1. Introduction
The convergence of artificial intelligence and machine learning with medical imaging [
1] is one of the most revolutionary advances in modern healthcare, bringing with it unprecedented potential to improve diagnostic precision, patient outcomes, and democratize access to expert medical knowledge. As global healthcare systems struggle with mounting pressures for fast and effective diagnosis, especially in low-resource environments, AI-driven medical imaging platforms have become essential solutions for meeting these demands. The intersection of cutting-edge deep learning frameworks, innovative image processing algorithms, and explainable AI methods has forged a new standard in medical imaging with the potential to transform the way clinicians diagnose, classify, and plan treatment for disease.
Recent progress in automated stroke detection has shown the tremendous potential of machine learning systems in helping to combat one of the major causes of death and disability across the globe. Salman et al. created the Hemorrhage Evaluation and Detector System for Underserved Populations (HEADS-UP) [
2], a cloud-based machine learning system with a specific application in the detection of intracranial hemorrhage from computed tomography (CT) scans in resource-poor healthcare settings. Leveraging Google Cloud Vertex AutoML with the Radiological Society of North America dataset of 752,803 labeled images, the system attained impressive performance levels of 95.80% average precision, 91.40% precision, and 91.40% recall. The approach utilized highly advanced image preprocessing strategies that merged pixel values from brain, subdural, and soft tissue windows into RGB images, illustrating the way in which data preparation can dramatically improve binary hemorrhage classification performance. Through four rounds of experimentation, the researchers determined that larger training sample size combined with proper image preprocessing had the potential to increase the performance of binary intracranial hemorrhage classification, with the most effective strategy using combined equally weighted RGB images alongside heavily weighted red channel pixel values.
The development of deep learning architectures for stroke detection has seen progressively more advanced approaches, with hybrid models showing remarkable performance potential. Hossain et al. [
3] proposed a new ViT-LSTM model that combines Vision Transformer and Long Short-Term Memory networks for stroke detection and classification in CT images, with accuracy of 94.55% on their main BrSCTHD-2023 dataset from Rajshahi Medical College Hospital and 96.61% on the Kaggle brain stroke dataset. Their method tackled class imbalance problems using innovative data augmentation techniques and included explainable AI methods such as attention maps, SHAP, and LIME to promote clinical interpretability. The model outperformed conventional CNNs and individual ViT models by leveraging spatial feature extraction strengths of Vision Transformers and temporal pattern recognition abilities of LSTM networks. The research focused on primary data collection from a particular regional hospital highlighted the need for localized datasets for mitigating specific clinical complications of stroke patients in underserved communities.
Complementary studies investigating computational efficiency in stroke detection have unveiled the key trade-off between accuracy and deployability in clinical environments. Abdi et al. [
4] proposed a compact CNN model with 97.2% validation accuracy using substantially fewer computational demands of 20.1 million parameters and 76.79 MB memory footprint, marking a 25% parameter and 76% memory decrease compared to state-of-the-art hybrid solutions. Their solution exhibited 89.73% accuracy on an external multi-center dataset comprising 9900 CT images, showcasing the potential as well as difficulties of generalizability in heterogeneous clinical environments. The study introduced the first use of LIME, occlusion sensitivity, and saliency maps for explainable stroke detection in CT imaging, offering clinicians a clear understanding of AI-driven diagnostic outputs. The work stressed the significance of high performance balanced with computational efficiency for real-time applicability in resource-limited clinical environments.
The clinical importance of rapid stroke identification is made even more apparent when placed in the context of the overall picture of acute stroke management, as presented by Patil et al. [
5] in their review of detection, diagnosis, and treatment methodologies for acute ischemic stroke. Ischemic strokes comprise about 85% of all strokes, with large vessel occlusions occurring in 11–29% of acute ischemic stroke cases, thus making timely and accurate identification crucial for best patient outcomes. The field triage stroke scales presently used clinically have considerable limitations with false-positive rates of 50–65% and peak accuracy of only 79% with National Institutes of Health Stroke Scale scores of 11 or more. These limitations clearly point to the pressing need for more advanced diagnostic methodologies offering dependable, rapid evaluation capability both in hospital and pre-hospital environments.
The creation of portable diagnostic technologies is an especially promising direction for the augmentation of stroke detection capabilities, with utility in underserved populations and remote clinical environments. The Strokefinder MD100, created by Medfield Diagnostics, is an example of a microwave-based technology used to detect intracranial hemorrhage with 100% sensitivity and 75% specificity within around 45 s, illustrating the possibility of fast, non-invasive evaluation tools deployable across a range of clinical settings. Likewise, the Lucid™ Robotic System uses transcranial Doppler ultra-sound technology to discriminate patients with large vessel occlusions, with 100% sensitivity and 86% specificity in the discrimination of affected patients from healthy controls, and 91% sensitivity with 85% specificity in patients with suspected stroke. The VIPS-based Visor
® device is another novel strategy, applying volumetric impedance phase shift spectroscopy for the detection of bioimpedance changes related to cerebral edema, with 93% sensitivity and 92% specificity in the discrimination of severe stroke from minor strokes resulting from large vessel occlusions [
5].
State-of-the-art machine learning methods for stroke prognosis and outcome pre-diction have also shown great potential for supporting clinical decision-making beyond the phase of initial diagnosis. Wang et al. [
6] created interpretable machine learning models for the diagnosis of cognitive and motor disorder levels in patients with stroke by MRI analysis, with 92.0% accuracy for cognitive disorder classification with Random Forest using fusion features and 82.5% accuracy for motor disorder classification with Linear Discriminant Analysis using radiomics features. Their method used radiomics and deep features extracted from 3D brain MRI OAx T2 Propeller sequences, which were fused with clinical and demographic data for the prediction of Fugl-Meyer and Mini-Mental State Examination levels. The study used SHAP methodology to ensure explainability at the cohort and individual patient levels, showing that age, baseline symptoms, brain atrophy markers, and early ischemic core features were the most significant predictors of post-stroke functional outcomes.
The application of sophisticated deep learning architectures in medical image classification has yielded remarkable results across multiple domains, with success demonstrated in brain pathology detection and classification. Patel et al. [
7] investigated the effectiveness of EfficientNetB0 for brain stroke classification using CT scans, achieving exceptional performance with 97% classification accuracy, 96% precision, 97% recall, and 97% F1-score. Their methodology incorporated comprehensive data preprocessing and augmentation techniques, including rotations, translations, zooms, and flips to enhance model generalization capabilities, while employing careful hyperparameter optimization to achieve optimal performance. The study’s comparison with other state-of-the-art approaches demonstrated the superiority of the EfficientNetB0 architecture for this specific application, establishing it as a highly effective tool for automated brain stroke classification that could significantly assist clinicians in making rapid and accurate diagnostic decisions.
Predictive modeling of severe stroke complications has become a key application domain, with neural networks proving to be more effective than conventional regression methods. Foroushani et al. [
8] proposed an automatic deep learning system for the pre-diction of malignant cerebral edema following ischemic stroke, leveraging Long Short-Term Memory neural networks to enable 100% recall and 87% precision for the identification of patients who would die with midline shift or require hemicraniectomy. Their method used volumetric information automatically extracted from standard CT imaging, such as cerebrospinal fluid volumes and hemispheric CSF volume ratios, fed through an imaging pipeline that processed both baseline and 24-h follow-up im-aging. The LSTM model proved substantially better than regression models and the validated EDEMA score, with an area under the precision-recall curve of 0.97 versus 0.74 for regression-based methods. The study’s use of SHAP methodology for explainability of the model identified hemispheric CSF ratio and 24-h NIHSS score to be the most predictive features for the development of malignant edema.
Machine learning applications in global stroke outcome prediction have shown the potential for synthesizing varied clinical and imaging data sources to boost prognostic performance. Jabal et al. [
9] created interpretable machine learning models for pre-interventional three-month functional outcome prediction post-mechanical thrombectomy in acute ischemic stroke patients, with 84% area under the receiver operating characteristic curve using Extreme Gradient Boosting with chosen features. Their method involved automated feature extraction from non-contrast CT and CT angiography images, leveraging traditional and novel imaging biomarkers as well as clinical features consistently available during emergency assessment. The study’s use of Shapley Additive Explanation methodology showed that age, baseline NIHSS score, side of occlusion, extent of brain atrophy, early ischemic core, and collateral circulation deficit volume were the most significant classifying features for the prediction of good functional outcomes. This study illustrated the feasibility of developing interpretable ML models that could offer useful decision support in patient triaging and management in acute stroke treatment.
Transfer learning methods have been especially useful in medical imaging applications where small datasets frequently limit model training and development efficacy. Connie et al. [
10] developed the use of transfer learning with VGGFace models for predicting health from facial features with 97% accuracy in classifying healthy versus sick individuals using a novel method that integrated VGGFace with convolutional neural networks as a combined feature extractor. They froze VGGFace layers while fine-tuning CNN parts to derive purified facial features relevant to different health conditions, illustrating how transfer learning can effectively resolve the prevalent issue of small medical imaging datasets while preserving high classification accuracy. The study demonstrated the prospect of non-invasive health examination methods that could augment conventional diagnostic methods, especially in environments where conventional medical imaging may not be easily accessible.
The advent of XAI in medical imaging has overcome one of the greatest obstacles to clinical deployment of AI systems: the “black box” characteristic of deep learning models that prevent clinicians from interpreting and trusting automated diagnostic decisions. Brima and Atemkeng [
11] created an extensive framework for comparing several saliency techniques, including Integrated Gradients, XRAI, GradCAM, GradCAM++, and ScoreCAM, for medical image analysis applications. Their results indicated that ScoreCAM, XRAI, GradCAM, and GradCAM++ consistently yielded focused and clinically meaningful attribution maps that clearly delineated pertinent anatomical structures and disease markers, improving model transparency and engendering clinical trust. Visual examinations indicated that these techniques were able to successfully identify potential biomarkers, reveal model biases, and offer useful insights into input feature to prediction relationships, greatly furthering the aim of making AI systems more interpretable and trustworthy for clinical applications.
Recent developments in 2024–2025 have further solidified the transformative potential of AI in stroke care across multiple domains. Kim et al. [
12] conducted a pivotal multicenter study demonstrating the efficacy of deep learning-based software for automated large vessel occlusion (LVO) detection in CT angiography, achieving a sensitivity of 85.8% and specificity of 96.9% across 595 ischemic stroke patients. Their reader assessment study revealed that AI assistance significantly improved the diagnostic sensitivity of early-career physicians by 4.0%, with an area under the receiver operating characteristic curve improvement of 0.024 compared to readings without AI assistance. The study notably demonstrated that the software achieved a sensitivity of 69% for detecting isolated MCA-M2 occlusions, which are traditionally challenging even for experienced clinicians, and showed exceptional performance within the endovascular therapy time window with 89% sensitivity and 96.5% specificity for patients imaged within 24 h of symptom onset.
Comprehensive reviews of AI applications in stroke care have highlighted the exponential growth and diversification of research in this field. Heo [
13] conducted an extensive scoping review examining 505 original studies on AI applications in ischemic stroke, categorizing them into outcome prediction, stroke risk prediction, diagnosis, etiology prediction, and complication and comorbidity prediction. The review revealed that outcome prediction was the most explored category with 198 articles, followed by stroke diagnosis with 166 articles, demonstrating the field’s maturation across the entire stroke care continuum. The analysis showed significant advancement in AI models for large vessel occlusion detection with clinical utility, automated imaging analysis and lesion segmentation streamlining acute stroke workflows, and predictive models addressing stroke-associated pneumonia and acute kidney injury for improved risk stratification and resource allocation.
The establishment of comprehensive longitudinal stroke datasets has emerged as a critical foundation for advancing AI research in stroke care. Riedel et al. [
14] introduced the ISLES’24 dataset, providing the first real-world longitudinal multimodal stroke dataset consisting of 245 cases with comprehensive acute CT imaging, follow-up MRI after 2–9 days, and longitudinal clinical data up to three-month outcomes. This dataset includes vessel occlusion masks from acute CT angiography and delineated infarction masks in follow-up MRI, offering unprecedented possibilities for investigating infarct core expansion over time and developing machine learning algorithms for lesion identification and tissue survival prediction. The multicenter nature of the dataset, spanning institutions in Germany and Switzerland with diverse scanner models and manufacturers, enables the development of robust and generalizable stroke lesion segmentation algorithms that reflect real-world stroke scenarios.
Systematic evaluation of AI stroke solutions has provided comprehensive insights into the current state of commercial and research-based platforms. Al-Janabi et al. [
15] conducted a systematic review of 31 studies examining AI applications for stroke detection, focusing primarily on acute ischemic stroke and large vessel occlusions with platforms including Viz.ai, RapidAI, and Brainomix
®. The review demonstrated that these AI tools consistently achieved high levels of diagnostic accuracy, with RapidAI showing 87% sensitivity for large vessel occlusion detection, Viz.ai demonstrating significant reductions in door-to-needle time from 132.5 to 110 min, and Brainomix
® achieving sensitivity ranging from 58.6% to 98% depending on the specific application. The analysis revealed that AI platforms have proven particularly valuable in reducing treatment times, improving workflow efficiency, and providing standardized technological systems for detecting LVOs and determining ASPECT scores.
The clinical feasibility of automated lesion detection on non-contrast CT has achieved significant milestones with large-scale validation studies. Heo et al. [
16] developed and externally validated automated software for detecting ischemic lesions on non-contrast CT using a modified 3D U-Net model trained on paired NCCT and diffusion-weighted imaging data from 2214 patients with acute ischemic stroke. The model achieved 75.3% sensitivity and 79.1% specificity in external validation of 458 subjects, with NCCT-derived lesion volumes correlating significantly with follow-up DWI volumes (ρ = 0.60,
p < 0.001). Importantly, the study demonstrated clinical relevance by showing that lesions greater than 50 mL were associated with reduced favorable outcomes and higher hemorrhagic transformation rates, while radiomics features improved hemorrhagic transformation prediction beyond clinical variables alone, achieving an area under the receiver operating characteristic curve of 0.833 versus 0.626 for clinical variables alone.
In this study, the research has great potential for implementation in emergency departments and resource-limited settings, where access to specialist neurological expertise is frequently unavailable. By facilitating quick and reliable stroke assessment, the system can help healthcare professionals make time-critical decisions, especially regarding the administration of tPA within the tight therapeutic window. The combination of high-performance classification algorithms with explainability capabilities guarantees that clinicians are not just presented with precise outcomes but also interpretable insights that can reinforce diagnostic confidence. Additionally, the fact that a clinician-friendly interface is provided improves the usability of the system.
2. Materials and Methods
This research uses a high-quality cranial CT imaging dataset [
17] specially curated for the Artificial Intelligence in Healthcare competition, held under the TEKNOFEST 2021 technology festival in Istanbul, Türkiye. The Turkish Health Institutes (TÜSEB) organized the competition with the support of the Ministry of Health and the General Directorate of Health Information Systems. The main goal of the competition was to create machine learning solutions for the automatic detection and classification of stroke from CT images, a particularly critical task in clinical decision support wherein diagnostic accuracy and speed are very important.
The dataset itself consists of a total of 6653 cross-sectional CT brain images acquired in the axial plane. These images were retrospectively collected over the course of 2019 and 2020 via Türkiye’s national e-Pulse and Teleradiology System infrastructure from patients admitted to emergency departments with acute neurological symptoms clinically suggestive of stroke, in accordance with standard acute stroke imaging protocols. Using diagnosis-related codes and strict filtering criteria, relevant cases were identified and anonymized in accordance with data protection standards. A total of seven experienced radiologists were tasked with manually reviewing, annotating, and validating the dataset, ensuring high clinical reliability and consistency across all image labels.
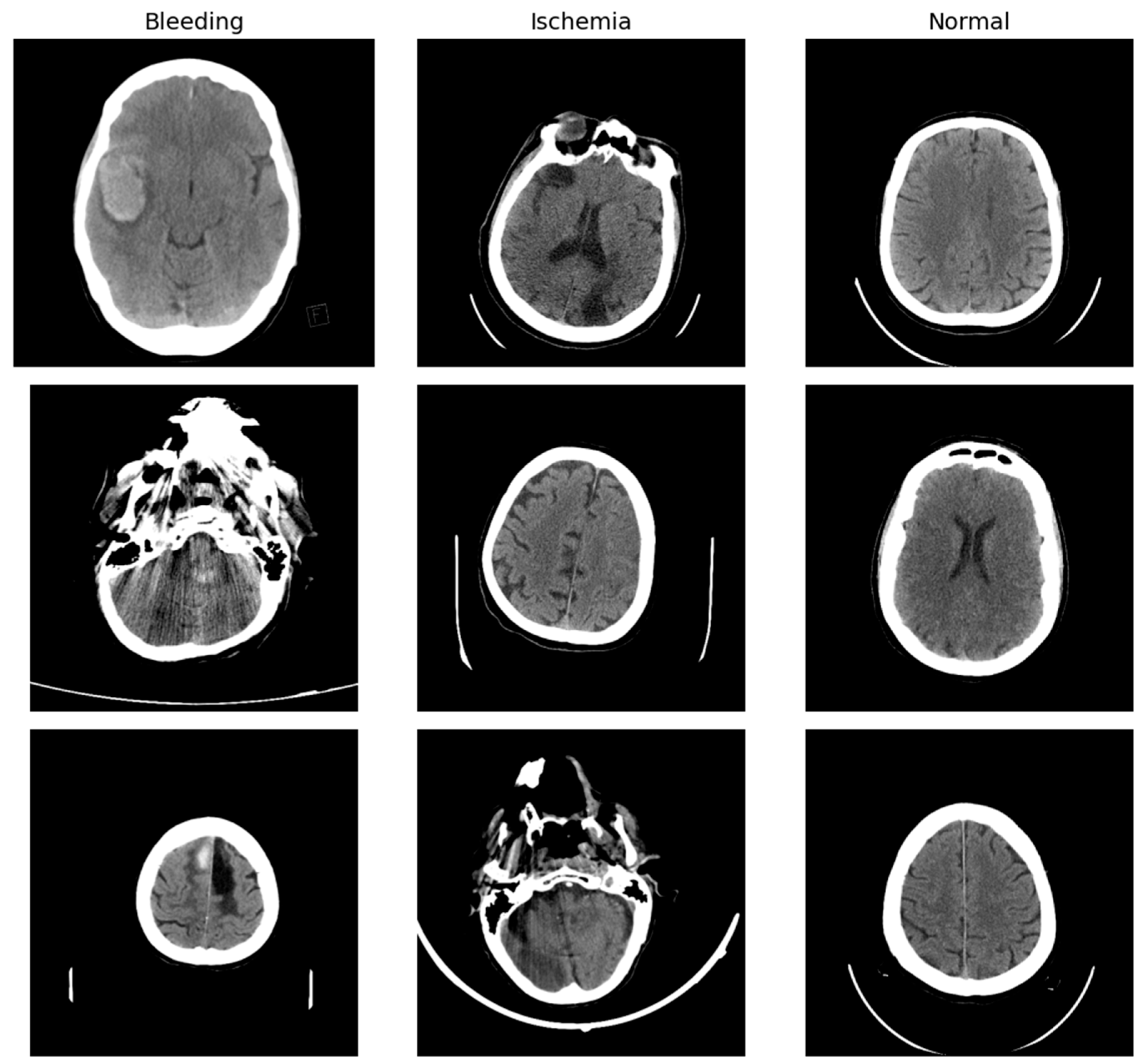
Each CT scan slice in the dataset is labeled with one of three diagnostic categories: normal (indicating no evidence of stroke), ischemic stroke, or hemorrhagic stroke due to intracranial bleeding. Among the 6653 images, 4428 are labeled as normal, 1131 as ischemic stroke, and 1094 as hemorrhagic stroke (
Figure 1). This relatively imbalanced distribution, especially the predominance of normal scans, reflects the clinical reality in emergency radiology where non-stroke cases are common among individuals presenting with neurological symptoms. The dataset is hosted openly by the Turkish Ministry of Health and is cited with its publication in the Eurasian Journal of Medicine [
17].
To prepare the dataset for machine learning pipelines, a systematic series of data engineering and preprocessing procedures was carried out. First, the data set was reorganized at the file system level, where image samples were collected into a common directory structure consistent with typical image classification libraries like PyTorch 1.10’s ImageFolder. The original class-wise folders—i.e., ‘Bleeding’, ‘Ischemia’, and ‘Normal’—were normalized to a lowercase, hyphenated convention: ‘bleeding’, ‘ischemia’, and ‘normal’, respectively, in line with naming conventions that minimize case-sensitivity errors and enhance cross-platform compatibility.
Next, all image files were programmatically checked for integrity and discarded if they were non-conforming or corrupted. File formats were normalized to .jpg or .png, and image sizes were optionally resized or padded to consistent sizes (e.g., 224 × 224 pixels) using libraries such as OpenCV or PIL, to enable downstream batching and model input compatibility. Metadata, if any, was stripped or anonymized to keep the focus on image-based features. This whole pipeline provided a reproducible, structured, and model-agnostic dataset, poised for efficient consumption by deep learning architectures.
Image preprocessing was carried out using a bespoke transformation pipeline that balanced the twin goals of preserving clinically relevant features and promoting model generalization through data augmentation. All images were first resized to 224 × 224 pixels to align with the input requirements of standard deep convolutional neural networks, those pre-trained on natural image datasets such as ImageNet. The resizing operation ensures aspect ratio preservation and spatial dimension uniformity for the entire dataset.
To enhance the robustness of the model to differences in acquisition conditions and patient anatomy, the training images were subjected to a series of stochastic transformations. These comprised random horizontal and vertical flipping, as well as modest rotations up to fifteen degrees. Horizontal flipping uses the bilateral symmetry of the brain to improve generalization across lesion lateralization. Vertical flipping, although not representative of typical acquisition orientations, was included as an aggressive augmentation strategy to promote robustness in a low-data setting. Affine transformations were additionally applied to mimic subtle patient positioning shifts, and color jittering was introduced to modulate brightness and contrast, albeit within a controlled fashion to prevent distortion of clinical features. Given that CT images are intrinsically grayscale, they were transformed into three-channel grayscale images to match the expected input format of pretrained networks but otherwise retained their original intensity-based structure. Post-augmentation, all pixel values were normalized with a mean and standard deviation of 0.5 for all channels. This normalization step maps pixel values to a standardized range of roughly −1 to 1, which is advantageous for training stability.
After the images had been preprocessed, the dataset was divided into training, validation, and testing subsets. The division was performed based on a stratified random sampling approach to maintain the original class distribution in each subset, a necessary step to avoid model bias. Seventy percent of the images were allocated to the training set, fifteen percent to the validation set, and fifteen percent to the test set. To achieve reproducibility, a fixed random seed was set when creating the splits.
The ensuing distribution of images within the three classes in each subset is provided in the following table (
Table 1). The training set comprised 765 cases of hemorrhagic stroke, 791 ischemic cases, and 3099 normal cases. There were 179 hemorrhagic stroke images, 176 ischemic images, and 642 normal images in the validation set. In the test set, there were 149 hemorrhagic stroke cases, 163 ischemic cases, and 686 normal cases. These numbers highlight the class imbalance in the dataset, and particularly the relatively high frequency of normal scans in relation to pathological cases. The stratification process, nonetheless, maintained this imbalance consistently across all three subsets of data, enabling the model to learn and be tested under real-world diagnostic distribution conditions.
By going through these preprocessing and data handling processes, the dataset was successfully converted from a raw clinical repository to a well-structured and well-balanced corpus for deep learning purposes. This groundwork permitted consistent training conditions, reduced data-related biases, and provided a basis for solid model assessment.
3. Experimental Setup
To construct a robust deep learning model for the classification of stroke types from CT brain scans, a staged training strategy founded on transfer learning and a ResNet-18 backbone was used. This architecture was chosen because it offered a balance of model depth, parameter efficiency, and performance, particularly well-suited for medical imaging tasks where training data is limited. The ResNet-18 model was initialized with weights pretrained on the ImageNet dataset, allowing the network to utilize feature representations learned from a large and diverse corpus of natural images. The final fully connected classification layer was redefined to produce three output probabilities corresponding to the target diagnostic categories: bleeding, ischemia, and normal.
The training procedure was conducted in two stages (
Figure 2). During the first phase, the whole network was fine-tuned, with all layers, including the pretrained ones, being allowed to adjust their weights. This end-to-end training was conducted with a moderate learning rate of 1 × 10
−4, chosen to guarantee stable convergence without degrading the representational capabilities of the pretrained base. The model was trained for 10 epochs with the Adam optimizer and categorical cross-entropy loss function. The loss was calculated as the negative log-likelihood of the predicted probability of the true class label and is the standard choice for multi-class classification with softmax outputs. Training was conducted on a T4 GPU with a batch size of 32 per optimization step.
To further enhance performance, a fine-tuning phase was conducted that re-cycled the model weights obtained in the initial training. During this phase, a smaller learning rate of 1 × 10−5 was utilized to allow for more subtle and refined weight adjustments. Fi-ne-tuning for an additional five epochs was conducted. This second phase permitted the model to continue adapting to the subtleties of the medical imaging task without overfitting or reversing the generalizable features learned in the initial phase.
To alleviate the problem of class imbalance, specifically the overrepresentation of normal cases, we included a weighted cross-entropy loss function during fine-tuning. The weight for each class was computed inversely proportional to its frequency in the training dataset. This approach placed higher penalty on misclassifying the minority classes (hemorrhagic stroke and ischemia), thus pushing the model to have balanced performance in all classes and avoid bias in the learned decision boundaries.
During both phases of training, the performance of the model was constantly checked with a validation set from the same distribution as the training set. Both accuracy and loss measures were noted at each epoch to assess training development. At the last epoch of fine-tuning, the model had a validation accuracy of 96% and training accuracy of 98%, reflecting successful learning and generalization. These best-performing model weights during this phase were saved and subsequently used for testing on the held-out test set.
To determine the model’s ability to generalize, it was tested on the independent test set and calculated a set of standard classification metrics. These were overall accuracy and class-specific precision, recall, and F1-score, all calculated from the confusion matrix. For each test image, the predicted class label was assigned using the argmax of the softmax probability outputs, corresponding to the class with the highest predicted probability. The model performed at a test accuracy of 95%, with strong performance throughout all three diagnostic classes. Alongside scalar metrics, a full classification report was created using the scikit-learn library and presented a breakdown of precision, recall, and F1-score for each class in isolation.
To aid in the interpretation of these results, a visual heatmap of the classification report was generated. This visualization allowed for an intuitive understanding of performance asymmetries and reinforced the model’s robustness, particularly in differentiating ischemic from hemorrhagic stroke presentations.
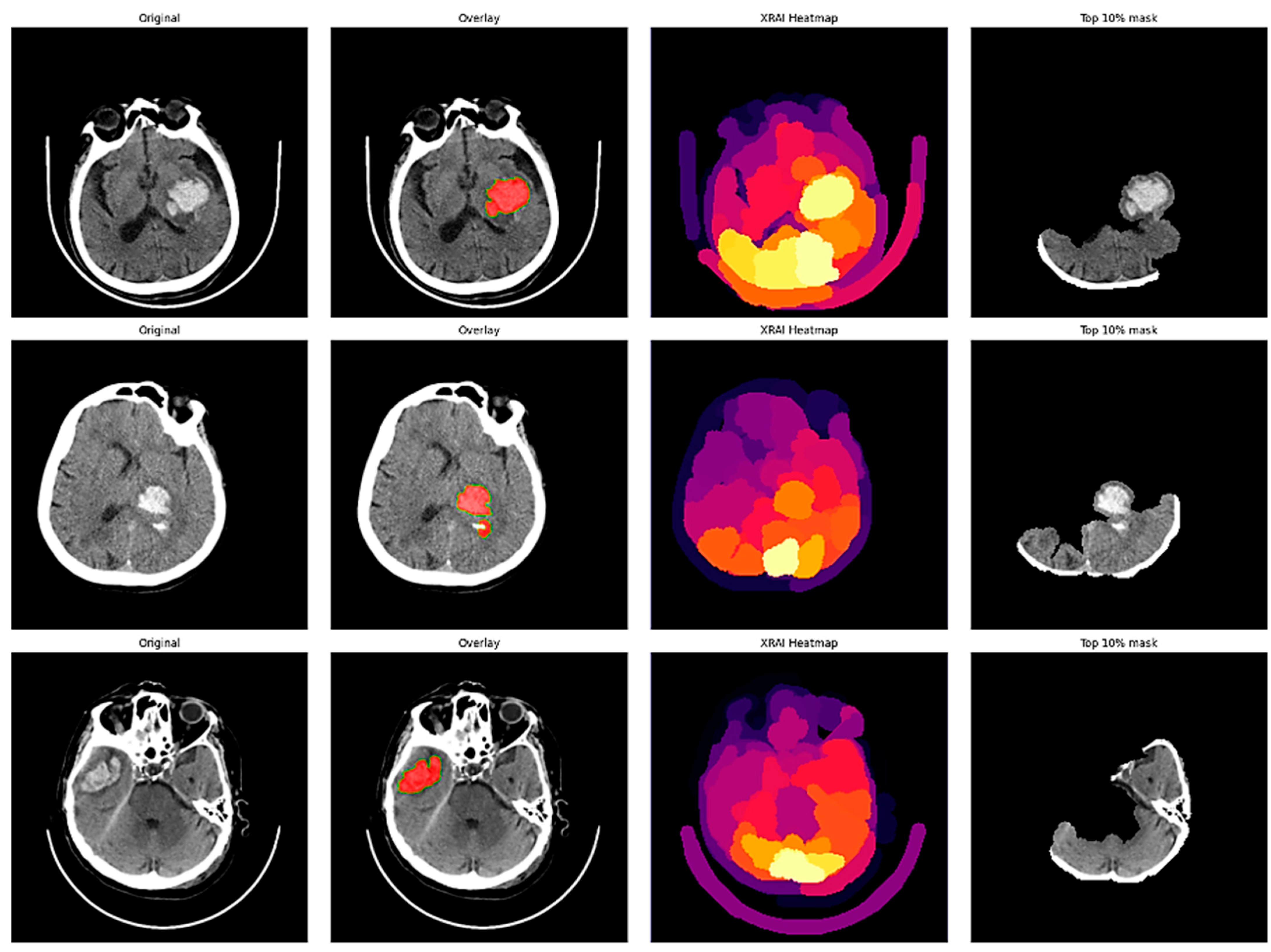
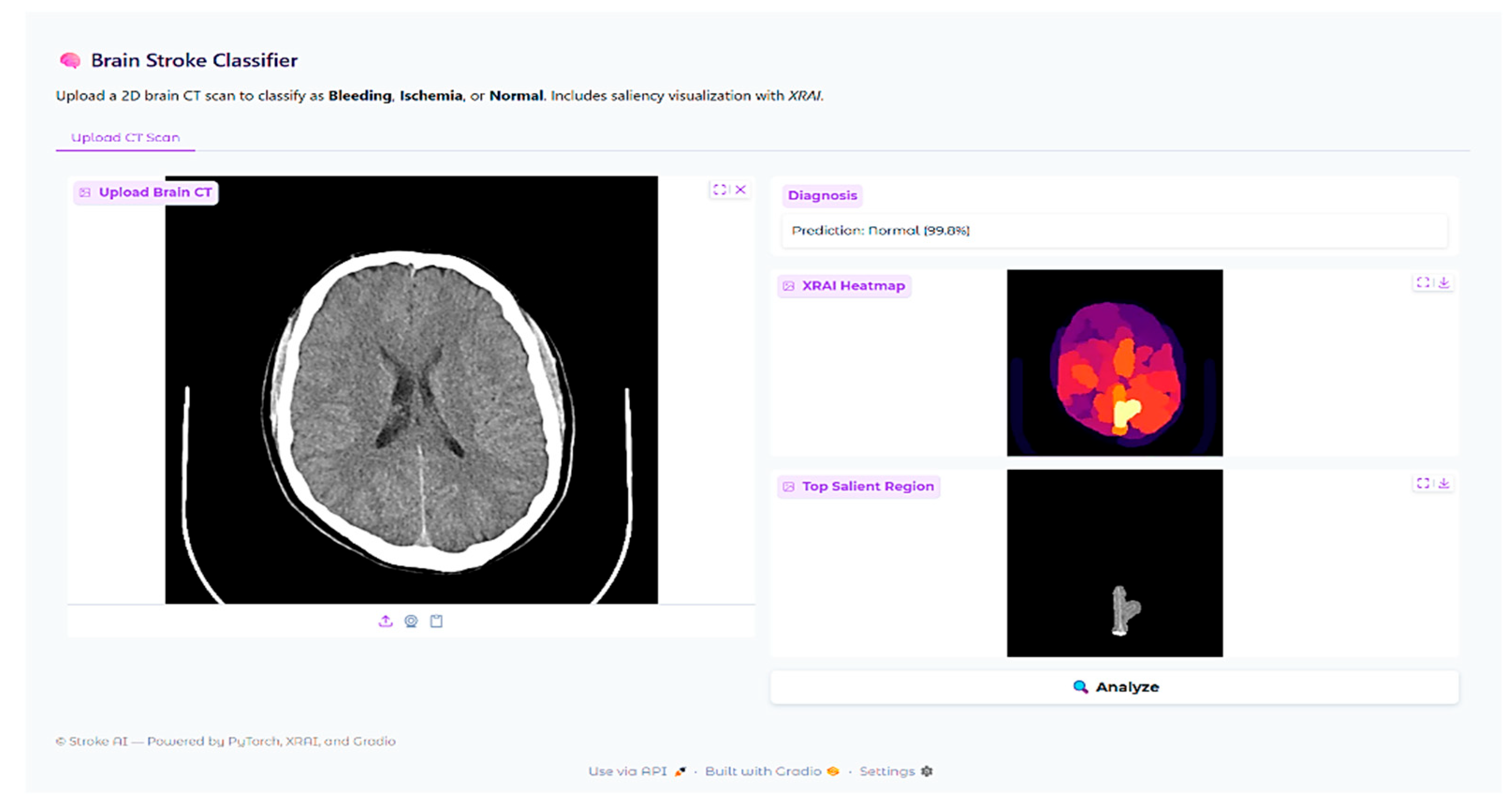
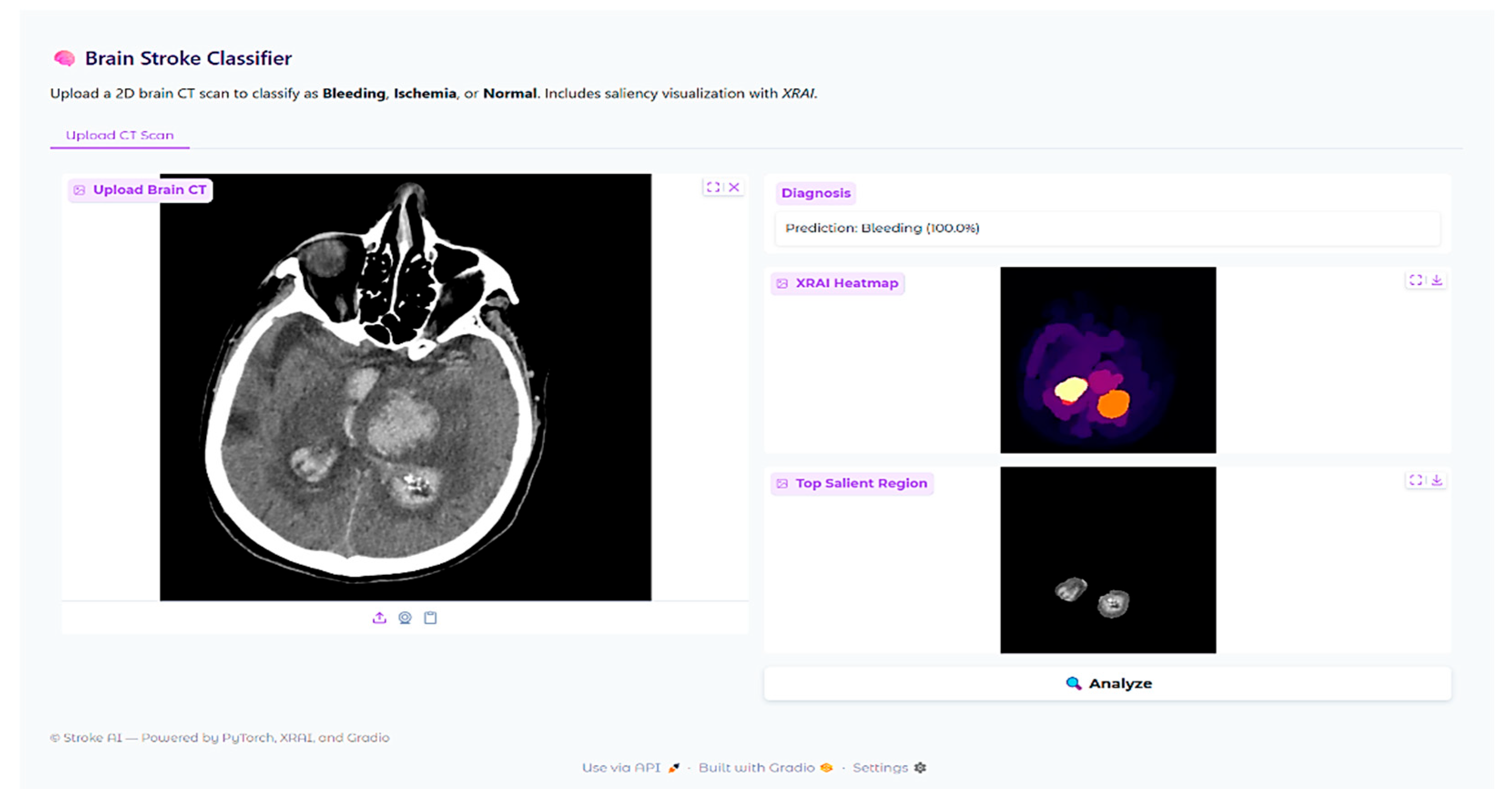
In addition to scalar performance metrics, explainable AI was incorporated to better understand model decision-making. The XRAI technique is a region-based attribution method that provides visual explanations for deep neural network predictions by identifying the most important image regions that drive the model’s decisions. Unlike gradient-based methods that operate pixel-wise, XRAI aggregates attributions in semantically meaningful regions and is therefore particularly suitable for medical imaging applications where understanding spatial context of pathologic features is crucial for clinical validation.
The XRAI deployment was paired with the fine-tuned ResNet-18 model with the saliency library. An explainability pipeline was created to pass CT brain images through the same preprocessing transformations as during training, such as resizing to 224 × 224 pixels, converting to RGB from grayscale, and normalizing. The model’s prediction for each input image was retrieved, and XRAI attribution maps were created by calculating input–output gradients for the predicted class. The attribution scores were plotted as heatmaps with the “inferno” colormap, with lighter regions representing larger attribution values. A thresholded visualization was also prepared by considering the top 10% of the attribution scores to highlight the most important regions identified by the model.
4. Results
4.1. Model Training Performance
ResNet-18 was trained in the two-stage process as explained in the experimental setup. The training process is shown in
Figure 3, where initial training and fine-tuning stages are plotted in accuracy graphs on training and validation sets. In the initial training stage (epochs 1–10), the model showed steep learning with training accuracy rising from around 80% to 95% by epoch 8 before leveling off. The validation accuracy also showed the same trend with almost 93% by the completion of the first training process, with some fluctuations between epochs 3 and 6.
Fine-tuning phase, epochs 11 to 15, saw the continuation of the model’s performance development. Training accuracy increased from 95% to about 98%, and validation accuracy from 93% to 96%. The concurrent development of the training and validation curves during these phases is a hallmark of successful learning without deep overfitting. The relatively small gap between training and validation accuracy is a sign of the great generalization ability of the trained model. This implies that the model not only learned from the new data but also managed to preserve its capability to generalize well with new unseen examples.
4.2. Test Set Performance
Following the completion of both the initial training and fine-tuning stages, the performance of the model was tested on a held-out test set to determine its ability to generalize. The final model achieved an overall test accuracy of 95% when evaluated on the independent test set, demonstrating very good generalization ability to unseen data. The classification performance is depicted in
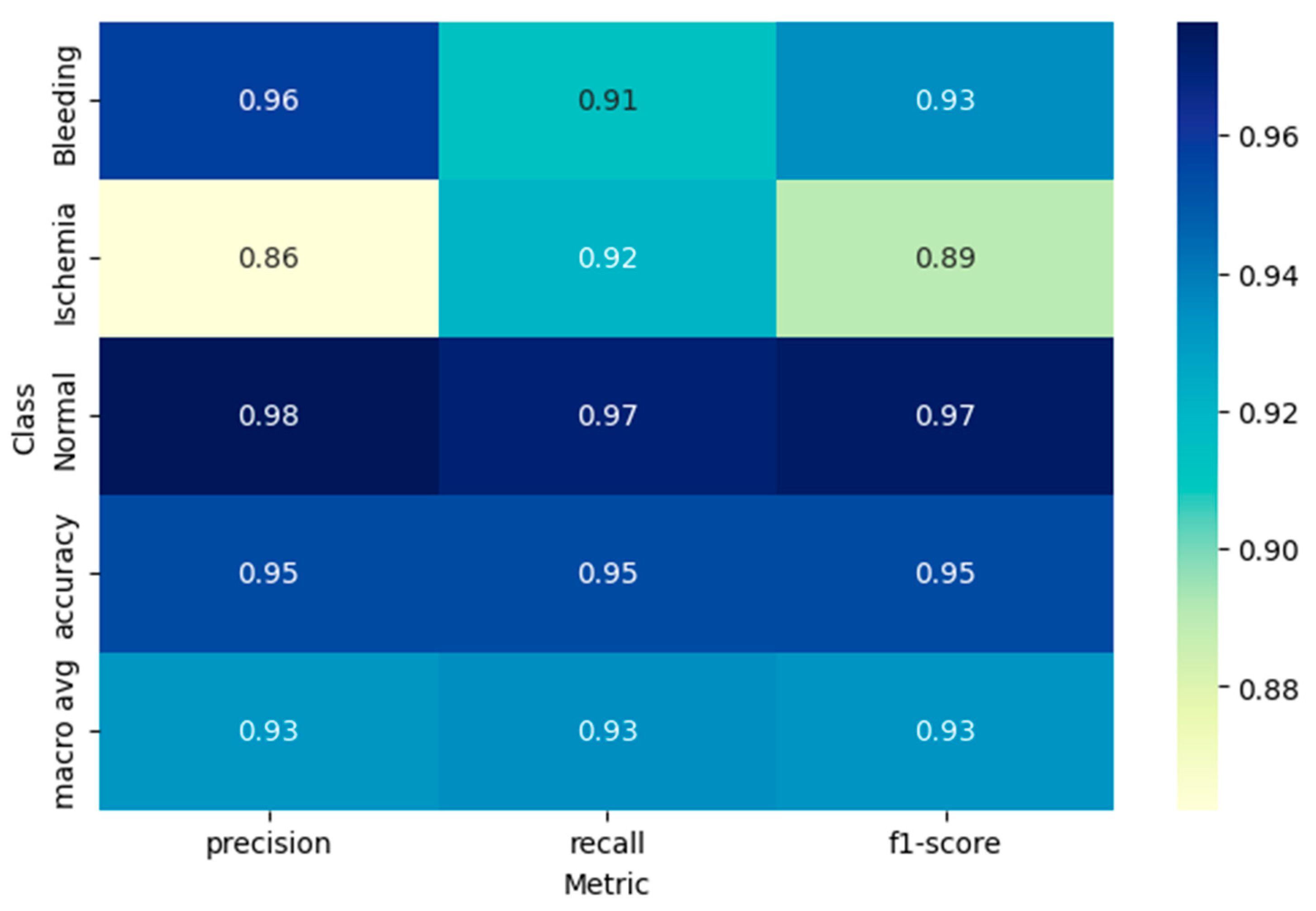
Figure 4, showing the heatmap of the classification report with precision, recall, and F1-score metrics for each diagnostic class.
For the hemorrhagic stroke cases, which are the hemorrhagic stroke cases, the model had a precision of 0.96, recall of 0.91, and F1-score of 0.93. This means when the model predicted hemorrhagic stroke, it was right 96% of the time and identified 91% of all the true hemorrhagic stroke cases in the test set correctly. The high accuracy also suggests a low rate of false positive predictions for hemorrhagic stroke, which is clinically important because misclassifying normal or ischemic cases as hemorrhagic could lead to incorrect treatment decisions.
The ischemic stroke class had accuracy 0.86, recall 0.92, and F1-score 0.89. The model accurately predicted 92% of true ischemic cases, although it was slightly less precise than for hemorrhagic cases. The rate of recall is especially crucial in the diagnosis of ischemic strokes since failure to identify these cases can postpone essential treatments like thrombolytic therapy.
Classification of normal cases achieved the best performance metrics with precision equal to 0.98, recall equal to 0.97, and F1-score equal to 0.97. Better performance for normal cases illustrates the ability of the model to distinguish between pathological and normal brain imaging, which is crucial for reducing false positives in clinical practice.
The macro-averaged F1-score, precision, and recall were 0.93 across all the classes, reflecting balanced performance across the diagnostic classes. The weighted average F1-score = 0.95, precision = 0.96, and recall = 0.95, considering class distribution, reflect high model performance overall.
4.3. Confusion Matrix Analysis
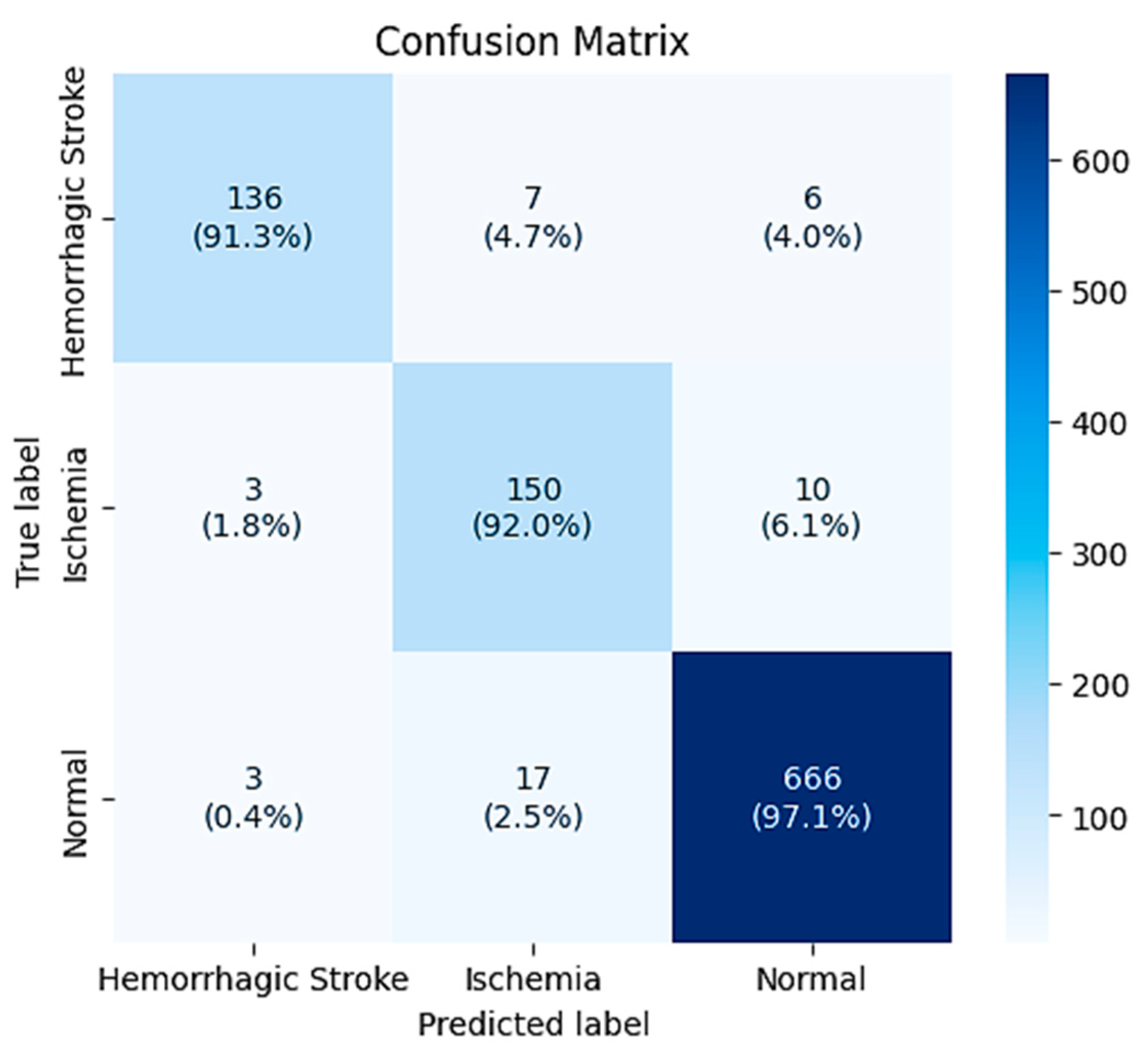
The confusion matrix (
Figure 5) provides an overview of the classification performance of the model on the provided 998 test instances. The matrix shows the distribution of correct classifications along the diagonal and misclassifications on the off-diagonal entries.
In cases of hemorrhagic stroke (n = 149), 136 cases (91.3%) were correctly classified, and 7 were misclassified as ischemia and 6 as normal. Hemorrhagic stroke-ischemia confusion accounts for 4.7% of hemorrhagic stroke cases and could reflect difficult cases with confounding imaging characteristics or pathologic characteristics that are atypical.
Ischemic cases (n = 163) comprised 150 correct diagnoses (92.0%), 3 misclassified as hemorrhagic stroke, and 10 as normal. The low incidence of ischemia-to-hemorrhagic stroke misclassification (1.8%) is clinically desirable, since this kind of error can result in inappropriate treatment. Nevertheless, 6.1% of ischemic cases were diagnosed as normal and therefore could potentially delay interventions that are needed.
In all instances, classification accuracy was optimal, as demonstrated by correct classification of 666 of 686 cases (97.1%). Misclassification of just 3 normal cases as hemorrhagic stroke and 17 as ischemia was seen, corresponding to false positive rates of 0.4% and 2.5%, respectively. The low false positive rate for hemorrhagic stroke is clinically important in that it prevents unnecessary fear of hemorrhagic stroke in patients with no abnormalities.
The overall trend in the confusion matrix is that the model is most confident when separating normal from pathological cases, and that the hardest classifications are between the two types of strokes. This finding is consistent with clinical expectations, in that the differentiation between ischemic and hemorrhagic stroke is commonly difficult even for experienced radiologists, particularly in subtle or early pathology.
4.4. Class-Specific Performance Analysis
Measurement metrics illustrate significant trends for the model’s diagnostic performance across different types of strokes. The increased effectiveness seen in the hemorrhagic instances (bleeding) is likely reflective of the distinct imaging characteristics seen with acute hemorrhage, expressed as hyperdense regions on CT scans and providing a stark visual contrast to normal brain tissue. This distinctive appearance improves the ability for reliable automated detection and classification.
The modestly lower performance of ischemic stroke detection is explained by the more subtle imaging manifestations of acute ischemia, especially within the initial hours after stroke onset when CT changes can be minimal. Ischemic areas usually manifest as hypodense regions with loss of gray-white matter differentiation, which can be more difficult to automatically detect compared with the overt hyperdensity of hemorrhage.
The excellent performance shown in normal cases proves the model has successfully learned the ability to identify the typical features of normal brain anatomy and can consistently separate pathologic from physiological imaging findings. This function is extremely important for clinical applications because it guarantees the system will not return an excessive amount of false positives with the potential to undermine clinician confidence or detract from workflow efficiency.
6. Conclusions
An explainable AI and clinical decision support-enabled stroke classification system based on deep learning with 95% test accuracy was developed. ResNet-18 performed well across diagnostic categories, with especially good hemorrhagic stroke detection (precision 0.96, recall 0.91) due to the characteristic hyperdense CT appearance of acute bleeding. Slightly reduced performance for ischemic stroke (0.86) is in line with clinical practice in that subtle early ischemic changes may be present on CT scan, but high recall (0.92) significantly reduces missed diagnoses acutely.
XRAI explainability experiments revealed both encouraging strengths and serious weaknesses. While attribution maps generally correlated well with radiologist annotations in hemorrhage cases, there were instances with concerning patterns of mislocalization, e.g., highlighting skull-based regions rather than the actual lesions. In ischemic strokes, the model consistently favored high-contrast anatomical borders over subtle parenchymal abnormalities, with reliance on anatomical landmarks rather than pathological tissue characteristics. These findings demonstrate that the most critical issue is this: high classification accuracy does not guarantee clinically acceptable decision-making processes. The observed discrepancies may reflect both the model’s learned features and the suitability of the XRAI attribution method for medical imaging tasks.
The single-center data used in this study may not capture the heterogeneity of imaging protocols and patient populations globally and would need to be externally validated across institutions. The explainability analysis, although comprehensive, was based on a single attribution scheme and would be enhanced by calling upon validation using multiple methods to instill higher confidence in observed trends.
This study highlights the potential and limitations of creating clinically applicable AI systems for medical diagnosis. Although technical performance is promising, the results regarding explainability demonstrate the key value of testing AI reasoning processes beyond conventional accuracy assessment, especially in high-risk medical uses where an understanding of the underlying rationale for diagnostic judgments is essential to assure patient safety and facilitate physician trust.
The web-based deployment also offers important clinical utility to emergency departments and resource-limited settings for acute diagnostic support when neurology experts are not available. It allows for crucial treatment decisions, such as tPA administration within therapeutic time windows. The real-time processing capability and intuitive interface design allow integration into existing clinical workflows while maintaining transparency with visual explanations. While explainability analysis offered valuable insights into the model’s decision-making processes, some limitations were encountered that should be further investigated before more extensive clinical deployment. The integration of classification performance, interpretability analysis, and clinical interface design constitutes a valuable road map for the development of medically applicable AI systems. Future work should be directed toward further improving the correspondence between features and clinical reasoning, investigating additional directions of explainability, and conducting prospective, multi-center hospital validation studies. Such studies are essential to evaluate the system’s performance within real-world clinical workflows, confirm generalizability across diverse patient populations and imaging protocols, and ensure safe and effective deployment in the healthcare setting.