Influence of Post-Milking Treatment on Microbial Diversity on the Cow Teat Skin and in Milk
Abstract
:1. Introduction
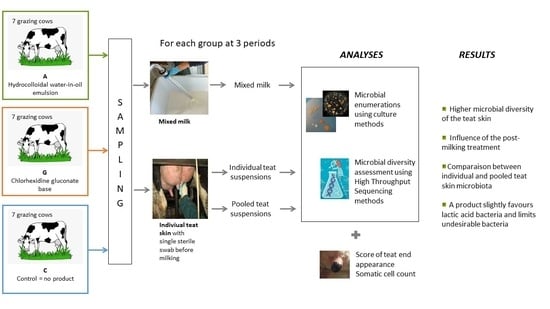
2. Materials and Methods
2.1. Experimental Design
2.2. Sampling
2.3. Appearance of Teat-End and Udder Health
2.4. Analyses
2.5. Statistical Analyses and Bioinformatics
3. Results
3.1. Teat Condition and Somatic Cell Counts
3.2. Microbial Enumeration Using Culture Methods
3.3. Microbial Diversity Assessment Using High Throughput Sequencing Methods
3.4. Phylogenetic Profiles of Teat and Milk Microbial Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doyle, C.J.; Gleeson, D.; O’Toole, P.W.; Cotter, P.D. Impacts of seasonal housing and teat preparation on raw milk microbiota: A high-throughput sequencing study. Appl. Environ. Microbiol. 2017, 83, e02694-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skeie, S.B.; Håland, M.; Thorsen, I.M.; Narvhus, J.; Porcellato, D. Bulk tank raw milk microbiota differs within and between farms: A moving goalpost challenging quality control. J. Dairy Sci. 2019, 102, 1959–1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oikonomou, G.; Addis, M.F.; Chassard, C.; Nader-Macias, M.E.F.; Grant, I.; Delbès, C.; Bogni, C.I.; Le Loir, Y.; Even, S. Milk Microbiota: What Are We Exactly Talking about? Front. Microbiol. 2020, 11, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montel, M.C.; Buchin, S.; Mallet, A.; Delbès-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef]
- Verdier-Metz, I.; Gagne, G.; Bornes, S.; Monsallier, F.; Veisseire, P.; Delbès-Paus, C.; Montel, M.C. Cow Teat Skin, a Potential Source of Diverse Microbial Populations for Cheese Production. Appl. Environ. Microbiol. 2012, 78, 326–333. [Google Scholar] [CrossRef] [Green Version]
- Frétin, M.; Martin, B.; Rifa, E.; Verdier-Metz, I.; Pomiès, D.; Ferlay, A.; Montel, M.C.; Delbès, C. Bacterial community assembly from cow teat skin to ripened cheeses is influenced by grazing systems. Sci. Rep. 2018, 8, 200. [Google Scholar] [CrossRef] [Green Version]
- Doyle, C.J.; Gleeson, D.; O’Toole, P.W.; Cotter, P.D. High-throughput metataxonomic characterization of the raw milk microbiota identifies changes reflecting lactation stage and storage conditions. Int. J. Food Microbiol. 2017, 255, 1–6. [Google Scholar] [CrossRef]
- Monsallier, F.; Verdier-Metz, I.; Agabriel, C.; Martin, B.; Montel, M.C. Variability of microbial teat skin flora in relation to farming practices and individual dairy cow characteristics. Dairy Sci. Technol. 2012, 92, 265–278. [Google Scholar] [CrossRef] [Green Version]
- Gleeson, D.; O’Brien, B.; Flynn, J.; O’Callaghan, E.; Galli, F. Effect of pre-milking teat preparation procedures on the microbial count on teats prior to cluster application. Ir. Vet. J. 2009, 62, 461. [Google Scholar] [CrossRef] [Green Version]
- Fitzpatrick, S.R.; Garvey, M.; Flynn, J.; Jordan, K.; Gleeson, D. Are some teat disinfectant formulations more effective against specific bacteria isolated on teat skin than others? Acta Vet. Scand. 2019, 61, 21. [Google Scholar] [CrossRef]
- Singh, O.; Gupta, D.K.; Singh, R.S.; Sharma, S.; Bansal, B.K. Comparison of Post Milking Teat Disinfection Alone With Pre and Post Milking Teat Disinfection in the Prevention of Bovine Mastitis. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 1467–1474. [Google Scholar] [CrossRef]
- Fitzpatrick, S.R.; Garvey, M.; Jordan, K.; Flynn, J.; O’Brien, B.; Gleeson, D. Screening commercial teat disinfectants against bacteria isolated from bovine milk using disk diffusion. Vet. World 2019, 12, 629–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skowron, K.; Sękowska, A.; Kaczmarek, A.; Grudlewska, K.; Budzyńska, A.; Białucha, A.; Gospodarek-Komkowska, E. Comparison of the effectiveness of dipping agents on bacteria causing mastitis in cattle. Ann. Agric. Environ. Med. 2019, 26, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ren, Y.; Xi, X.X.; Huang, W.; Zhang, H. A novel lactobacilli-based teat disinfectant for improving bacterial communities in the milks of cow teats with subclinical mastitis. Front. Microbiol. 2017, 8, 1782. [Google Scholar] [CrossRef] [PubMed]
- Reeves, S.K.; Borchers, M.R.; Bewley, J.M. Evaluating Teat Skin Condition in Response to Phenoxyethanol as a Post-Milking Teat Disinfectant on Lactating Dairy Cows. J. Dairy Vet. Anim. Res. 2017, 6, 291–296. [Google Scholar] [CrossRef]
- Zened, A.; Forano, E.; Delbès, C.; Verdier-Metz, I.; Morgavi, D.; Popova, M.; Ramayo-Caldas, Y.; Bergonier, D.; Meynadier, A.; Marie-Etancelin, C. Les microbiotes des ruminants: État des lieux de la recherche et impacts des microbiotes sur les performances et la santé des animaux. INRAE Prod. Anim. 2021, 33, 249–260. [Google Scholar] [CrossRef]
- Kamal, R.M.; Bayoumi, M.A. Efficacy of premilking and postmilking teat dipping as a control of subclinical mastitis in Egyptian Dairy cattle. Int. Food Res. J. 2015, 22, 1037–1042. [Google Scholar]
- Lago, A.; Bruno, D.R.; Lopez-Benavides, M.; Leibowitz, S. Short communication: Efficacy of glycolic acid-based and iodine-based postmilking barrier teat disinfectants for prevention of new intramammary infections in dairy cattle. J. Dairy Sci. 2016, 99, 7467–7472. [Google Scholar] [CrossRef] [Green Version]
- Sieber, R.L.; Farnsworth, R.J. Prevalence of chronic teat-end lesions and their relationship to intramammary infection in 22 herds of dairy cattle. J. Am. Vet. Med. Assoc. 1981, 178, 1263–1267. [Google Scholar]
- Irobi, J.; Schoofs, A.; Goossens, H. Genetic identification of Candida species in HIV-positive patients using the polymerase chain reaction and restriction fragment length polymorphism analysis of its DNA. Mol. Cell. Probes 1999, 13, 401–406. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Mills, D.A. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 2013, 79, 2519–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theil, S.; Rifa, E. rANOMALY: AmplicoN wOrkflow for Microbial community AnaLYsis. F1000Research 2021, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.P.; King, S.H.; Lewis, M.J.; Torre, P.M.; Matthews, K.R.; Dowlen, H.H. Efficacy of Chlorhexidine as a Postmilking Teat Disinfectant for the Prevention of Bovine Mastitis During Lactation. J. Dairy Sci. 1990, 73, 2230–2235. [Google Scholar] [CrossRef]
- Hillerton, J.E.; Cooper, J.; Morelli, J. Preventing bovine mastitis by a postmilking teat disinfectant containing acidified sodium chlorite. J. Dairy Sci. 2007, 90, 1201–1208. [Google Scholar] [CrossRef] [Green Version]
- Martins, C.M.M.R.; Pinheiro, E.S.C.; Gentilini, M.; Benavides, M.L.; Santos, M.V. Efficacy of a high free iodine barrier teat disinfectant for the prevention of naturally occurring new intramammary infections and clinical mastitis in dairy cows. J. Dairy Sci. 2017, 100, 3930–3939. [Google Scholar] [CrossRef]
- Morrill, K.M.; Scillieri Smith, J.C.; Dann, H.M.; Gauthier, H.M.; Ballard, C.S. Evaluation of powdered 0.5% chlorhexidine acetate-based postmilking teat dip compared with a foamed 1% iodine-based postmilking teat dip under cold weather conditions in northern New York. J. Dairy Sci. 2019, 102, 2507–2514. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, M.D.; Larsen, H.D. The Effect of Post Milking Teat Dip and Suckling on Teat Skin Condition, Bacterial Colonisation, and Udder Health. Acta Vet. Scand. 1998, 39, 443–452. [Google Scholar] [CrossRef]
- Wattenburger, K.; Schmidt, R.; Placheta, L.; Middleton, J.R.; Adkins, P.R.F. Evaluation of 4 different teat disinfection methods prior to collection of milk samples for bacterial culture in dairy cattle. J. Dairy Sci. 2020, 103, 4579–4587. [Google Scholar] [CrossRef]
- Pantoja, J.C.F.; Correia, L.B.N.; Rossi, R.S.; Latosinski, G.S. Association between teat-end hyperkeratosis and mastitis in dairy cows: A systematic review. J. Dairy Sci. 2020, 103, 1843–1855. [Google Scholar] [CrossRef]
- Monsallier, F.; Couzy, C.; Chatelard-Chauvin, C.; Bouton, Y.; Feutry, F.; Verdier-Metz, I.; Hulin, S.; Montel, M.C. Accompagner les producteurs laitiers pour orienter les équilibres microbiens des laits en faveur de la qualité des fromages au lait cru. Innov. Agron. 2016, 49, 267–279. [Google Scholar]
- Verdier-Metz, I.; Boudon, M.C.; Fournier, F.; Pradel, P.; Monsallier, F.; Montel, M.C. Effect of post dipping treatment after milking on teat and milk microbiota. In Proceedings of the Science of Artisan Cheese, Online, 18 August 2014; Neal’s Yard Dairy; North Cadbury Court: Somerset, UK. [Google Scholar]
- Widyastuti, Y.R.; Febrisiantosa, A. The Role of Lactic Acid Bacteria in Milk Fermentation. Food Nutr. Sci. 2014, 5, 435–442. [Google Scholar] [CrossRef] [Green Version]
- Blaya, J.; Barzideh, Z.; LaPointe, G. Interaction of starter cultures and nonstarter lactic acid bacteria in the cheese environment 1. J. Dairy Sci. 2017, 101, 3611–3629. [Google Scholar] [CrossRef] [PubMed]
- Kable, M.E.; Srisengfa, Y.; Laird, M.; Zaragoza, J.; McLeod, J.; Heidenreich, J.; Marco, M.L. The core and seasonal microbiota of raw bovine milk in tanker trucks and the impact of transfer to a milk processing facility. MBio 2016, 7, e00836-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breitenwieser, F.; Doll, E.V.; Clavel, T.; Scherer, S.; Wenning, M. Complementary Use of Cultivation and High-Throughput Amplicon Sequencing Reveals High Biodiversity Within Raw Milk Microbiota. Front. Microbiol. 2020, 11, 1557. [Google Scholar] [CrossRef]
- Kamimura, B.A.; Cabral, L.; Noronha, M.F.; Baptista, R.C.; Nascimento, H.M.; Sant’Ana, A.S. Amplicon sequencing reveals the bacterial diversity in milk, dairy premises and Serra da Canastra artisanal cheeses produced by three different farms. Food Microbiol. 2020, 89, 103453. [Google Scholar] [CrossRef]
- Callon, C.; Duthoit, F.; Delbès, C.; Ferrand, M.; Le Frileux, Y.; De Crémoux, R.; Montel, M.C. Stability of microbial communities in goat milk during a lactation year: Molecular approaches. Syst. Appl. Microbiol. 2007, 30, 547–560. [Google Scholar] [CrossRef]
- Mallet, A.; Guéguen, M.; Kauffmann, F.; Chesneau, C.; Sesboué, A.; Desmasures, N. Quantitative and qualitative microbial analysis of raw milk reveals substantial diversity in fl uenced by herd management practices. Int. Dairy J. 2012, 27, 13–21. [Google Scholar] [CrossRef]
- Delavenne, E.; Mounier, J.; Asmani, K.; Jany, J.-L.L.; Barbier, G.; Le Blay, G. Fungal diversity in cow, goat and ewe milk. Int. J. Food Microbiol. 2011, 151, 247–251. [Google Scholar] [CrossRef]
- Hohmann, M.F.; Wente, N.; Zhang, Y.; Krömker, V. Bacterial load of the teat apex skin and associated factors at herd level. Animals 2020, 10, 1647. [Google Scholar] [CrossRef]
- Andrews, T.; Neher, D.A.; Weicht, T.R.; Barlow, J.W. Mammary microbiome of lactating organic dairy cows varies by time, tissue site, and infection status. PLoS ONE 2019, 14, e0225001. [Google Scholar] [CrossRef] [Green Version]
- Coulon, J.B.; Dauver, F.; Garel, J.P. Facteurs de variation de la numération cellulaire du lait chez des vaches laitières indemnes de mammites cliniques. Prod. Anim. 1996, 9, 133–139. [Google Scholar] [CrossRef]
- Olde Riekerink, R.G.M.; Barkema, H.W.; Stryhn, H. The effect of season on somatic cell count and the incidence of clinical mastitis. J. Dairy Sci. 2007, 90, 1704–1715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmoslemany, A.M.; Keefe, G.P.; Dohoo, I.R.; Wichtel, J.J.; Stryhn, H.; Dingwell, R.T. The association between bulk tank milk analysis for raw milk quality and on-farm management practices. Prev. Vet. Med. 2010, 95, 32–40. [Google Scholar] [CrossRef] [PubMed]

| A | G | C | SEM | p-Value | |
|---|---|---|---|---|---|
| Before experiment (P1) | 1.38 | 1.64 | 1.50 | 0.093 | 0.541 |
| Beginning of experiment (P2) | 1.62 a,b | 1.73 b | 1.25 a | 0.067 | 0.006 |
| Middle of experiment | 1.64 b | 2.00 b | 1.07 a | 0.077 | <0.001 |
| End of experiment (P3) | 1.56 b | 2.17 c | 1.13 a | 0.077 | <0.001 |
| Log(SCC/1000) | A | G | C | SEM | p-Value |
|---|---|---|---|---|---|
| Before experiment (P1) | 1.66 | 1.99 | 1.83 | 0.064 | 0.081 |
| Beginning of experiment (P2) | 1.57 a | 1.88 b | 1.80 a,b | 0.043 | 0.008 |
| Middle of experiment | 1.84 a | 1.91 a | 2.31 b | 0.058 | 0.001 |
| End of experiment (P3) | 2.00 | 2.00 | 2.25 | 0.053 | 0.084 |
| Group 1 | Period 2 | p-Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Microbial Populations | A | G | C | P1 | P2 | P3 | SEM | Group | Period | Group × Period |
| Milk 3 (n = 27) | ||||||||||
| Total mesophilic bacteria | 3.55 | 3.87 | 3.80 | 3.90 | 3.57 | 3.75 | 0.19 | 0.24 | 0.30 | 0.12 |
| Gram-negative bacteria | 2.73 | 2.50 | 2.66 | 2.36 | 2.80 | 2.74 | 0.20 | 0.50 | 0.08 | 0.98 |
| Lactic acid bacteria | 2.88 a | 2.35 b | 2.57 a,b | 2.22 b | 2.93 a | 2.65 a,b | 0.19 | 0.05 | 0.005 | 0.54 |
| Ripening population | 2.68 | 2.41 | 2.47 | 1.93 b | 2.96 a | 2.66 a | 0.12 | 0.11 | <0.001 | 0.27 |
| Yeasts | 2.99 a | 2.42 b | 2.65 b | 2.41 b | 2.77 a | 2.88 a | 0.10 | <0.001 | 0.001 | 0.03 |
| Moulds | 0.77 | 0.22 | 0.64 | 0.16 b | 1.06 a | 0.42 b | 0.23 | 0.07 | <0.001 | 0.20 |
| Pooled teat suspensions 4 (n = 27) | ||||||||||
| Total mesophilic bacteria | 4.92 | 5.09 | 5.26 | 4.16 c | 5.88 a | 5.23 b | 0.19 | 0.22 | <0.001 | 0.59 |
| Gram-negative bacteria | 3.87 | 4.14 | 4.37 | 2.56 b | 4.93 a | 4.90 a | 0.28 | 0.23 | <0.001 | 0.79 |
| Lactic acid bacteria | 3.14 | 2.50 | 2.64 | 2.15 b | 3.47 a | 2.65 a,b | 0.38 | 0.24 | 0.01 | 0.09 |
| Ripening population | 4.13 | 4.34 | 4.08 | 3.27 b | 4.73 a | 4.54 a | 0.39 | 0.77 | <0.001 | 0.15 |
| Yeasts | 2.12 | 2.07 | 2.13 | 1.67 b | 2.22 a,b | 2.45 a | 0.97 | 0.01 | 0.77 | |
| Moulds | 1.34 | 1.55 | 1.64 | 1.35 | 1.82 | 1.40 | 0.44 | 0.13 | 0.55 | |
| Individual teat suspensions 4 (n = 189) | ||||||||||
| Total mesophilic bacteria | 4.77 | 4.81 | 4.91 | 3.66 c | 5.71 a | 5.11 b | 0.11 | 0.43 | <0.001 | 0.03 |
| Gram-negative bacteria | 3.38 b | 3.70 a,b | 3.78 a | 1.78 b | 4.63 a | 4.44 a | 0.17 | 0.04 | <0.001 | 0.44 |
| Lactic acid bacteria | 2.72 | 2.48 | 2.51 | 1.64 b | 3.30 a | 2.77 a | 0.17 | 0.30 | <0.001 | 0.02 |
| Ripening population | 4.00 | 3.96 | 4.19 | 3.13 c | 4.68 a | 4.33 b | 0.11 | 0.08 | <0.001 | <0.001 |
| Yeasts | 1.71 | 1.72 | 1.77 | 1.14 b | 1.93 a | 2.14 a | 0.86 | <0.001 | 0.57 | |
| Moulds | 1.34 | 1.33 | 1.52 | 1.07 c | 1.71 a | 1.40 b | 0.12 | <0.001 | 0.34 | |
| Group | Period | Group × Period 1 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A | G | C | p-Value | P1 | P2 | P3 | p-Value | p-Value | |
| 16S (Bacteria) | |||||||||
| Chao1 | |||||||||
| Milk | 79.1 | 90.0 | 98.2 | 0.66 | 65.1 b | 114.2 a | 82.9 b | 0.003 | 0.02 |
| Individual Teat | 110.2 | 115.8 | 109.0 | 0.55 | 118.9 a | 124.9 a | 92.3 b | <0.001 | <0.001 |
| Pooled Teat | 130.5 | 116.9 | 100.6 | 0.11 | 134.6 | 120.2 | 85.0 | 0.05 | 0.05 |
| Shannon | |||||||||
| Milk | 3.1 | 3.4 | 3.4 | 0.16 | 3.2 | 3.5 | 3.2 | 0.09 | 0.20 |
| Individual Teat | 3.0 | 3.1 | 3.0 | 0.51 | 3.0 ab | 3.1 a | 2.9 b | 0.02 | 0.002 |
| Pooled Teat | 3.3 | 3.2 | 3.1 | 0.55 | 3.2 | 3.3 | 3.2 | 0.53 | 0.40 |
| ITS2 (Fungi) | |||||||||
| Chao1 | |||||||||
| Milk | 15.9 | 19.6 | 15.7 | 0.09 | 14.3 | 20.8 | 15.6 | 0.06 | 0.07 |
| Individual Teat | 43.9 | 43.3 | 43.2 | 0.99 | 27.9 c | 60.6 a | 43.8 b | <0.001 | <0.001 |
| Pooled Teat | 58.0 | 51.5 | 25.6 | 0.20 | 31.5 | 68.4 | 26.5 | 0.06 | 0.21 |
| Shannon | |||||||||
| Milk | 1.2 | 1.2 | 1.3 | 0.61 | 1.3 | 1.3 | 1.2 | 0.46 | 0.35 |
| Individual Teat | 2.4 | 2.6 | 2.6 | 0.36 | 1.9 b | 3.1 a | 2.6 a | 0.0010 | 0.02 |
| Pooled Teat | 2.6 | 2.4 | 2.0 | 0.55 | 2.0 | 3.0 | 1.7 | 0.14 | 0.48 |
| Abundance (%) | ||||
|---|---|---|---|---|
| Teat | ||||
| ASV | Species | Milk | Pool | Individual |
| Bacteria (16S rRNA) | Actinobacteria | |||
| 46fe2c89eaf45507201f73d05f7dd682 | Kocuria salsicia | 1.00 | − | 0.00 |
| 4c0a7b78bc28cd297d940358c90fca11 | Kocuria salsicia_varians | 1.92 | 0.08 | 0.09 |
| Bacteroidetes | ||||
| 45a0f682d1b59ec4c012c6edc5a8593f | Chryseobacterium haifense | 1.05 | 0.02 | 0.05 |
| Firmicutes | ||||
| 88a4abe161399fb663f20a3249edd2c6 | Aerococcus urinaeequi_viridans | 1.67 | 3.58 | 3.48 |
| bdcf0a98c3a746ae82f10262eb2cc273 | Clostridioides difficile_bartlettii | 2.62 | 6.11 | 6.08 |
| 260ed04a6ad8d8fb320e4803f51ee938 | Clostridium disporicum_saudiense | 0.76 | 1.65 | 1.66 |
| 662e2046cdeb3803a324da7c7d4b55ed | Clostridium disporicum_saudiense | 0.28 | 1.14 | 1.04 |
| 4dbaad38558ae942a1e56ae96ba14f4a | Exiguobacterium aestuarii | 0.04 | 1.59 | 2.65 |
| 2535950da4f5ce2e05509946880e1912 | Lactococcus lactis | 7.05 | 0.05 | 0.07 |
| 19c50dacd1bcf20c7ee29330b0565eb6 | Pediococcus stilesii_acidilactici | 1.36 | 0.00 | 0.01 |
| 851262e388f9eda58b0f5016acc737fe | Pediococcus stilesii_acidilactici | 1.03 | 0.00 | 0.01 |
| de38e8e17d9c8f260f6dbb99561881ab | Paeniclostridium sordellii | 4.99 | 10.40 | 9.92 |
| e0c59b317f7537c66626d5f49ea44a89 | Romboutsia sedimentorum | 1.63 | 3.19 | 3.45 |
| ff2c64ded51bdfc792648dca80a3d375 | Romboutsia timonensis | 11.13 | 23.12 | 21.65 |
| 09e64bf0e07c9a7959b90f5bb78d63b2 | Staphylococcus aureus | 1.09 | 0.21 | 0.26 |
| 3ea7d63cf8427d340ff7a71adf338fee | Staphylococcus haemolyticus_petrasii | 0.29 | 1.30 | 1.73 |
| 42697dcb9c518dcc6bf2e48c1ae1b276 | Staphylococcus petrasii | 6.13 | 2.73 | 3.07 |
| ad306fc9d2bd79082dc8e044dfe8588d | Streptococcus uberis_porcinus | 1.01 | 0.01 | 0.02 |
| 56deff739fb6bfbc421cccbba6170ad6 | Turicibacter sanguinis | 2.14 | 5.06 | 4.52 |
| Proteobacteria | ||||
| 2d711873254cf9212d36b48150030c42 | Acinetobacter albensis_lwoffii | 0.01 | 1.08 | 0.58 |
| 239d5c60225d2e64c348234e9a85de3b | Acinetobacter haemolyticus | 0.01 | 1.65 | 1.43 |
| b7176533054c4a045208143afd50f67b | Acinetobacter indicus | 0.32 | 6.01 | 3.23 |
| 3239a6358dec42f7d5288124e6639ebd | Moraxella osloensis | 5.42 | 0.64 | 1.87 |
| 2e127c4643317603c1522a72a25ab663 | Pseudomonas alcaliphila_chengduensis_toyotomiensis_oleovorans | 1.02 | 1.57 | |
| 542870b5c028dea2cf7adbddcf28f104 | Pseudomonas indoloxydans | 0.08 | 4.37 | 6.17 |
| f2a166e1c7a8de0a617183907f36e3ff | Ralstonia pickettii | 9.52 | 0.22 | 0.44 |
| Fungi (ITS) | Moulds | |||
| be901ea53977fe923f96a7495b0bd739 | Ampelomyces quisqualis | 0.00 | 1.16 | 0.03 |
| daf19ea76f64fac5bdbd4ee6db820021 | Cladosporium crousii_pini-ponderosae_colombiae | 0.04 | 3.68 | 4.41 |
| 619fa4027595729b90c2b18fb38ff24a | Cladosporium subcinereum_antarcticum_phlei_macrocarpum | 0.03 | 3.02 | 2.22 |
| 17f87672bb942ca8b3596354625b1542 | Claviceps humidiphila | 0.01 | 1.48 | 1.89 |
| aeb121cc9a64ee4fe15af0b356649b27 | Claviceps macroura | 0.00 | 2.66 | 0.96 |
| 714228661ee55d498566562d4d782b62 | Claviceps macroura | - | 1.89 | 1.02 |
| 6c95e6c5792d5e1af4b184f43b6eb6b6 | Claviceps pazoutovae_monticola | 0.00 | 1.34 | 0.83 |
| 67602484504bca4f1a2105f6c222cf0d | Epicoccum phragmospora_Nothophoma macrospora | 0.05 | 2.10 | 2.82 |
| 5ca556568551faf004e83e42c1d1c765 | Neoascochyta cylindrispora_desmazieri | 0.01 | 0.51 | 1.26 |
| 38ab9a64ce7271afcf3101ac717dc661 | Neosetophoma phragmitis | 0.02 | 2.11 | 3.07 |
| 0a8cc784caae3436b2a2f56df6cec4c0 | Neosetophoma phragmitis | 0.00 | 1.67 | 3.33 |
| 5663634563309440e3ca3696c23088f6 | Penicillium fuscoglaucum_caseifulvum_commune | - | 1.28 | 0.00 |
| b672393eff9c6c00f455ea2f5a3acfcf | Preussia persica | 0.01 | 1.22 | 0.82 |
| 3fb396dc4a7b06fbd95897929a06723a | Pseudoconiothyrium broussonetiae | 0.00 | 0.73 | 1.11 |
| a4234fd305f00f140744be4bb2e543c1 | Pseudopithomyces rosae | 0.03 | 1.39 | 1.99 |
| 63baa23ebd09803e4a135175228373e6 | Pyrenochaetopsis microspora_leptospora | - | 0.85 | 1.03 |
| 5c6b15eadc471efbba1b5044b740272c | Thelebolus spongiae_ellipsoideus | 0.00 | 1.97 | 2.26 |
| 734cc9d4e22dae3261bac2e7a68c972e | Ustilago nunavutica | 0.00 | 1.27 | 2.74 |
| 82a81b8fe817daccea54156e773630ac | Ustilago nunavutica | 0.01 | 0.58 | 1.42 |
| 5beac8cb91c1c4f4e0a45c78e5e9d0af | Vacuiphoma oculihominis_Neodidymella thailandicum | - | 1.31 | 2.20 |
| cf14789892a27b7ecafb127f44bc2130 | Vishniacozyma victoriae | 0.00 | 0.79 | 1.22 |
| fcd5aded7ad3e8947a2ac66434368f80 | Xenopyrenochaetonopsis | - | 1.16 | 0.12 |
| Yeasts | ||||
| 66d5d96e85ffe8985a9a7957c8b6d932 | Candida inconspicua | 28.97 | 0.19 | 0.59 |
| d51af0a89a56f6289e89e6858ca54857 | Candida inconspicua _Pichia cactophila | 32.61 | 0.62 | 0.81 |
| b8672e965ae0ef9638e7424195de16d5 | Candida pseudoglaebosa | 3.11 | 0.11 | 0.04 |
| ddc456b20cd74e9247f05913562029b0 | Candida santamariae var. membranifaciens | 3.42 | 5.23 | 1.81 |
| 2ffbdc8725b87e1923c46eeb2e9e8c50 | Cutaneotrichosporon curvatum | 0.09 | 2.72 | 2.28 |
| c8a80e78d59949c51837339037dad7a3 | Cutaneotriphosporon curvatum_cyanovorans | 0.10 | 0.42 | 1.10 |
| 53193ba9d7fda4ce594b108dae39bc83 | Debaryomyces prosopidis_vindobonensis_fabryi | - | 7.52 | 0.07 |
| 0a5c97aafaa956e98ed3b0984ed1c8b2 | Geotrichum silvicola | 26.63 | 3.72 | 3.04 |
| 47597a8681f2304dd2d2a753350eb50a | Kluyveromyces lactis | 1.36 | 0.57 | 1.76 |
| 5feaa83003fb3b799ee8ffa513d6f08c | Malassezia restricta | 0.00 | 1.54 | 0.01 |
| 6e4adb61e7eeda079bf2af3dd2806138 | Sporobolomyces ruberrimus | 0.00 | 1.19 | 1.04 |
| 42dc2fa2227e97ab47f9619dbfff3159 | Trichosporon aquatile | 0.12 | 0.57 | 1.11 |
| cc81bbf3a7e847b76b28c7359999852b | Wickerhamiella shivajii | 0.85 | 19.18 | 14.10 |
 (0.00) Lowest value.
(0.00) Lowest value.| (a) Bacterial Taxa | Period | Treatments Compared | Statistical Methods 1 | Treatment with Highest abundance 2 |
|---|---|---|---|---|
| Milk | ||||
| Staphylococcus aureus | P1 | A_vs._G | 1 | G |
| Romboutsia timonensis | P1 | C_vs._G | 1 | G |
| Staphylococcus aureus | P1 | A_vs._C | 1 | C |
| Pediococcus stilesii_acidilactici | P2 | A_vs._G | 1,2 | A |
| Pediococcus stilesii_acidilactici | P2 | A_vs._G | 2 | A |
| Moraxella osloensis | P2 | A_vs._C | 1 | A |
| Kocuria salsicia | P2 | A_vs._C | 1 | A |
| Kocuria salsicia_varians | P2 | A_vs._C | 1 | A |
| Ralstonia pickettii | P2 | A_vs._C | 1,3 | C |
| Pooled teat suspension | ||||
| Staphylococcus petrasii | P1 | C_vs._G | 1 | G |
| Turicibacter sanguinis | P1 | C_vs._G | 1 | C |
| Staphylococcus petrasii | P3 | C_vs._G | 1 | C |
| Clostridioides difficile_bartlettii | P3 | A_vs._C | 1 | A |
| Individual teat suspension | ||||
| Acinetobacter haemolyticus | P1 | A_vs._G | 1 | G |
| Acinetobacter indicus | P1 | A_vs._G | 1,3 | G |
| Acinetobacter haemolyticus | P1 | A_vs._C | 2 | C |
| Acinetobacter indicus | P1 | A_vs._C | 2,3 | C |
| Exiguobacterium aestuarii | P2 | A_vs._G | 3 | G |
| Paeniclostridium sordellii | P2 | C_vs._G | 3 | G |
| Romboutsia sedimentorum | P2 | C_vs._G | 3 | G |
| Romboutsia timonensis | P2 | C_vs._G | 3 | G |
| Clostridioides difficile_bartlettii | P3 | A_vs._C | 3 | A |
| Plaeniclostridium sordellii | P3 | A_vs._C | 3 | A |
| Romboustia sedimentorum | P3 | A_vs._C | 3 | A |
| Romboustia timonensis | P3 | A_vs._C | 3 | A |
| Exiguobacterium aestuarii | P3 | A_vs._G | 1,3 | G |
| Acinetobacter indicus | P3 | C_vs._G | 1 | G |
| Clostridioides difficile_bartlettii | P3 | C_vs._G | 1,3 | G |
| Pseudomonas alcaliphila_chengduensis_toyotomiensis_oleovorans | P3 | A_vs._C | 3 | C |
| Pseudomonas alcaliphila_chengduensis_toyotomiensis_oleovorans | P3 | C_vs._G | 3 | C |
| Pseudomonas indoloxydans | P3 | C_vs._G | 3 | C |
| Staphylococcus haemolyticus_petrasii | P3 | C_vs._G | 3 | C |
| Staphylococcus petrasii | P3 | C_vs._G | 3 | C |
| (b) Fungal Taxa | Period | Treatments Compared | Statistical Methods 1 | Treatment with Highest Abundance 2 |
| Milk | ||||
| Candida inconspicua | P1 | A_vs._C | 1 | A |
| Candida inconspicua _Pichia cactophila | P1 | A_vs._C | 1 | A |
| Kluyveromyces lactis | P1 | A_vs._C | 1 | C |
| Kluyveromyces lactis | P1 | C_vs._G | 1 | C |
| Candida inconspicua | P2 | A_vs._G | 1 | A |
| Candida inconspicua _Pichia cactophila | P2 | A_vs._G | 1 | A |
| Candida inconspicua | P2 | A_vs._C | 1 | A |
| Candida inconspicua _Pichia cactophila | P2 | A_vs._C | 1 | A |
| Candida santamariae var. membranifaciens | P2 | A_vs._G | 1 | G |
| Candida santamariae var. membranifaciens | P2 | A_vs._C | 1 | C |
| Kluyveromyces lactis | P3 | A_vs._G | 1 | G |
| Candida santamariae var. membranifaciens | P3 | A_vs._G | 1 | G |
| Pooled teat suspension | ||||
| Xenopyrenochaetonopsis 89% | P2 | A_vs._G | 1 | A |
| Ampelomyces quisqualis | P2 | A_vs._C | 1 | C |
| Ampelomyces quisqualis | P2 | C_vs._G | 1 | C |
| Xenopyrenochaetonopsis 89% | P2 | C_vs._G | 1 | C |
| Cutaneotrichosporon curvatum | P3 | A_vs._C | 1 | A |
| Neosetophoma phragmitis | P3 | A_vs._C | 1 | A |
| Debaryomyces prosopidis_vindobonensis_fabryi | P3 | A_vs._C | 1 | A |
| Vacuiphoma oculihominis_Neodidymella thailandicum | P3 | A_vs._C | 1 | A |
| Epicoccum phragmospora_Nothophoma macrospora_Verrucoconiothyrium eucalyptigenum | P3 | A_vs._C | 1 | A |
| Sporobolomyces ruberrimus | P3 | A_vs._C | 1 | A |
| Pseudopithomyces rosae | P3 | A_vs._C | 1 | A |
| Cladosporium crousii_pini-ponderosae_colombiae | P3 | A_vs._C | 1 | A |
| Penicillium fuscoglaucum_caseifulvum_commune | P3 | A_vs._C | 1 | C |
| Individual teat suspension | ||||
| Wickerhamiella shivajii | P1 | A_vs._C | 3 | A |
| Claviceps humidiphila | P1 | A_vs._G | 1 | G |
| Kluyveromyces lactis | P1 | C_vs._G | 1,3 | G |
| Candida santamariae var. membranifaciens | P1 | C_vs._G | 1 | G |
| Geotrichum silvicola | P1 | A_vs._C | 3 | C |
| Claviceps humidiphila | P1 | A_vs._C | 1,3 | C |
| Geotrichum silvicola | P3 | C_vs._G | 1 | C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verdier-Metz, I.; Delbès, C.; Bouchon, M.; Pradel, P.; Theil, S.; Rifa, E.; Corbin, A.; Chassard, C. Influence of Post-Milking Treatment on Microbial Diversity on the Cow Teat Skin and in Milk. Dairy 2022, 3, 262-276. https://doi.org/10.3390/dairy3020021
Verdier-Metz I, Delbès C, Bouchon M, Pradel P, Theil S, Rifa E, Corbin A, Chassard C. Influence of Post-Milking Treatment on Microbial Diversity on the Cow Teat Skin and in Milk. Dairy. 2022; 3(2):262-276. https://doi.org/10.3390/dairy3020021
Chicago/Turabian StyleVerdier-Metz, Isabelle, Céline Delbès, Matthieu Bouchon, Philippe Pradel, Sébastien Theil, Etienne Rifa, Agnès Corbin, and Christophe Chassard. 2022. "Influence of Post-Milking Treatment on Microbial Diversity on the Cow Teat Skin and in Milk" Dairy 3, no. 2: 262-276. https://doi.org/10.3390/dairy3020021
APA StyleVerdier-Metz, I., Delbès, C., Bouchon, M., Pradel, P., Theil, S., Rifa, E., Corbin, A., & Chassard, C. (2022). Influence of Post-Milking Treatment on Microbial Diversity on the Cow Teat Skin and in Milk. Dairy, 3(2), 262-276. https://doi.org/10.3390/dairy3020021