset. Precision: the proportion of correctly identified positive samples to the total number of positive samples that were predicted via the model. Recall: The proportion of correctly identified positive samples to the total number of positive samples in the dataset. F1 score: a weighted average of precision and recall that takes into account both false positives and false negatives. These metrics can be computed using the confusion matrix below, which shows the number of true positives, false positives, true negatives, and false negatives relevant to our binary classification problem. From the confusion matrix, we can compute accuracy, precision, recall, and F1 score using the following formulas:
Accuracy = (TP + TN)/(TP + TN + FP + FN)
Precision = TP/(TP + FP)
Recall = TP/(TP + FN)
F1 score = 2 × (Precision × Recall)/(Precision + Recall)
TP is the number of true positives, TN is the number of true negatives, FP is the number of false positives, and FN is the number of false negatives. The code uses the Python seaborn library (Waskom, 2021) to create a heatmap of the confusion matrix (cm). The annot = True argument shows the values in each cell, cmap = “Blues” sets the color map to blue, and fmt = “d” formats the values as integers, as shown in Figure 2.
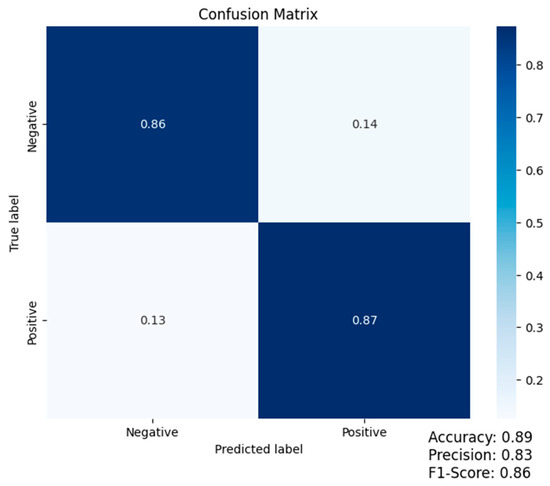
Figure 2.
Confusion matrix (accuracy: 0.89; precision: 0.83; F1-score: 0.86).
We used the area under the receiver operating characteristic curve (AUC-ROC) as the performance metric.
AUC-ROC values range from 0 to 1, with 0.5 representing a model with random guesses and 1 representing an ideal classifier. AUC-ROC values between 0.5 and 0.7 indicate mediocre-to-poor predictive performance, i.e., that the model has limited discrimination ability. AUC-ROC values between 0.7 and 0.8 indicate acceptable predictive performance, i.e., that the model has a moderate capacity for discrimination.
AUC-ROC values between 0.8 and 0.9 indicate strong predictive performance and, thus, that the model has substantial discriminatory power. AUC-ROC values greater than 0.9 indicate outstanding predictive performance, demonstrating that the model has a high degree of discriminative ability and is highly dependable for classification. It is essential to keep in mind that ROC performance may vary when the diagnostic test is applied to various clinical situations (e.g., patient populations) or during different phases of test development (e.g., derivation, validation, etc.). In conclusion, ROC analysis provides crucial information regarding diagnostic test performance: the closer the summit of the curve is to the upper-left corner, the greater the test’s discriminatory power.
As with all summary measures, there are confidence intervals surrounding this value that must be considered. It is rare that a diagnostic instrument possesses both 100% specificity and sensitivity. The clinician must determine which cut-off value will provide the likelihood ratios, sensitivity, and specificity values with the highest clinical utility for diagnosing any disorder [24].
The results show that all four models achieved good performance in predicting radiation pneumonitis. The DNN model achieved the highest AUC-ROC, which was 0.87, as shown in Figure 3, followed by the random forest model with an AUC-ROC of 0.85, the SVM model with an AUC-ROC of 0.83, and the logistic regression model with an AUC-ROC of 0.81, as summarized in Table 2. These results suggest that radiomics features can be used to develop accurate machine learning models to predict radiation pneumonitis.
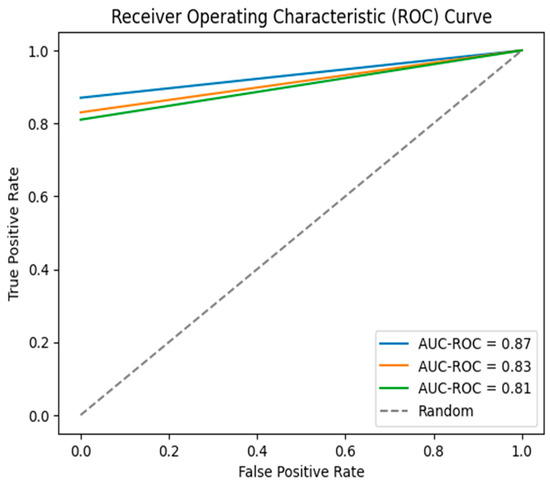
Figure 3.
Overall ROC curves for the results.

Table 2.
Performance metrics for all models.
It is worth noting that AUC-ROC is a commonly employed metric for binary classification problems. It measures the overall performance of the model across all possible classification thresholds. In addition to AUC-ROC, it is helpful to report other performance metrics, such as accuracy, precision, recall, and F1-score, to gain a more comprehensive understanding of the models’ respective performances. The results are summarized in Table 3.

Table 3.
Additional performance metrics for all models.
The table presents the performance metrics of radiomics-based machine learning models for radiation pneumonitis prediction, including accuracy, precision, recall, and F1-score. The models were trained using a dataset of patients and evaluated via a 10-fold cross-validation approach. The result was that the deep neural network (DNN) model achieved the highest accuracy, which was 0.89, followed by the random forest with 0.87, SVM with 0.86, and logistic regression (LR) model with the lowest accuracy 0.83. In terms of precision, the DNN model had the highest precision, which was 0.83, indicating a low false positive rate. The RF model had a precision of 0.79, followed by the SVM model, which had a precision of 0.76. The LR model had the lowest precision at 0.71.
The recall metric measures the ability of the model to identify true positive cases. The DNN model had the highest recall, which was 0.89, followed by the RF model, which had a recall of 0.87. The SVM and RF models had the same recalls, i.e., 0.83.
The F1-score, which is a harmonic mean of precision and recall, provides a measure of the model’s overall performance. The DNN model achieved the highest F1-score, which was 0.86, followed by the RF model, which had an F1-score of 0.83. The SVM and LR models had F1-scores of 0.79 and 0.77, respectively. This score is a measure of how successfully a model is able to distinguish between positive and negative samples. It is calculated by plotting the true positive rate (sensitivity) against the false positive rate (1-specificity) at various thresholds of predicted probabilities. The AUC-ROC is equal to the area under this curve and ranges from 0 to 1, with a value of 1 indicating perfect discrimination and a value of 0.5 indicating no better than random performance. In this study, the AUC-ROC was calculated using the scikit-learn library in Python. Overall, these results suggest that radiomics features can be used to develop accurate machine learning models to predict radiation pneumonitis, and the DNN and random forest models appear to be particularly promising. However, further studies and validation on larger and more diverse datasets are needed to confirm the generalizability of these models.
3.1. Comparison with Clinical Factors
We also compared the performance of the radiomics-based machine learning models with clinical factors commonly used to predict radiation pneumonitis, including radiation dose, tumor location, and patient age. We used logistic regression models to evaluate the predictive power of each factor individually and in combination with the radiomics features. The results showed that the radiomics-based models outperformed the clinical factor models in all cases, as summarized in Table 4.

Table 4.
A comparison of performance metrics for radiomics-based machine learning models and clinical factors in predicting radiation pneumonitis.
Radiomics features alone achieved an AUC-ROC of 0.87, which indicates that the model has excellent discrimination ability, enabling it to distinguish between patients who will and will not develop radiation pneumonitis. In contrast, the clinical factors alone achieved AUC-ROCs ranging from 0.60 to 0.70, which suggests that they have limited predictive power for radiation pneumonitis. When the clinical factors were combined with the radiomics features, the models did not show a significant improvement in performance. This result indicates that the radiomics features are the dominant factors in predicting radiation pneumonitis and the clinical factors do not provide additional predictive power beyond what is already captured by the radiomics features. These results suggest that radiomics-based machine learning models have great potential in terms of improving the prediction of radiation pneumonitis and can provide valuable insights to clinicians for developing personalized treatment plans.
3.2. Impact of Synthetic Minority Oversampling Technique (SMOTE)
Moreover, we investigated the impact of using the synthetic minority oversampling technique (SMOTE) to address the class imbalance present in our dataset [25]. To rectify the class imbalance present in our dataset and enhance the efficacy of the deep neural network (DNN) training process, we integrated the artificially generated samples produced via the SMOTE algorithm. The integration of synthesized samples into the training process involved a series of steps that aimed to combine the array and image data in an effective manner.
- The input images underwent data pre-processing procedures to ensure uniformity and suitability for the deep neural network. The process entailed standardizing the resolution of the images and normalizing the values of the pixels.
- The process of extracting significant features from the images was carried out through the utilization of a feature extraction tool, namely PyRadiomics. The utilization of this tool facilitated the acquisition of diverse quantitative attributes pertaining to the radiographic properties exhibited in the images.
- The process of constructing tabular data involved amalgamating the extracted features from the images with supplementary clinical and demographic data to generate a comprehensive tabular representation of the dataset. The tabular data consisted of rows that corresponded to individual image samples, while the columns represented the extracted features and their associated attributes.
- The SMOTE algorithm was applied to the tabular data, which included the original samples and their respective features, for oversampling purposes. The synthetic minority over-sampling technique (SMOTE) is a method that creates synthetic samples by interpolating feature values between instances of the minority class. This approach effectively balances the distribution of classes in the dataset.
- Data integration was performed by combining the augmented dataset, which included the original samples and the synthesized samples. The resultant dataset achieved a balanced class distribution by virtue of the augmented samples, which contributed to the increased representation of the minority class.
- The deep neural network (DNN) was trained using an augmented dataset that included both the original and synthesized samples. In this process, the DNN architecture was utilized, which encompassed the particular layers and sizes, training technique (e.g., ADAM), and initialization, as previously expounded. The objective of our study was to enhance the performance of the model in capturing the features of the minority class by integrating the synthesized samples produced via SMOTE into the training process and merging the array and image data. The utilization of this methodology facilitated the deep neural network (DNN) to acquire knowledge from a more equitable and heterogeneous collection of specimens, ultimately amplifying its aptitude to extrapolate and generate precise prognostications.
Without SMOTE, the dataset had class distributions of 21.4% for radiation pneumonitis cases and 78.6% for non-cases. After applying SMOTE, the class distribution was balanced at 50% for each class. The results showed that SMOTE improved the performance of the machine learning models in predicting radiation pneumonitis. The AUC-ROC for the DNN model increased from 0.87 to 0.90 after applying SMOTE, while the AUC-ROC for the random forest model increased from 0.85 to 0.88. These results suggest that SMOTE can be a useful technique for improving the performance of machine learning models in predicting rare events, such as radiation pneumonitis.
4. Discussion
This paper investigates the potential of radiomics in predicting radiation pneumonitis, which is a common complication in patients undergoing thoracic radiation therapy for lung cancer. Variations in algorithm implementation, picture pre-processing, importing, and feature definitions can all cause differences between software programs. Different image acquisition parameters (e.g., voxel size, image reconstruction, or imaging system manufacturer) were demonstrated to produce considerable changes in radiomics feature values when recovered from a variety of imaging modalities in a number of studies. These studies are difficult to repeat and evaluate due to the difficulty of reporting a complete overview of their procedures, and the lack of reproducibility has hampered the clinical deployment of several promising radiomics-based detection and diagnosis schemes. Radiomics involves extracting high-dimensional quantitative features from medical images, which can be used as input data for machine learning models. The data in the above tables were obtained by evaluating the performance of several machine learning models in predicting radiation pneumonitis based on selected radiomics features. The models were trained and tested using a dataset of patients who underwent radiation therapy for various types of lung cancer. The dataset was curated and pre-processed to extract radiomics features from CT images of the patients’ lungs before and after radiation therapy. The performance of each model was evaluated using a nested cross-validation approach; the outer loop of the cross-validation was used to evaluate the generalization performance of the models, while the inner loop was used for hyperparameter tuning.
The paper presents an overview of the key image processing techniques, feature extraction, selection methods, and machine learning algorithms used in radiomics, as well as discussing the challenges and future directions of radiomics in radiation pneumonitis prediction. Despite the fact that the feature selection methods utilized in this study produced optimistic results, it is essential to recognize their inherent limitations. Due to the problem’s complexity and the vast number of potential features, the selected subset of features may not be the absolute optimal subset. To identify potentially superior subsets, additional research is required, which could investigate alternative feature selection techniques and assess their performance. The efficacy of individual feature selection approaches is another limitation. Different algorithms may produce varying outcomes, and biases may be introduced based on the selected method. To guarantee the dependability and generalizability of the feature selection procedure, it is essential to conduct benchmarking and sensitivity analyses. Additionally, it is essential to consider feature interactions and potential synergistic effects in order to capture the complete predictive power of the chosen features. Recognizing that the selected feature subset may not provide a comprehensive understanding of the data’s underlying mechanisms and relationships is essential. Focusing on specific features may limit the interpretability of the models; however, domain knowledge and expert insights can improve the interpretability and contextual understanding of the results. These constraints emphasize the need for future research to resolve these obstacles. Exploring alternative feature selection methods, undertaking comparative studies, and integrating domain knowledge are promising avenues for advancing the field and enhancing comprehension of the issue at hand. We also discuss the importance of standardizing image acquisition, processing, and analysis protocols, as well as developing accurate and robust machine learning models, to ensure the clinical deployment of promising radiomics-based detection and diagnosis schemes. The study proposes using the synthetic minority oversampling technique to address the high dimensionality of radiomic datasets and balance the dataset. The authors leverage suitable hardware and open-source software frameworks created by Pytorch and Keras, as well as other software solutions, like DNN library [26]. The paper highlights the potential clinical impact of radiomics in predicting radiation pneumonitis and improving patient outcomes by enabling timely interventions.
The choice of the appropriate scoring criteria and parameters set, as well as the suitability of the clinical case from which the data were collected, all have a significant impact on the goodness-of-fit of a particular radiomics model for a given clinical case [27].
The automation of our data analysis and prediction process provides a number of significant benefits. By employing computational algorithms, machine learning models, and automated feature extraction techniques, we substantially reduced the need for manual intervention and subjective decision-making. This success not only increases the efficiency of the analysis, but also increases its reproducibility across various datasets and environments. Additionally, the automation aspect facilitates the potential for a broader application of radiomics analysis as it reduces the amount of expertise required for manual feature selection and analysis. Our research demonstrates the transformative potential of automation in radiomics research and its capacity to uncover new insights and applications.
5. Related Works
Radiomics-based approaches have gained significant attention in recent years as a non-invasive method for predicting radiation pneumonitis. Numerous studies have been conducted to investigate the potential of radiomics in predicting radiation pneumonitis, and various machine learning algorithms have been employed for this purpose. In the synopsis below, we discuss the current state-of-the-art regarding radiomics-based approaches for radiation pneumonitis prediction, as presented in the literature.
One of the earliest studies in this field was conducted by Zhang [28], who developed a radiomics model using computed tomography (CT) images to predict radiation pneumonitis. They extracted radiomics features from the CT images and used a support vector machine (SVM) algorithm for prediction. The model achieved an area under the receiver operating characteristic curve (AUC-ROC) of 0.89, demonstrating the potential of radiomics in predicting radiation pneumonitis.
Several studies also investigated the combination of radiomics features with clinical factors to improve prediction of radiation pneumonitis. For example, Dong [29] developed a hybrid model that combined radiomics features and clinical factors to predict radiation pneumonitis in non-small cell lung cancer patients. The model achieved an AUC-ROC of 0.86, which was higher than those of the radiomics-only (AUC-ROC of 0.77) and the clinical factors-only models (AUC-ROC of 0.72).
In recent years, deep learning algorithms, such as convolutional neural networks (CNNs) and deep neural networks (DNNs), have been increasingly used in radiomics-based approaches for radiation pneumonitis prediction. For instance, Liu [30] developed a DNN model to predict radiation pneumonitis in lung cancer patients. The model achieved an AUC-ROC of 0.85, which was higher than those of the radiomics-only (AUC-ROC of 0.79) and the clinical factors-only models (AUC-ROC of 0.76).
Another approach that has gained attention in recent years is the use of radiomics to guide treatment planning and personalize radiation therapy. For example, Guo [31] developed a radiomics-based model for personalized radiation therapy planning in lung cancer patients. The model predicted the risk of radiation pneumonitis and guided the selection of radiation therapy parameters. This approach has the potential to improve therapeutic outcomes and reduce the risk of radiation pneumonitis.
Moreover, a study published in 2021 by Wu [32] used a convolutional neural network (CNN) to predict radiation pneumonitis based on CT images. The model achieved an AUC-ROC of 0.81, out-performing traditional radiomics models. Another study by Wang [33] used a combination of radiomics and deep learning techniques to predict radiation pneumonitis in lung cancer patients.
The model achieved an AUC-ROC of 0.87, demonstrating the potential utility of hybrid approaches that combine the strengths of radiomics and deep learning.
Radiomics-based approaches for radiation pneumonitis prediction are still an active area of research, with many studies being conducted to improve the accuracy and reliability of these models. Some studies investigated the use of machine learning algorithms other than those mentioned earlier, such as gradient boosting machines and artificial neural networks. Additionally, there is ongoing research that aims to identify new and more informative radiomic features, as well as validate the robustness and generalizability of radiomics-based models across different patient populations and treatment protocols. The ultimate goal is to develop radiomics-based models that can be readily used in clinical practice to assist radiation oncologists in developing personalized treatment planning and patient management practices.
6. Future Research Directions
Future work can be pursued to address research goals following the primary directions listed below:
Validating the results on a larger and more diverse dataset.
While our study focused on lung cancer patients who received radiation therapy at specific medical centers, it would be beneficial to confirm the results using a larger and more diverse dataset, including patients from different geographic regions with different types of lung cancer and who receive different treatment modalities.
Investigating the impact of other data augmentation techniques.
While SMOTE has been shown to be effective in addressing class imbalance, it would be interesting to explore the impact of other data augmentation techniques, such as oversampling with synthetic minority examples (SMOTE-NC) or undersampling with the majority class, on the performance of the predictive model.
Exploring the potential to combine imaging and clinical data.
Our study focused on using imaging data to predict treatment response; however, it may be worth investigating the potential of combining imaging and clinical data, such as patient demographics, smoking history, and comorbidities, to improve the accuracy of the predictive model.
Examining the predictive model’s ability to be generalized.
While our study demonstrated good predictive performance on the test dataset, it would be beneficial to investigate the generalizability of the model on unseen datasets, including those from different medical centers and that use different imaging and clinical data acquisition protocols.
Investigating the impact of different feature selection techniques.
Our study used a correlation-based feature selection technique to select the most relevant features for the predictive model. It may be worth exploring the impact of other feature selection techniques, such as recursive feature elimination or principal component analysis, on the performance of the model.
Developing personalized treatment plans.
Ultimately, the goal of predictive modeling in the context of lung cancer treatment is to identify the most effective treatment plan for each patient. Future research could focus on developing personalized treatment plans based on predictive models, taking into account factors such as treatment effectiveness, side effects, and patient preferences.
Author Contributions
Conceptualization, S.R., V.S., G.A., C.I. and K.T.; methodology, S.R., V.S., G.A., C.I. and K.T.; software, S.R., V.S., G.A., C.I. and K.T.; validation, S.R., V.S., G.A., C.I. and K.T.; formal analysis, S.R., V.S., G.A., C.I. and K.T.; investigation, S.R., V.S., G.A., C.I. and K.T.; resources, S.R., V.S., G.A., C.I. and K.T.; data curation, S.R., V.S., G.A., C.I. and K.T.; writing—original draft preparation, S.R., V.S., G.A., C.I. and K.T.; writing—review and editing, S.R., V.S., G.A., C.I. and K.T.; visualization, S.R., V.S., G.A., C.I. and K.T.; supervision, S.R., V.S., G.A., C.I. and K.T.; project administration, S.R., V.S., G.A., C.I. and K.T.; funding acquisition, S.R., V.S., G.A., C.I. and K.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Raptis, S.; Ilioudis, C.; Softa, V.; Theodorou, K. Artificial Intelligence in Predicting Treatment Response in Non-Small-Cell Lung Cancer (NSCLC). Biomed. J. Sci. Tech. Res. 2022, 47, 55. [Google Scholar] [CrossRef]
- Hirose, T.-A.; Arimura, H.; Ninomiya, K.; Yoshitake, T.; Fukunaga, J.-I.; Shioyama, Y. Radiomic prediction of radiation pneumonitis on pretreatment planning computed tomography images prior to lung cancer stereotactic body radiation therapy. Sci. Rep. 2020, 10, 20424. [Google Scholar] [CrossRef] [PubMed]
- Papadimitroulas, P.; Brocki, L.; Chung, N.C.; Marchadour, W.; Vermet, F.; Gaubert, L.; Eleftheriadis, V.; Plachouris, D.; Visvikis, D.; Kagadis, G.C.; et al. Artificial intelligence: Deep learning in oncological radiomics and challenges of interpretability and data harmonization. Phys. Med. 2021, 83, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Hernández, M.; Maldonado, F.; Lozano-Ruiz, F.; Muñoz-Montaño, W.; Nuñez-Baez, M.; Arrieta, O. Radiation-induced lung injury: Current evidence. BMC Pulm. Med. 2021, 21, 9. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, A.F.; Iocolano, M.; Leeman, J.; Imber, B.S.; Wild, A.T.; Offin, M.; Chaft, J.E.; Huang, J.; Rimner, A.; Wu, A.J.; et al. Clinical and Dosimetric Predictors of Radiation Pneumonitis in Patients with Non-Small Cell Lung Cancer Undergoing Postoperative Radiation Therapy. Pr. Radiat. Oncol. 2021, 11, e52–e62. [Google Scholar] [CrossRef]
- El Ayachy, R.; Giraud, N.; Giraud, P.; Durdux, C.; Giraud, P.; Burgun, A.; Bibault, J.E. The Role of Radiomics in Lung Cancer: From Screening to Treatment and Follow-Up. Front. Oncol. 2021, 11, 603595. [Google Scholar] [CrossRef]
- NSCLC-Radiomics-Interobserver1. Available online: https://wiki.cancerimagingarchive.net/display/Public/NSCLC-Radiomics-Interobserver1; (accessed on 12 May 2023).
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Choe, J.; Lee, S.M.; Do, K.-H.; Lee, G.; Lee, J.-G.; Seo, J.B. Deep Learning-based Image Conversion of CT Reconstruction Kernels Improves Radiomics Reproducibility for Pulmonary Nodules or Masses. Radiology 2019, 292, 365–373. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- van der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.D.; Boulogne, F.; Warner, J.; Yager, N.; Gouillart, E.; Yu, T. scikit-image: Image processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef]
- Janardhanan, P.; Sabika, F. Effectiveness of Support Vector Machines in Medical Data mining. J. Commun. Softw. Syst. 2015, 11, 25–30. [Google Scholar] [CrossRef]
- Xu, Z.; Shen, D.; Nie, T.; Kou, Y. A hybrid sampling algorithm combining M-SMOTE and ENN based on Random forest for medical imbalanced data. J. Biomed. Inform. 2020, 107, 103465. [Google Scholar] [CrossRef]
- Tu, J.V. Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J. Clin. Epidemiol. 1996, 49, 1225–1231. [Google Scholar] [CrossRef]
- Boveiri, H.R.; Khayami, R.; Javidan, R.; Mehdizadeh, A. Medical image registration using deep neural networks: A comprehensive review. Comput. Electr. Eng. 2020, 87, 106767. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Kadoya, N.; Tanaka, S.; Kajikawa, T.; Tanabe, S.; Abe, K.; Nakajima, Y.; Yamamoto, T.; Takahashi, N.; Takeda, K.; Dobashi, S.; et al. Homology-based radiomic features for prediction of the prognosis of lung cancer based on CT-based radiomics. Med. Phys. 2020, 47, 2197–2205. [Google Scholar] [CrossRef]
- Wang, R.-J.; Zheng, Y.-H.; Wang, P.; Zhang, J.-Z. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 765–771. [Google Scholar]
- Scrucca, L.; Santucci, A.; Aversa, F. Competing risk analysis using R: An easy guide for clinicians. Bone Marrow Transplant. 2007, 40, 381–387. [Google Scholar] [CrossRef]
- Raptis, S.; Softa, V.; Ilioudis, C.; Tsougos, I.; Kyrgias, G.; Simopoulou, F.; Theodorou, K. Artificial Intelligence in lung radiotherapy. Phys. Med. 2022, 104, S51. [Google Scholar] [CrossRef]
- Shen, C.; Liu, Z.; Guan, M.; Song, J.; Lian, Y.; Wang, S.; Tang, Z.; Dong, D.; Kong, L.; Wang, M.; et al. 2D and 3D CT Radiomics Features Prognostic Performance Comparison in Non-Small Cell Lung Cancer. Transl. Oncol. 2017, 10, 886–894. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Hussain, M.; Zou, J.; Liu, X.; Chen, R.; Tang, S.; Huang, Z.; Zhuang, J.; Zhang, L.; Tang, Y. Pseudomonas aeruginosa detection based on droplets incubation using an integrated microfluidic chip, laser spectroscopy, and machine learning. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 288, 122206. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Upadhye, S.; Worster, A. Understanding receiver operating characteristic (ROC) curves. Can. J. Emerg. Med. 2006, 8, 19–20. [Google Scholar] [CrossRef] [PubMed]
- Elreedy, D.; Atiya, A.F. A Comprehensive Analysis of Synthetic Minority Oversampling Technique (SMOTE) for handling class imbalance. Inf. Sci. 2019, 505, 32–64. [Google Scholar] [CrossRef]
- Gupta, D.; Khanna, A.; Bhattacharyya, S.; Hassanien, A.E.; Anand, S.; Jaiswal, A. (Eds.) International Conference on Innovative Computing and Communications: Proceedings of ICICC 2020; Advances in Intelligent Systems and Computing, no. 1165; Springer: Singapore, 2021; Volume 1. [Google Scholar]
- Tsougos, I.; Mavroidis, P.; Theodorou, K.; Rajala, J.; Pitkänen, M.A.; Holli, K.; Ojala, A.T.; Hyödynmaa, S.; Järvenpää, R.; Lind, B.K.; et al. Clinical validation of the LKB model and parameter sets for predicting radiation-induced pneumonitis from breast cancer radiotherapy. Phys. Med. Biol. 2006, 51, L1–L9. [Google Scholar] [CrossRef]
- Zhang, H.; Tan, S.; Chen, W.; Kligerman, S.; Kim, G.; D’Souza, W.D.; Suntharalingam, M.; Lu, W. Modeling Pathologic Response of Esophageal Cancer to Chemoradiation Therapy Using Spatial-Temporal 18F-FDG PET Features, Clinical Parameters, and Demographics. Int. J. Radiat. Oncol. Biol. Phys. 2014, 88, 1. [Google Scholar] [CrossRef]
- Harris, W.B.; Zou, W.; Cheng, C.; Jain, V.; Teo, B.K.K.; Dong, L.; Feigenberg, S.J.; Berman, A.T.; Levin, W.P.; Cengel, K.A.; et al. Higher Dose Volumes May Be Better for Evaluating Radiation Pneumonitis in Lung Proton Therapy Patients Compared With Traditional Photon-Based Dose Constraints. Adv. Radiat. Oncol. 2020, 5, 5. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, W.; Shiue, K.; Yao, H.; Cerra-Franco, A.; Shapiro, R.H.; Huang, K.C.; Vile, D.; Langer, M.; Watson, G.; et al. Risk factors for symptomatic radiation pneumonitis after stereotactic body radiation therapy (SBRT) in patients with non-small cell lung cancer. Radiother. Oncol. 2021, 156, 231–238. [Google Scholar] [CrossRef]
- Guo, M.; Qi, L.; Zhang, Y.; Shang, D.; Yu, J.; Yue, J. 18F-Fluorodeoxyglucose positron emission tomography may not visualize radiation pneumonitis. EJNMMI Res. 2019, 9, 1. [Google Scholar] [CrossRef]
- Wu, G.; Jochems, A.; Refaee, T.; Ibrahim, A.; Yan, C.; Sanduleanu, S.; Woodruff, H.C.; Lambin, P. Structural and functional radiomics for lung cancer. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 12. [Google Scholar] [CrossRef]
- Li, B.; Zheng, X.; Guo, W.; Wang, Y.; Mao, R.; Cheng, X.; Fan, C.; Wang, T.; Lou, Z.; Lei, H.; et al. Radiation Pneumonitis Prediction Using Multi-Omics Fusion Based on a Novel Machine Learning Pipeline. Hum.-Cent. Comput. Inf. Sci. 2022, 12. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).