1. Introduction
As a major disease that seriously threatens human health, cancer is characterized by high incidence rate and mortality. Tongue cancer, as the most aggressive subtype of oral cancer, originates from the malignant biological behaviors of heterogeneous epithelial cell proliferation and stromal remodeling [
1,
2]. Histologically, tongue cancer is primarily composed of squamous cell carcinoma nests containing keratin pearls, accompanied by extensive inflammatory cell infiltration and neovascularization in the stroma. Its five-year local recurrence rate reaches 30% to 50%. Despite hematoxylin-eosin (HE) staining combined with biopsy remaining the gold standard for diagnosis, the global shortage of pathologists has significantly prolonged diagnostic timelines [
3,
4]. The progress of oral cancer diagnosis is significantly delayed in low- and middle-income countries [
5].
Notably, during the progression of tongue squamous cell carcinoma, microscopic structural alterations occur, including nuclear enlargement (increased nuclear-cytoplasmic ratio), collagen fiber disorganization (anisotropic changes), and extracellular matrix density abnormalities (refractive index fluctuations). These changes distinctly influence tissue optical scattering properties, which can be clearly visualized through polarization imaging [
6]. Polarization imaging techniques, by analyzing the vector interaction between light and tissues, enable subcellular-level structural information acquisition without labeling, offering unique advantages in delineating tumor margins and detecting early carcinogenesis [
7].
Beyond polarization imaging, a diverse array of quantitative optical imaging techniques has emerged as powerful tools for non-invasive, label-free characterization of tissue microstructure and function, gaining significant traction in digital pathology for neoplastic lesion detection. These techniques exploit different light-tissue interactions to extract rich, quantitative biomarkers. Quantitative Phase Imaging (QPI), including digital holographic microscopy and tomography, measures optical path length delays induced by tissue refractive index variations and cellular morphology, enabling high-contrast visualization of subcellular features and dry mass distribution relevant to neoplasia [
8,
9]. Spectral Imaging (Multispectral/Hyperspectral Imaging) captures tissue reflectance or fluorescence across multiple wavelengths, revealing biochemical composition (e.g., hemoglobin oxygenation, metabolic states) and spatial distribution of endogenous chromophores that can distinguish healthy from diseased tissue [
10,
11].
Optical Coherence Tomography (OCT) utilizes low-coherence interferometry to provide depth-resolved, micron-scale cross-sectional images of tissue microstructure, analogous to ultrasound but using light, widely used for assessing epithelial and stromal changes in cancers [
12,
13].
Among these, Mueller matrix microscopy (MMM) provides a comprehensive characterization of tissue microstructures, making it increasingly prevalent in cancer detection research [
14,
15].
Cancer, as a major threat to global health, is characterized by high morbidity and mortality, imposing severe physical, psychological, and socioeconomic burdens on patients and their families [
16,
17]. Current clinical cancer detection methods vary widely [
18]. Blood tests, for instance, detect tumor markers such as carcinoembryonic antigen (CEA) and alpha-fetoprotein (AFP) [
19,
20]; however, their specificity and sensitivity are not absolute, often leading to false positives or negatives due to benign conditions. Imaging modalities like X-ray [
21], computed tomography (CT) [
22], magnetic resonance imaging (MRI) [
23], and ultrasound provide spatial and morphological tumor information but struggle to identify early-stage microlesions or determine pathological types [
24].
Only biopsy combined with histopathological diagnosis is universally recognized as the “gold standard” [
25]. However, traditional workflows require individual slides for each sample, necessitating frequent exchanges during large-scale screenings or drug trials, significantly hindering efficiency [
26,
27]. For the challenge, tissue chip technology integrates hundreds of tissue samples into a single substrate, enabling simultaneous processing and analysis. This innovation drastically reduces time and costs, particularly in drug efficacy evaluations where multiple samples are tested concurrently [
28,
29]. Combined with rapid detection technologies, tissue chip holds transformative potential for high-throughput cancer screening.
Meanwhile, artificial intelligence (AI), as a representative of emerging productive forces, promises fully automated pathological diagnostics. Deep learning, a cutting-edge machine learning paradigm, constructs multi-layered neural networks to autonomously extract hierarchical features from vast datasets, demonstrating remarkable efficacy in medical imaging and histopathology. Despite the abundance of stained slide archives from traditional biopsies, their effective utilization remains underexplored. With the rise of AI-driven digital pathology, establishing a polarized light imaging database from existing stained slides could aid pathologists in lesion diagnosis.
In this study, we propose a novel tongue cancer detection method based on MMM. Using this technology, we acquired Mueller matrices from normal tongue tissues and stage II, III, and IV tongue cancers. A dataset of 881 samples with six types of Mueller matrix parameter images was constructed from a tongue cancer tissue chip. Four classic CNN models—AlexNet, ResNet50, DenseNet121, and VGGNet16—were systematically evaluated for detection efficacy under different parameter combinations. Experimental results demonstrated that DenseNet121 achieved a detection accuracy of 98.48% when input with combined parameters (equivalent waveplate fast-axis azimuth, retardance, depolarization, diattenuation, orientation angle, and purity). Compared to full-parameter and individual-element input schemes, this combination improved accuracy by 2.69% and 0.48%, respectively. Our findings provide a theoretical foundation for developing high-precision optical pathology diagnostic systems.
3. Display of Partial Muller Matrix Parameter Images
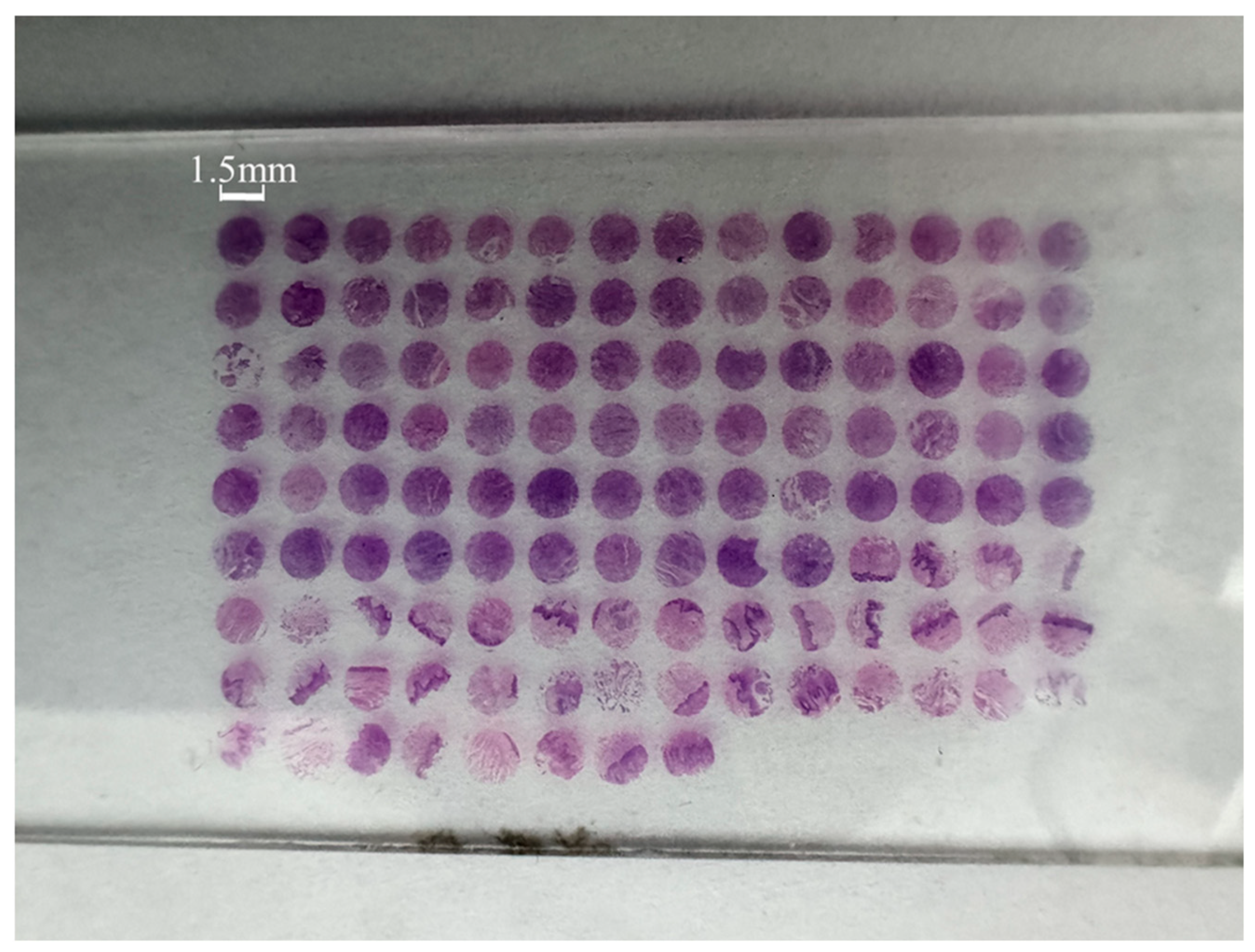
Figure 2 shows four samples in the tissue chip.
Figure 2a represents normal tongue tissue,
Figure 2b represents stage II tongue cancer tissue,
Figure 2c represents stage III tongue cancer tissue, and
Figure 2d represents stage IV tongue cancer tissue. The original microscopic images were collected by a commercial microscope (Olympus BX53M) with an objective magnification of 50×. Through the original microscopic images of the tongue tissue, it can be observed that as the tumor progresses, there are some changes in the microscopic images of each stage. However, distinguishing these changes requires pathologists to carefully observe the tissue samples to identify abnormal areas. This process is not only time-consuming but also prolongs the diagnosis time due to the limited number of pathologists.
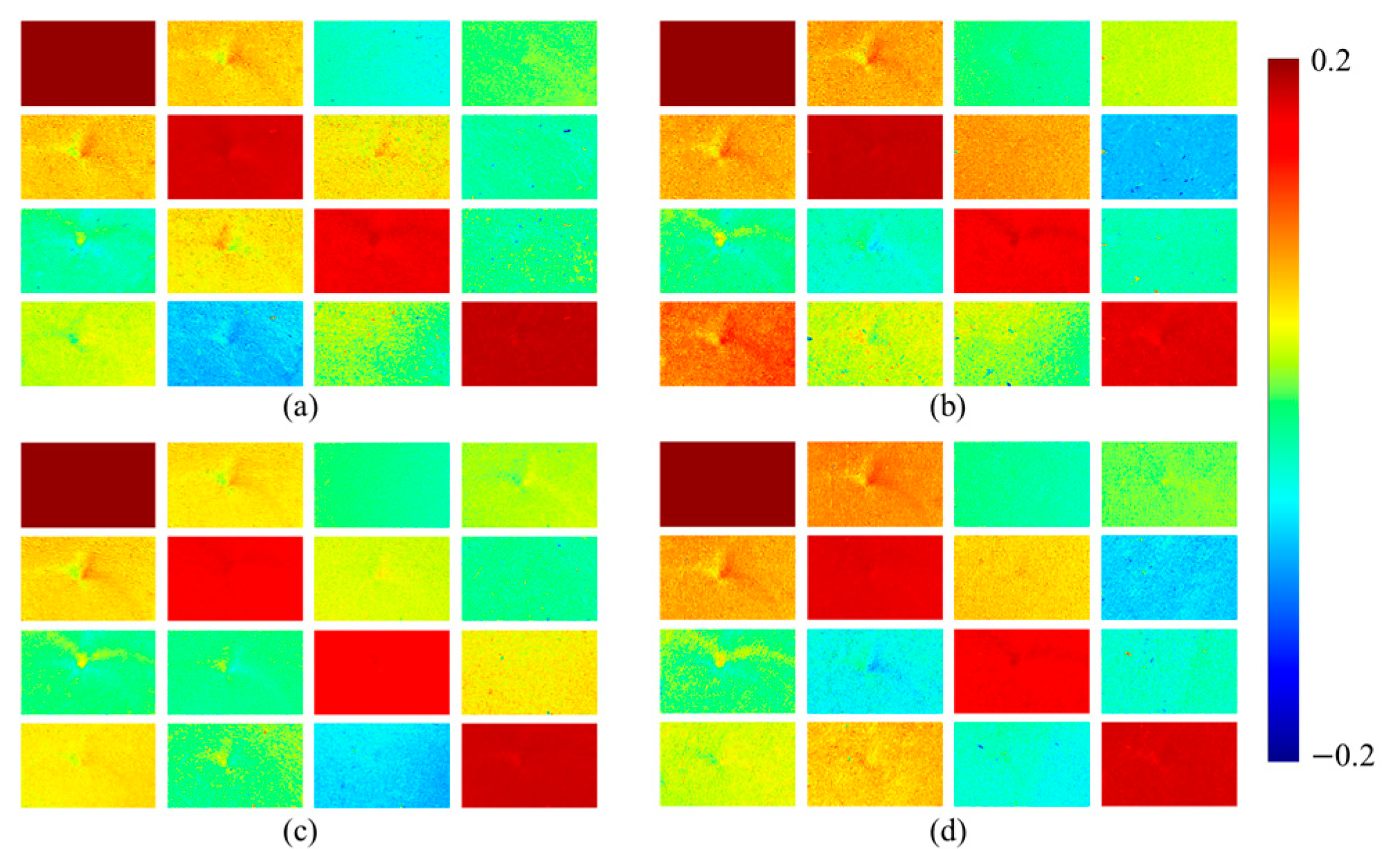
Figure 3 shows the pseudo color images of Mueller matrix elements corresponding to the four tissue samples in
Figure 2. The values of the pseudo color images exhibit a clear spatial distribution between cancer tissue and normal tissue, with positive values (warm colors) representing enhanced linear birefringence and negative values (cold colors) reflecting phase delay orientation shift. The matrix elements
and
primarily characterize the birefringence properties of the sample. As shown in the figure, the matrix elements
and
exhibit noticeable differences in values between cancerous and normal tissues. The values differences between stage II and stage III tongue cancer tissues are minimal, whereas stage IV tissue shows significantly stronger values.
For , there is little difference in values between normal tissue and stage II cancer tissue. However, the values become more pronounced in stage III and IV tissues. This is attributed to cancer progression, during which nuclear volume increases, collagen fibers become increasingly disordered, and the extracellular matrix exhibits greater abnormality, which can significantly affect tissue birefringence.
Although the and images of normal and cancer tissue cannot be visually distinguished, their specific features can be obtained through deep learning. However, these parameters exhibit an inverse correlation with tissue depolarization capacity. Therefore, it is necessary to add depolarization-related parameters into the Mueller matrix parameter image dataset. , , , and can reflect the diattenuation characteristics of the sample. Normal tongue tissue shows differences from cancer tissue at different stages, and deep learning can effectively generalize these diattenuation differences.
Although the Mueller matrix encompasses comprehensive polarization information of the sample, the physical interpretation of each individual element remains ambiguous. In order to obtain polarization parameters with interpretable physical meanings, the Mueller matrix purity introduced in
Section 2.2, the Mueller matrix polar decomposition introduced in
Section 2.3, and the Mueller matrix transformation introduced in
Section 2.4 are further analyzed in detail. Currently, the Mueller matrix parameters that characterize the polarization characteristics of cancerous tissues mainly include the equivalent waveplate fast axis azimuth angle (
), the phase delay (
), the depolarization (
Δ), the bidirectional attenuation (
D), the anisotropic orientation direction (
), and the Mueller matrix purity (
). After acquiring the Mueller matrix of the tongue cancer tissue chip, the Mueller matrix polar decomposition and transformation methods were used to obtain the Mueller matrix parameter images of all samples, and the
images of all samples were obtained using Equation (2), where the parameters derived from the Mueller matrix polar decomposition include
,
, Δ, and
D and the parameter derived from the Mueller matrix transformation is
.
Figure 4 shows the images of six Mueller matrix parameters for four types of tissue samples.
5. Results and Discussion
In the research field of medical image classification, four commonly used CNN models for medical image classification, AlexNet, ResNet50, DenseNet121 and VGGNet16, were used to test the classification accuracy of different combinations of Mueller matrix parameters for different types of tongue cancer tissues and normal tongue tissues. All four models were trained on eight NVIDIA H100 GPUs using the Aadm optimizer to minimize the cross entropy loss function. The learning rate was 0.0001, and the batch size for each GPU was 64. The number of iterations was 30. Model training uses partitioned and preprocessed training set data. To prevent overfitting, all models apply L2 regularization with a weight decay coefficient of 1 × 10−4.
The specific results of the training are presented in
Table 5. It is found that under these four CNN models, the parameter combination
corresponds to the highest level of classification accuracy, which proves that
,
,
,
,
, and
can all be effective parameters for detecting tongue cancer. Among them, within the framework of the DenseNet121 model, the classification accuracy of this parameter combination is particularly outstanding, reaching 98.48%.
Figure 5 shows in detail the variation of loss values with training iterations for the four CNN models under different combinations of detection parameters. Obviously, when using the parameter combination of
, the loss value is more stable.
In other related studies on medical image classification, various methods of inputting Mueller matrix parameters have demonstrated significant outcomes. For instance, a study utilized the method of inputting a comprehensive set of multiple Mueller matrix parameters, including the 16 original matrix elements, 6 polar decomposition parameters, and 6 transformation parameters, into a convolutional neural network (CNN) for the diagnosis of osteosarcoma. This approach achieved a final accuracy of 95.79% [
40]. Another study focused on classifying different types of breast cancer cells by employing 16 Mueller matrix element parameters as CNN inputs, resulting in a classification accuracy of 88.34% [
41]. Furthermore, in the field of dermatological diagnostics, the application of 16 Mueller matrix element parameters as inputs to a CNN for the classification of various skin diseases yielded an accuracy as high as 98% [
42]. Additionally, the use of Mueller matrix elements as CNN inputs for the detection of hepatitis B led to an accuracy rate of 90.9% [
43].
However, when comparing these results with the classification performance of tongue cancer tissue using the parameter combinations, it becomes evident that the accuracy achieved by the latter approach is superior. This observation highlights the potential of the parameter combinations in achieving higher diagnostic accuracy in the specific context of tongue cancer tissue classification. Such comparative analyses underline the importance of parameter selection in enhancing the diagnostic capabilities of CNN-based medical image classification models.
Due to the fact that the parameter combination
has the highest accuracy in tongue cancer detection among the four models, in order to further evaluate its true performance, this study combines resampling techniques and category loss weights to construct a model to explore the diagnostic efficacy of the parameter combination
. The category loss weight value is set to (normal: stage II: stage III: stage IV = 0.2820, 0.3038, 0.1997, 0.2145), and this weight is adjusted based on the number of resampled samples (the smaller the sample size, the greater the weight). Subsequently, the trained model is used to classify the images in the test dataset to evaluate the performance of the parameter combination
. Among 11,800 annotated test images, only 235 images were misclassified under the AlexNet model, 228 images were misclassified under the ResNet50 model, 130 images were misclassified under the DenseNet121 model, and 231 images were misclassified under the VGGNet16 model. The DenseNet model has the highest accuracy, about 98.90%. This once again confirms the effectiveness of deep learning polarization imaging in tongue cancer detection. To present the results more clearly, the relevant confusion matrix is shown in
Table 6,
Table 7,
Table 8 and
Table 9. In addition, classification reports are included in
Table 10,
Table 11,
Table 12 and
Table 13, which evaluate the accuracy, recall, and F1-score of each category as classification metrics. From the tables, it can be seen that these indicators maintain similar and large values among various categories, indicating that parameter combination
has good classification performance.
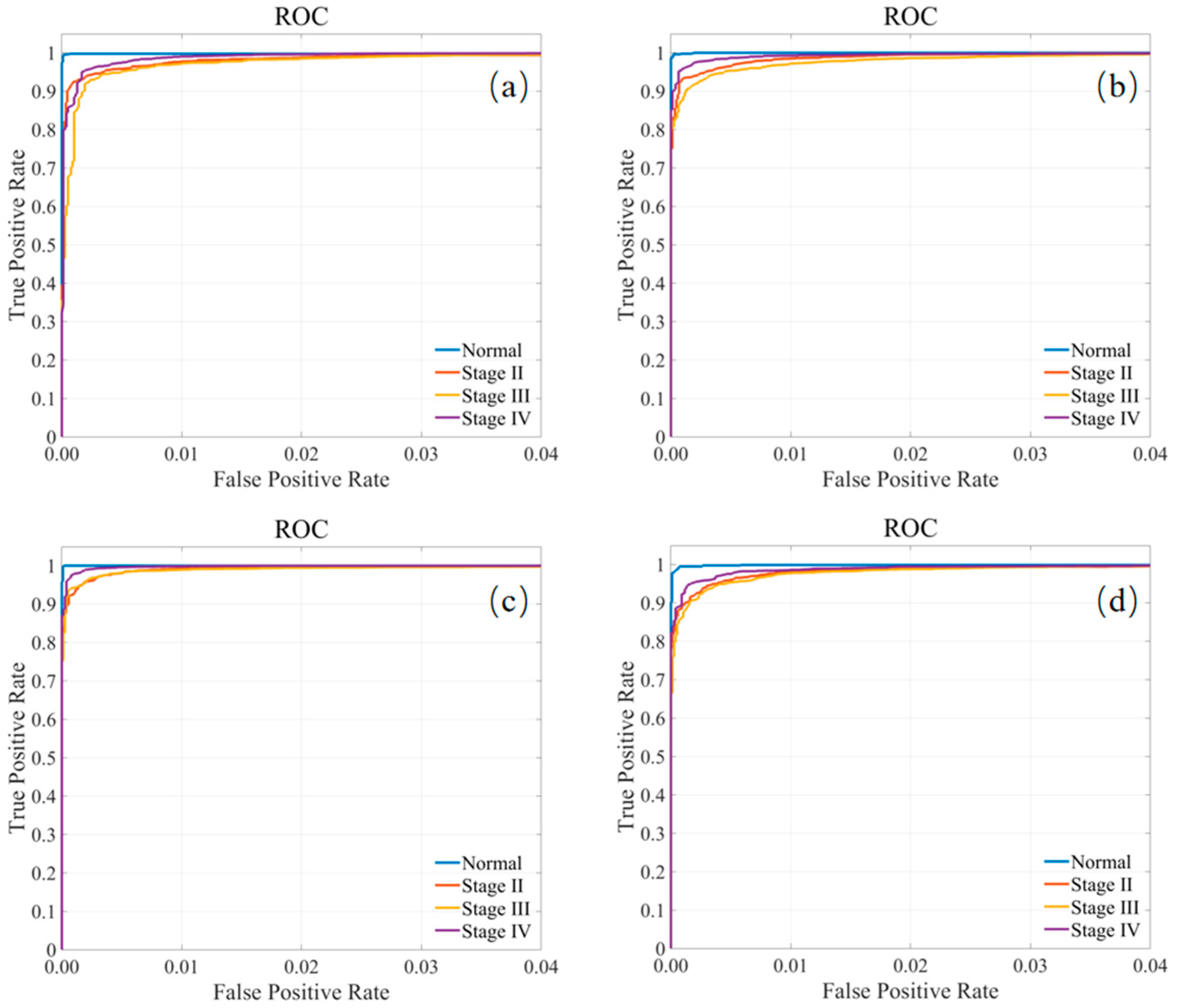
Finally, the receiver operating characteristic (ROC) curves for all tongue cancer tissue classes are shown in
Figure 6, again confirming the good classification capabilities of the model for all classes.
In addition, we also used Grad-CAM to display which regions of normal tongue tissue and stage II, III, and IV tongue cancer tissue images are most relevant to the model.
Figure 7a–d show images of normal tongue tissue and stage II, III, and IV tongue cancer tissue, respectively. Note that the left subimage of each figure is a microscopic image, while the right subimage corresponds to a Grad-CAM image. Normal tongue tissue is a highly ordered structure; therefore, in Grad-CAM images, differentiated mature, morphological regularity, orderly arranged cells, and clear stratification (epithelium, lamina propria, muscle) are the main features. The main features of Grad-CAM images in tongue cancer tissues are highly heterogeneous and malignant characteristics of cells (varying sizes and shapes, abnormal nuclei, active division), complete disorder and destruction of tissue structure, and loss of normal stratification and polarity. The most crucial thing is that cancer cells break through the limitations of the basement membrane and have the ability to infiltrate and grow, invading and destroying deep tissues (lamina propria, muscles). This uncontrolled invasive growth and destructive nature are essential features of cancer.
In summary, through the comparative analysis of multiple experimental results, the significant superiority of this diagnostic parameter combination in the field of medical image classification has been fully verified. In view of this, the combination of diagnostic parameters not only has extremely high application value in the diagnosis of tongue cancer tissue but also has great potential to expand to other meaningful pathological tissue classification work and is expected to provide more accurate and effective technical support for the field of medical diagnosis.








.jpg)


