Abstract
The de novo design and synthesis of peptide-based biocatalysts that can mimic the activity of natural enzymes is an exciting field with unique opportunities and challenges. In a natural enzyme, the active site is composed of an assembly of different amino acid residues, often coordinated with a metal ion. A metalloenzyme’s catalytic activity results from the dynamic and concerted interplay of various interactions among the residues and metal ions. Aiming to mimic such enzymes, simple peptide fragments, drawing structural inspiration from natural enzymes, can be utilized as a model. In our effort to mimic a metal-containing hydrolase, we designed peptide amphiphiles (PA) 1 and 2 with a terminal histidine having amide and acid functionalities, respectively, at its C-terminal, imparting differential ability to coordinate with Zn and Cu ions. The PAs demonstrate remarkable self-assembly behavior forming excellent nanofibers. Upon coordination with metal ions, depending on the coordination site the nanofibers become rigidified or weakened. Rheological studies revealed excellent mechanical properties of the hydrogels formed by the PAs and the PA–metal co-assemblies. Using such co-assemblies, we mimic hydrolase activity against a p-nitrophenyl acetate (p-NPA) substrate. Michaelis–Menten’s enzyme kinetic parameters indicated superior catalytic activity of 2 with Zn amongst all the assemblies.
1. Introduction
Nature precisely engineers a structurally complex system such as protein to perform complex functions with excellent conformational control, structural specificity, and feedback mechanisms [1,2]. Understanding nature’s approach to out-of-equilibrium self-assembly and mimicking it with supramolecular self-assembly in the laboratory offers several challenges, as it typically gets dictated by the free energy of the equilibrium process [3,4,5]. Amyloids, as well as other prion infection proteins, driven by nucleation-mediated self-assembly, encompass pathway complexity involving multiple on- and off-pathway aggregates [6]. These fibrillar networks and supramolecular assemblages possessing catalytic abilities create compartmentalized systems capable of self-replication, which might have functioned as the proto-enzymes in the early stages of life on Earth [7,8,9]. Following millions of years of evolutionary optimization, the resulting macromolecular enzymes that we find in nature have the exquisite properties of superior catalytic activity, substrate specificity, and recognition [1,10]. However, their intricacy also implies that engineering a synthetic enzyme mimic with tailored functionality and efficient activity is thoroughly difficult. Consequentially, utilizing fibrillar assembly with efficiency such as natural enzymes still remain an unattained target. The fact that natural enzymes are very sensitive to their physical and chemical environment presents further challenges in utilizing the enzymes in their original form in diverse chemical or biotechnological scenarios. Motivated by the rising industrial demands for greener alternatives, it is of urgent importance to develop efficient biocatalysts in the current scenario [11].
Structurally simple peptide fragments have been found to mimic the properties of amyloids and offer a promising alternative in the pursuit of designing de novo biocatalysts [12,13]. Their simplicity allows for a high degree of control over their sequence and structure and offers higher stability, and they can be functionalized with a variety of chemical groups, such as metal ions, organic cofactors, and reactive side chains [13]. Their self-assembly into various supramolecular structures, such as β-sheet fibrils or nanosheets, further enhances their catalytic properties by providing a confined environment for substrate binding and catalytic reaction [13], thereby creating robust and modular catalytic amyloids tailored for a specific activity. A metal-chelated histidine residue is ubiquitous in the metal-binding domains of biologically important proteins. The metal cofactor that we commonly find associated with these histidine residues is zinc, copper, or iron, among which zinc is the most prevalent [14,15,16]. The active sites of metalloenzymes such as carbonic anhydrases, carboxypeptidases, hydrolases, and structural motifs such as zinc fingers in transcription factors are a few examples of the diverse and substantial occurrence of the histidine–zinc pair in biology [14,17,18]. Similarly, the His-Cu pair also plays a vital role in the activity of several important metalloenzymes. In this regard, designing de novo metal-containing catalytic peptides has been an active area of research. His- terminal has been employed in such functional peptides to impart hydrolytic activity [19]. Gazit et al. have demonstrated that even a single amino acid, when grafted with zinc, can catalyze a hydrolysis reaction of p-NPA [20]. Similarly, Korendovych et al. have designed a copper-containing catalytic amyloid capable of phosphoester hydrolysis [21]. Stupp et al. and Ulijn et al. have shown that histidine-anchored peptide assemblies can be used to modulate the ester hydrolysis rate by tweaking the number and location of histidine residues on the peptide sequence [22]. The substrate specificity of such peptide fragments is augmented by co-assembling a triad of such sequences having different terminal residues, similar to the catalytic triads found in nature [23].
However, controlling the conformation and resulting properties of such amyloid-mimicking short peptides require an external trigger. In that regard, our group has not only demonstrated precise control over the 1D and 2D morphologies arising from the pathway-dependent assembly of the amphiphiles by living supramolecular polymerization but also developed strategies to sequester the self-assembly pathway using photochemical, redox cues [24,25,26]. We have developed several strategies for on-demand self-assembly–disassembly affecting not just the fibrillar morphology but rather to tweak optomechanical and bioelectronic properties such as piezo response [27,28]. Fibrillar assemblies resulting from the controlled self-assembly of such amyloid-inspired peptide amphiphiles have also been exploited in making stiff and interpenetrating hydrogels for designing biomimetic materials and tissue engineering [27,29,30]. Further, such peptide amphiphiles with different chiralities exhibit self-sorted and enzyme-responsive compartments [31]. In another work, we demonstrated that the morphology of the nanostructures arising from the self-assembly of catalytic amyloid, modulated by external thermal and photochemical cues, is in fact a determining parameter in their catalytic activity [32].
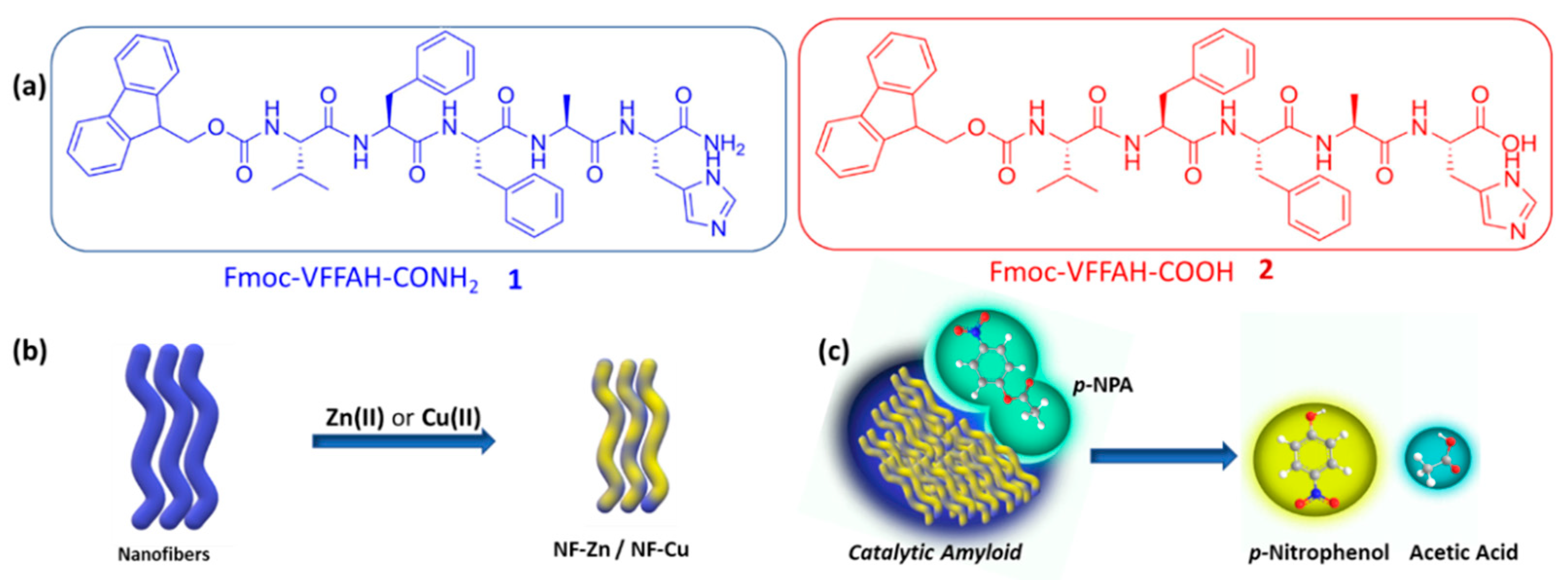
In this work, we envisioned the metal-coordinated terminal histidine to act as the catalytically active site in our effort to develop a hydrolase mimic. We altered the terminal functionality in the amphiphiles: 1 having an amide functionality and 2 having a carboxylic acid functionality. We anticipated augmented metal coordination and, therefore, the activated catalytic activity of 2 with respect to 1 (Scheme 1). The assembly of a peptide amphiphile is governed by supramolecular interactions, namely, hydrophobic interaction, π-π stacking, H-bonding, and electrostatic interactions. Our rational design of the amphiphiles 1 and 2 in this work directs specific importance on tuning the supramolecular interactions so that efficient self-assembly can be achieved, in view of the fact that the assembly behavior subsequently affects the catalytic properties as well. We attached a hydrophobic Fmoc- moiety at one periphery of an amyloid NVFFAC core acting as the nucleation center and a histidine residue at the other periphery, which would be the functionally active part of the enzyme mimic. The well-tuned balance of these interactions leads to the self-assembly of the amphiphiles into nanofibers (Scheme 1). The Zn(II) or Cu(II) coordinated amphiphiles also form metal-incorporated fibrils in a similar way. The assemblage of the peptide fibrils leads to the formation of hydrogels, and we concurrently explored the mechanical properties of the peptide hydrogels and the effects of metal ion incorporation, since the coordination with metal ions can dictate the self-assembly pathway of the peptide amphiphiles [33]. Furthermore, our rheological investigations reveal that the same co-assembly exhibits superior hydrogel strength amongst the other assemblies. The Fmoc-VFFAH-COOH (2) and Zn(II) fibrillar co-assembly demonstrates superior hydrolytic behavior over the other contenders against para-nitrophenyl acetate as a model substrate. We also establish the superiority in kinetic efficiency of this co-assembly as a potent hydrolase mimic by quantifying the enzymatic parameters from the Michaelis–Menten equation.
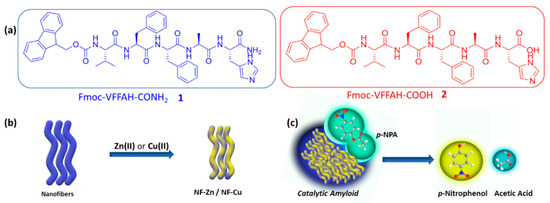
Scheme 1.
(a) Molecular structures of peptide amphiphile 1 and 2; (b) self-assembly of PA 1 and 2 to form nanofibers which, upon addition of Zn(II) or Cu(II), form metal-coordinated nanofibers (NF-Zn or NF-Cu); (c) hydrolysis of p-NPA catalyzed by aggregated amyloid nanofibers to form p-NP and acetic acid.
2. Materials and Methods
Fmoc- protected amino acids, piperazine, para-nitrophenylacetate (p-NPA) were purchased from Sigma, St. Louis, MO, USA; Diisopropyl carbodiimide (DIC), hexafluoroisopropanol (HFIP), and triisopropylsilane (TIS) were purchased from TCI, Thioflavin-T (ThT), Oxyma, Rink amide MBHA Resin, and His-Preloaded CTC Resin, were purchased from Sigma-Aldrich. Cu(II) chloride, Zn(II) nitrate hexahydrate, NH3 (aq.), Dimethylformamide (DMF), dichloromethane (DCM), HPLC grade acetonitrile, HPLC water, and ethanol were ordered from Merck.
2.1. Synthesis of Peptides 1 and 2
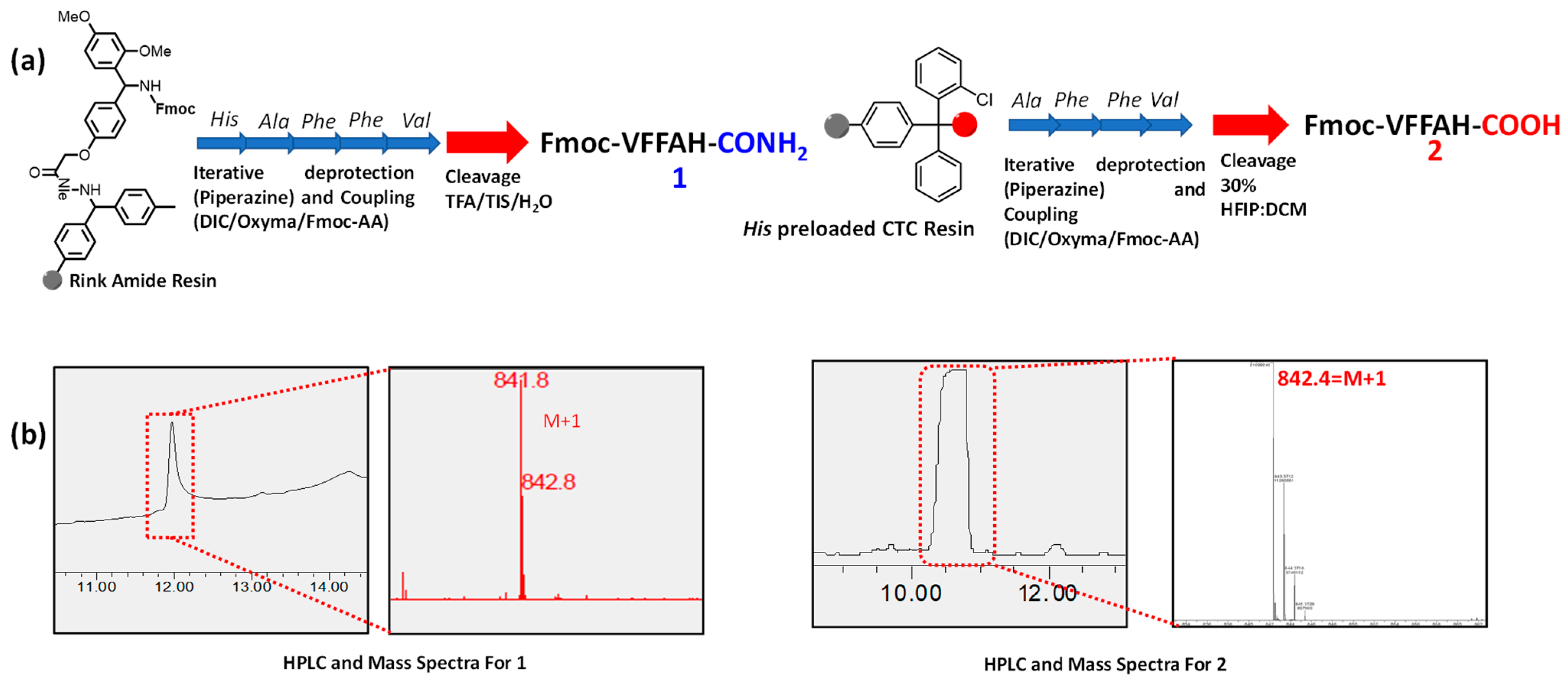
Peptides 1 and 2 were synthesized via solid-phase peptide synthesis technique using the CEM Liberty Blue automated microwave-assisted peptide synthesizer. For the synthesis of 1, Rink Amide MBHA resin was chosen to get a terminal amide functionality after cleavage from the resin. For the synthesis of 2, His- preloaded CTC resin yields a carboxylic acid group upon cleavage.
Fmoc-Rink amide MBHA resin was swelled in DMF, and His-preloaded CTC resin was swelled in DCM for 15 min. Fmoc-protected amino acids were weighed as per the desired scale of the reaction and dissolved in the required DMF solution. Deprotection of Fmoc group from the amino acid was achieved by using 10% piperazine in DMF. Required mass of piperazine was stirred in 10% ethanol in ethanol/DMF solvent mixture for complete solubilization. DIC and Oxyma in DMF were used as activators for the coupling reaction between acid and amine to form the peptide bonds. A cycle of coupling and deprotection steps was repeated to synthesize the required peptides anchored to the resin. Peptide 1 was cleaved from the resin by stirring the resin in a mixture of trifluoroacetic acid (TFA), triisopropylsilane (TIS), and water (95:2.5:2.5) for 1 h, and peptide 2 using a mixture of 30% HFIP and DCM for 3 h at room temperature. The resin was then filtered and the filtrate containing desired peptides was concentrated by evaporating the excess solution. The peptides were then precipitated from cold diethyl ether and subsequently air-dried to obtain the peptides as white powder which were characterized using HPLC and mass spectroscopy (Scheme 2).
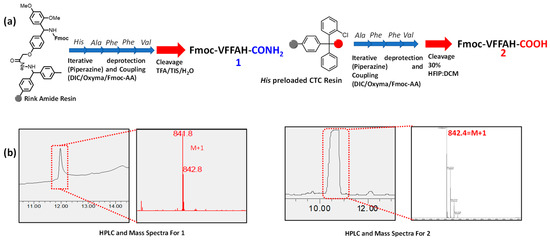
Scheme 2.
(a) Synthesis of PA 1 from Rink amide resin and 2 from His-preloaded CTC resin following the solid phase peptide synthesis protocol. (b) Characterization of PA 1 and 2 using HPLC and mass spectra.
2.2. HPLC and Mass Spectra
Reverse phase HPLC a of 1 and 2 was performed using Waters binary HPLC system by dissolving 1 mg of sample in 1 mL 50% acetonitrile/water, making the final concentration of the peptide 1 mM. A Nucleodur analytical column (C18 stationary phase, 5 μm, 4.6 × 250 mm) was used for the analysis. The sample was injected with an autosampler in gradient mode from 95% water with 0.1% formic acid as additive with 5% acetonitrile up to 5% water with 0.1% formic acid as additive with 95% acetonitrile over 15 min. Photodiode array (PDA) detector was used to detect the sample along with electrospray ionization mass spectrometry (ESI/MS) with Waters Acquity QDa detector.
2.3. Self-Assembly of the Peptide Amphiphiles
The peptide amphiphiles 1 and 2 were dissolved in a 1:1 mixture of ethanol and water. The composition was chosen in such a way that the peptides remain readily soluble. To allow for fibril growth, the solutions were left to incubate for 24 h at a temperature of 25 °C. In a similar way, we also prepared and incubated 1 mM peptide solutions in DMSO. To prepare co-assemblies of the peptides with metal ions, equivalent amounts of Zn(II) or Cu(II) ions were introduced to the 1 mM peptide solutions in ethanol/water or DMSO from 20 mM stock solutions of Zn(NO3)2·6H2O or CuCl2, respectively.
2.4. Circular Dichroism Spectra
The circular dichroism spectra were recorded using a JASCO J-1500 circular dichroism spectrometer (Easton, MD, USA). The peptide solution (0.1 mM) was added to a cuvette of the path length of 0.1 mm, and scanned in the wavelength range from 200 nm to 400 nm at a scan rate of 100 nm/min.
2.5. Thioflavin-T Binding Studies
The fluorescence emission spectra of Thioflavin-T were obtained using an FS5 spectrofluorometer from Edinburgh Instruments (Livingston, UK). Wavelength range was selected from 470 nm to 600 nm with a scan slit and offset slit of 1 mm each with a dwell time of 0.1 s at 25 °C. An increase in emission intensity at 485 nm suggests more organized assembly, i.e., β-sheet formation by the amyloids. An amount of 600 μL of 0.1 mM peptide solutions was taken into which 2 μL of ThT was added from a 10 mM stock solution, making the final concentration of the dye in the solution to be 0.03 mM.
2.6. Atomic Force Microscopy
An amount of 10 μL of the 0.1 mM peptide solutions was drop-casted on a clean silicon wafer, and the drops were blotted after 5 min. The wafers were air-dried at room temperature for 24 h before imaging. A Bruker Multimode 8 AFM system in tapping mode was used for imaging.
2.7. Scanning Electron Microscopy
An amount of 10 μL of 50 μM samples were drop-casted on silicon wafers, which were kept overnight at room temperature for air drying. SEM images were obtained using a JEOL JEM 2100 (Tokyo, Japan) scanning electron microscope at accelerating voltages not exceeding 15 kV.
2.8. XPS Study
To know the chemical states of the samples, X-ray photoelectron spectroscopy of the samples was performed on KAlpha plus XPS system by ThermoFisher Scientific instruments (Waltham, MA, USA) in an ultrahigh vacuum chamber (7 × 10−9 torr) using Al-Kα radiation (1486.6 eV). The samples were prepared by drop-casting 10 μL of 2 mM samples on silicon wafers, which were kept overnight at room temperature for air-drying.
2.9. Fourier Transform Infrared Spectroscopy
The FTIR spectra were recorded in the range of 1000 cm−1 to 2000 cm−1 in attenuated total reflectance (ATR) mode using Bruker Vertex 70 FTIR spectrophotometer (Billerica, MA, USA).
2.10. UV–Vis Spectroscopic Studies and Titration with Metal Ions
For obtaining the UV–Vis absorption spectra of the samples, 0.01 mM solutions were taken in a 1 mL quartz cuvette of a path length of 1mm, and the spectra were recorded in the range of 2150 nm to 400 nm using an Agilent Cary 60 UV–Vis spectrophotometer (Santa Clara, CA, USA). For titration studies with metals, the spectra were obtained using a TECAN Infinite 200Pro plate reader, by taking 200 μL 0.5 mM peptide solutions in ethanol/water in a transparent plate and varying the metal ion concentrations at 1, 0.75, 0.5 0.25, and 0.1 equivalents of metals.
2.11. Rheological Studies
Next, 1 w/v% peptide was dissolved in 500 μL 1:1 ethanol/water mixture, in which sodium phosphate buffer (SPB) was added so that the final concentration of SPB becomes 15 mM. Equimolar Zn(II) or Cu(II) ions were introduced from CuCl2 and Zn(NO3)2 ·6H2O stock solutions. Images of the hydrogels were captured using a digital camera. The mechanical characterizations of the gels thus prepared were done using an Anton Paar Modular Compact Rheometer 302 (Graz, Austria) with parallel plate geometry (25 mm plate diameter) equipped with a Peltier temperature control unit, to keep the temperature at 22 °C throughout the experiments. The gap between the plates was set to be 0.2 mm. Amplitude sweep measurements with 0.01% to 100% deformation at a constant angular frequency of 10 rad/s determined the linear viscoelastic region of the gels. Frequency sweep oscillatory rheology was performed over the frequency range from 0.1 rad/s to 100 rad/s at a constant shear strain of 1% in the linear viscoelastic region. The gels were prepared and incubated for 24 h prior to the measurements.
2.12. Kinetic Assays of Para-Nitrophenyl Acetate (p-NPA) Hydrolysis
The kinetics of p-NPA hydrolysis by 1, 2, 1 + Cu, 1 + Zn, 2 + Cu, and 2 + Zn at different concentrations of the substrate were studied using a TECAN Infinite 200Pro plate reader by recording the absorbance at 400 nm for 30 min in a transparent 96-well plate. Hydrolysis of p-NPA produces yellow-colored para-nitrophenolate ions as the product that shows strong absorption at 400 nm. An increase in absorbance at this wavelength indicates the accumulation of the hydrolysis product p-nitrophenol in the solution.
A 25 mM stock solution of p-NPA was made in acetonitrile, from which six batches of 1 to 10 μL were added to peptide solutions (0.25 mM) in the wells to maintain the final concentration of p-NPA from 0.125 to 1.25 mM with a final volume of 200 μL. The absorbance values for the peptide solutions in which p-NPA was not added were subtracted from the values obtained for the peptide solutions containing the p-NPA substrate. Thus, the initial rates of reaction (V0) were obtained. Using the obtained absorbance values at different time intervals, the initial slopes were calculated for different substrate concentrations of p-NPA at a fixed concentration of peptide (0.25 mM). These slope values, with respect to their substrate concentration values, were fitted in the Michaelis–Menten equation (V0 = Vmax [S]/(Km + [S]) to get the kinetic parameters—catalytic rate kcat, Michaelis–Menten’s constant KM, maximum velocity Vmax, and catalytic efficiency kcat/KM. The obtained values were further compared to those obtained from Lineweaver–Burk linearization method.
3. Results
The peptide amphiphiles 1 and 2 were designed by tethering a hydrophobic fluorenylmethyloxycarbonyl (Fmoc) group at the N-terminal and a histidine moiety at the C-terminal of an Aβ(1-42) inspired short peptide sequence NVFFAC. Amphiphiles 1 and 2, differing in the C-terminal functional group, were synthesized from the precursors following the solid-phase peptide synthesis (SPPS) protocol and characterized using HPLC-MS (Scheme 2). C-terminal amide containing peptide amphiphile 1 elutes at 12 min corresponding to an (M + 1) peak of 841, whereas owing to the carboxylic acid terminal PA 2 is more polar, and thus has a lesser retention time of 10.5 min with an (M + 1) peak of 842. This difference in polarity arising from the peripheral moiety also brings about several differences in the self-assembly pattern and metal binding ability influencing mechanical and functional properties.
3.1. Self-Assembly Behavior of the Peptide Amphiphiles
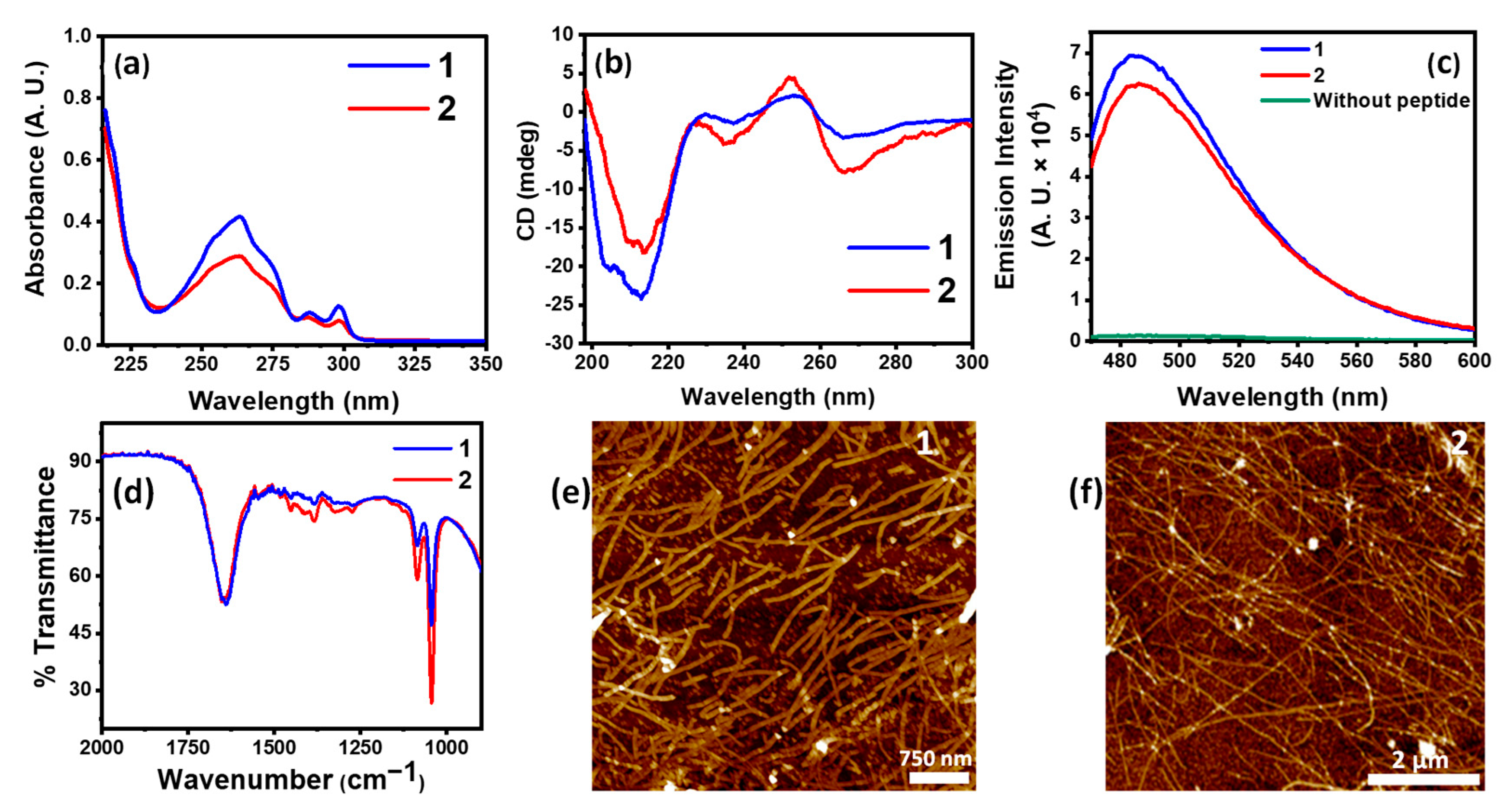
The core NVFFAC fragment in our amphiphiles is inspired by the aggregation-prone amyloid beta Aβ(1-42) in which this fragment plays a crucial role in its assembly behavior [34]. In this fragment, the two proximal phenylalanine residues containing aromatic rings provide an excellent surface for π-π stacking. The amide bonds between the residues provide superior H-bonding interaction. Overall, this short fragment acts as the nucleating core of the amphiphile, which, during assembly, acts as a “seed” onto which subsequent aggregation can occur. At its N-terminal, a hydrophobic fluorenylmethyloxycarbonyl (Fmoc) moiety has been attached which also provides π-π stacking interaction and imparts the necessary hydrophobicity to the amphiphiles. Solvent plays a crucial role to allow for solubility and efficient supramolecular interactions for optimum self-assembly. Using 50% EtOH as co-solvent in water, both the peptides were allowed to self-assemble. The perfect balance of these interactions resulted in the formation of nanofibers from the self-assembling peptide amphiphiles, as shown in Figure 1e,f. However, choosing a solvent such as DMSO, even at 10% v/v with water, rather hindered the supramolecular interactions such as H-bonding. AFM images show that, even at a low concentration of 0.001 mM, definite self-assembly into fibrillar morphologies takes place in ethanol–water, but not when 10% DMSO is used (Figure S1). This was also indirectly observed when co-assemblies of our PAs 1 and 2 with Cu/Zn showed a complete absence of fibrillar morphology (Figure S2) and, instead, formed nanoparticles of varying dimensions, indicating a lack of definite self-assembly. Thus, we selected 50% EtOH:water as our solvent of choice for further self-assembly studies. The UV–vis spectra of 1 and 2 showed a broad absorption band spanning from 230 nm to 270 nm (Figure 1a). Owing to the stacking of Fmoc-moieties in the assembled amphiphiles, the characteristic peak of Fmoc at 265 nm showed an increase in intensity in aged solutions when compared to the intensity observed in freshly prepared peptide solutions (Figure S3). The secondary structural information of the native assemblies in the solution was investigated using circular dichroism (CD). The CD spectrum of a peptide assembly in the region of 200–400 nm can reveal important information about its secondary structure, such as β-sheets, α-helices, or random coil orientation, each of which imparts a characteristic spectral signature. In the CD spectra of 1 and 2, the negative Cotton effect centered at 213 nm characteristic for β-sheets was observed. Additionally, induced CD bands due to Fmoc-stacking interactions were noted at 235 nm and 265 nm. It is noteworthy that the intensity of the induced bands is higher in the case of 2 as compared to 1, indicative of differences in their overall stacking pattern during assembly. Figure 1d displays bell-shaped emission curves of self-assembled peptides with the amyloid reporter fluorescent dye Thioflavin T (ThT). ThT displays enhanced emission at 490 nm when it binds to β-sheet rich structures such as amyloid aggregates. We observed an intense emission peaking at 490 nm for both 1 and 2 (Figure 1c). Here, the emission intensity for 1 was noted to be slightly higher than that of 2. This trend correlates with our observation from CD spectra (Figure 1b) which also indicates slightly stronger self-assembly for 1. The peptide backbone characterized by vibrational spectroscopy in Figure 1d describes the IR spectra of 1 and 2. The amide I and II vibrational modes which are indicative of secondary structural information of the peptides can be seen as a merged broad peak around 1600 cm−1 (Figure 1d). The morphological assessment of the nanostructures formed upon self-assembly was performed via atomic force microscopy (AFM) imaging. The amphiphiles in ethanol–water showed formation of nanofibers with a median height of 5 ± 1 nm (Figure 1e,f). However, it is to be noted that despite 1 displaying traits of stronger assembly, the fibers formed are relatively shorter and more disperse than the long strands formed by 2.
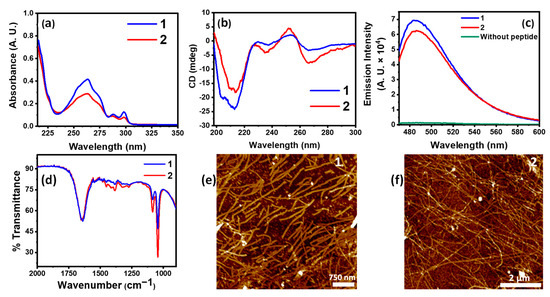
Figure 1.
(a) UV absorption spectra of 1 and 2 showing characteristic absorption bands for stacked Fmoc-moieties; (b) CD spectra of 1 and 2 showing β-sheet signature; (c) ThT fluorescence emission spectra of 1, 2, and unbound ThT; (d) IR absorption spectra of 1 and 2; (e) AFM image of 1; (f) AFM image of 2.
3.2. Effect of Metal Ions
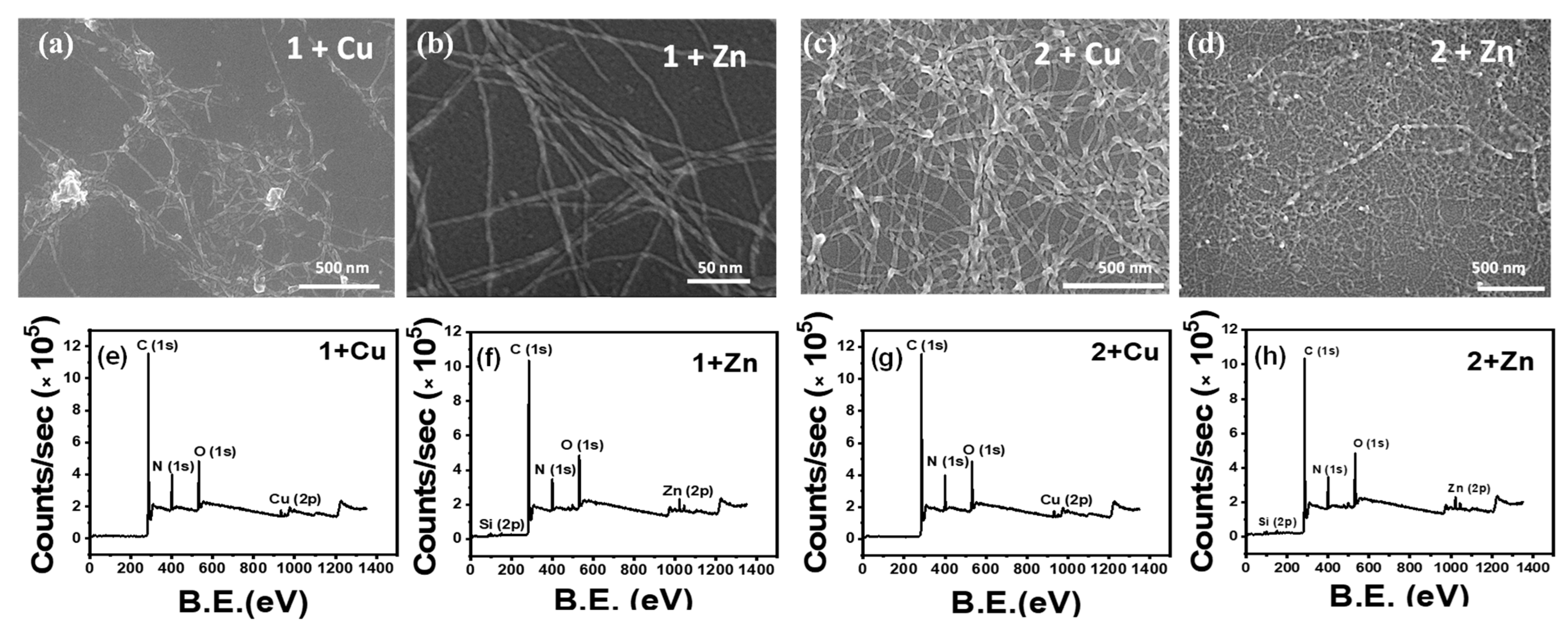
Amphiphiles 1 and 2 have a strong tendency to coordinate with metal ions due to the presence of the terminal imidazole moieties in histidine residue. Co-assembly of 1 with Zn/Cu ions shows histidine–metal complexation at the expense of axial stacking interactions responsible for fiber formation. SEM showed noticeably more dispersed short fibers in the case of metals-1 co-assembly indicating metal binding causing a reduction of the fibrillar network density. However, in the case of 2, it was observed to be reinforced (Figure 2a–d). This observation was also confirmed from the AFM images obtained for the samples (Figure S4). To further study the surface elemental composition of the metal–peptide assemblies, an XPS study was performed. The XPS Survey scan obtained confirmed the presence of the metal ions in the corresponding samples. In an attempt to quantify the effect of a metal addition on the β-sheet self-assembly, we studied the extent of ThT binding to metal-bound peptides compared to the native assemblies. However, both Zn and Cu ions were seen to quench the fluorescence of ThT (Figure S5). In an additional aim to determine the complexation ratio with the metal ions, UV–vis spectra at different metal equivalents were recorded; however, a significant change in the absorbance was not observed (Figure S6).
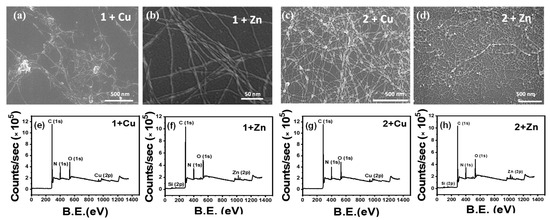
Figure 2.
SEM Image of (a) 1 + Cu, (b) 1 + Zn, showing scattered fibrils; SEM Image of (c) 2 + Cu, (d) 2 + Zn displaying remarkably close-knit network of fibrils; XPS elemental survey for (e) 1 + Cu, (f) 1 + Zn, (g) 2 + Cu, (h) 2 + Zn confirming presence of metal ions.
3.3. Gelation Properties
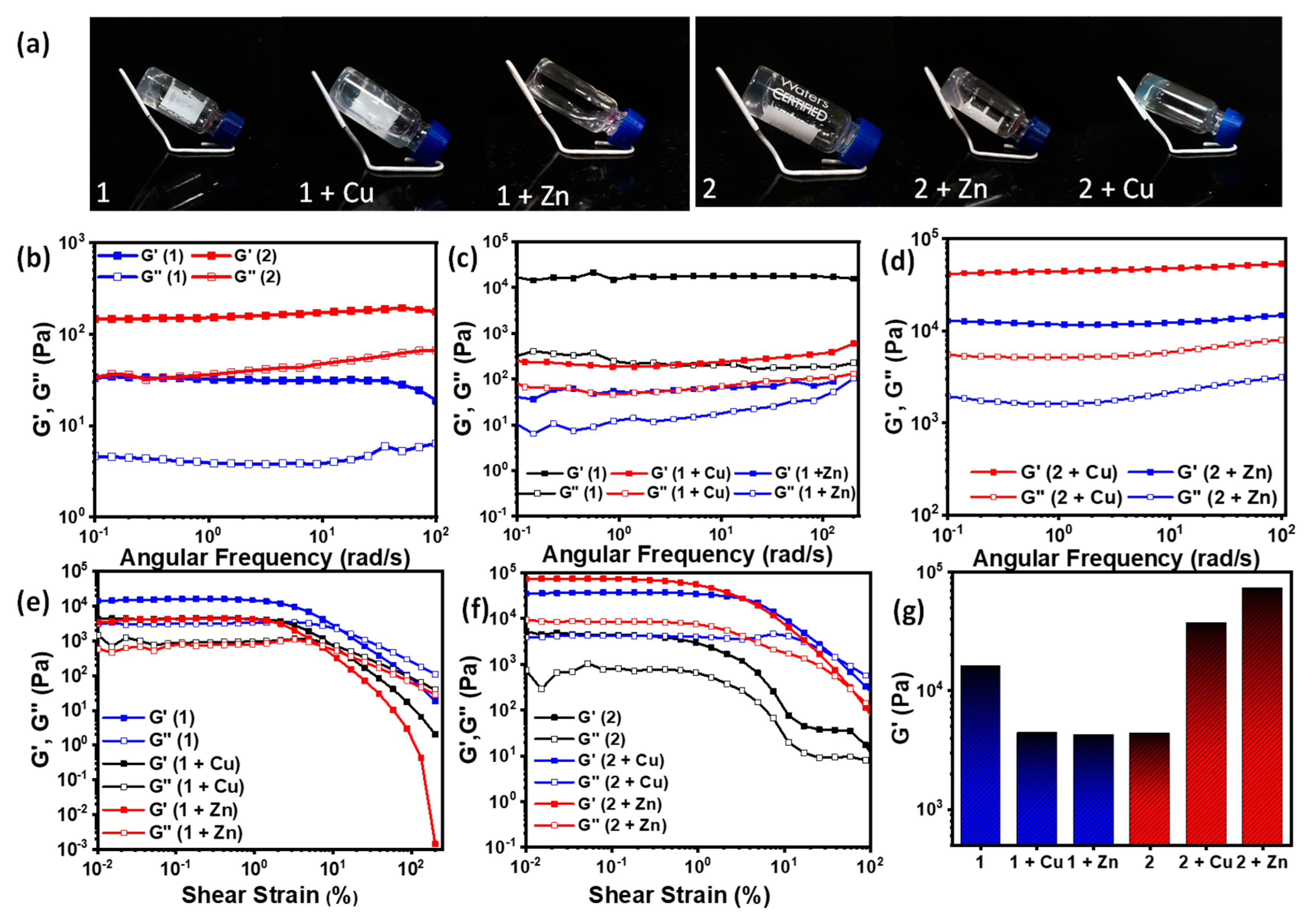
The gels were prepared at a concentration of 1% wt/v in ethanol/water, followed by the addition of sodium phosphate buffer (15 mM). Equivalent amounts of the metal ions were introduced from the respective stock solutions. The inter-fiber bridging by counter-ions led to uniform crosslinking resulting in the hydrogel networks. To check the self-standing ability of the various gels thus formed, the gels were prepared and incubated in a glass vial and thereafter held at an inverted position. Of the six gels, only the two gels formed by co-assembling the metal ions with 1, i.e., 1 + Cu and 1 + Zn, were found to be incapable of maintaining their vertical position when inverted (Figure 3a, Table 1).
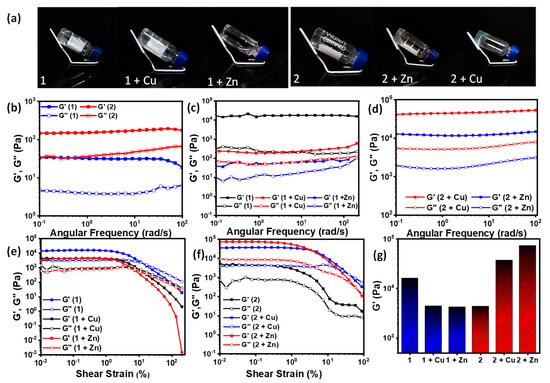
Figure 3.
(a) Digital images of the gels in inverted vials depicting their self-standing behavior; (b) frequency sweep study for 1 and 2; (c) frequency sweep study for 1 + Cu and 1 + Zn; (d) frequency sweep study for 2 + Cu and 2 + Zn; (e) amplitude sweep study for 1, 1 + Cu, and 1 + Zn; (f) amplitude sweep study for 2, 2 + Cu, and 2 + Zn; (g) comparative histogram of the storage moduli for the hydrogels.

Table 1.
Inverted vial test for the gels.
The mechanical properties of the gels were characterized by detailed rheological investigation, which included amplitude sweep and frequency sweep measurements. The rheology data showed some interesting details concerning the properties of the gels formed with and without metal ions (Figure 3b–f). In the frequency sweep studies, all the gels exhibited consistent storage moduli and frequency-independent behavior over the whole frequency range owing to hydrogel network formation. It was noted that the addition of Cu(II) and Zn(II) to 1 reduced the gel strength significantly, by three orders of magnitude, whereas in the case of 2, it was found that the metal ions in fact reinforced the gels remarkably, increasing the gel strength by an order of magnitude. The coordination of the metal ion from dual sites, i.e., the carboxylate center, as well as the imidazole ring in this case could be attributed as the reason. Among the six gels, 2 + Zn showed the highest gel strength. The gel strengths follow the order 2 + Zn > 2 + Cu > 2 > 1 > 1 + Zn > 1 + Cu (Figure 3g).
3.4. Catalysis
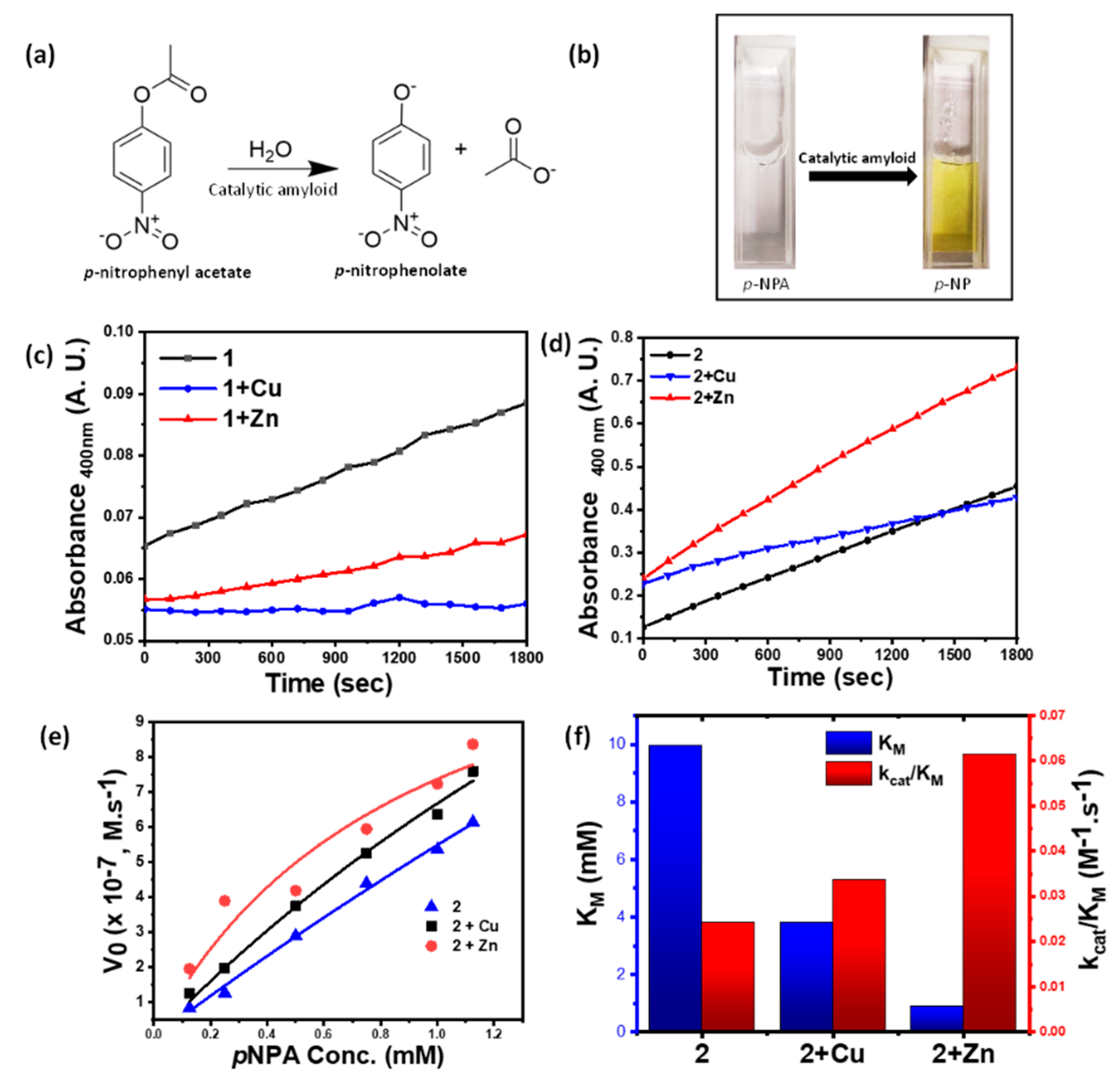
We envisaged the histidine-grafted metal-peptide assemblies as a biomimetic equivalent of the metal-histidine active sites of the hydrolase class of metalloenzymes. Thus, to study their catalytic behavior, we investigated their activity against the model substrate, para-nitrophenyl acetate (p-NPA), which upon hydrolysis generates a yellow-colored p-nitrophenolate ion (Figure 4a,b). The kinetics of the reaction, henceforth, can be followed easily via UV–Vis spectrophotometer. The gradual change in the kinetic studies of p-NPA hydrolysis revealed excellent catalytic activities of the metal-coordinated amphiphiles. To determine the comparative rates of reactions, kinetic studies against a specific p-NPA concentration of 0.25 mM were performed. Thus, 2 + Zn showed pronouncedly higher activity compared to 2 + Cu and 2. The catalytic activity of 1, 1 + Cu, or 1 + Zn was found to be significantly lower than the others (Figure 4c,d). Amongst the same metal-ion assembly pair, using co-solvent as EtOH or DMSO made a drastic reduction in the catalytic activity. The underlying fact of lack of efficient supramolecular polymerization in DMSO as co-solvent and inability to assemble into nanofibrous morphology has a huge spatio-temporal impact reflected in its enzymatic activity. This highlights the role of self-assembled nanofibers in catalyzing the substrate (Figure S7). Considering the lower catalytic activity for 1 and further suppression of activity upon metal coordination, we disregarded 1, 1 + Cu, and 1 + Zn for further catalytic studies. Here, owing to the absence of a -COOH group, the metal did not activate the catalyst.
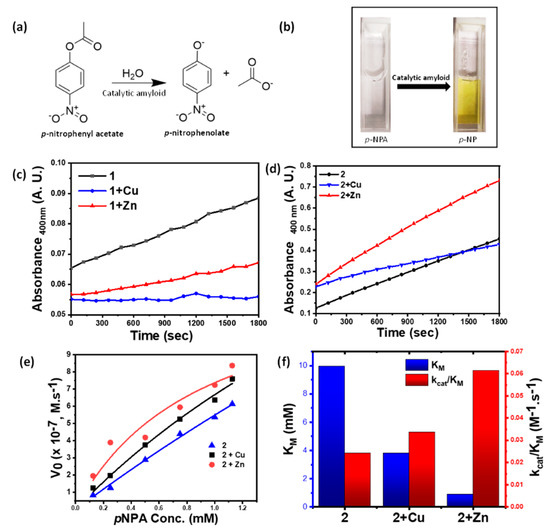
Figure 4.
(a) Hydrolysis reaction of p-NPA, catalyzed by metal-grafted amyloid fibrils; (b) change in color of p-NPA solution upon addition of nanofibers due to accumulation of hydrolysis product p-NP; (c) absorbance vs. time plot for 0.25 mM p-NPA concentration by 1, 1 + Cu, 1 + Zn (0.25 mM); (d) absorbance vs. time plot for 0.25 mM p-NPA concentration by 2, 2 + Cu, 2 + Zn (0.25 mM); (e) comparison of the initial rate of ester hydrolysis using 2, 2 + Cu and 2 + Zn (0.25 mM); (f) Michaelis–Menten’s constant (KM) and catalytic efficiency (kcat/KM) for 2, 2 + Cu and 2 + Zn.
In order to determine the catalytic parameters, a kinetic assay against varying substrate concentrations was performed. The slope values thus obtained were fitted in the Michaelis–Menten’s equation to calculate the kinetic parameters, the Michaelis–Menten’s constant (KM), along with the catalytic rate (kcat) and catalytic efficiency (kcat/KM), validating the hydrolase catalysis (Figure 4e,f and Table 2). A lower value of KM reflects a higher affinity of the substrates to the catalysts. Thus, 2 + Zn turned out to be the most active catalyst with a minimum KM value of 0.92 mM and a maximum kcat/KM of 61.57 mM−1 s−1, implying superior catalytic activity. The values obtained are found to match closely with those obtained from Lineweaver–Burk plots (Figure S8 and Table S1).

Table 2.
Catalytic parameters with different peptide assemblies.
This result draws parallel to the histidine-coordinated zincmediated catalytic activity of natural hydrolase enzymes. The efficient Lewis acidity and flexible coordination geometry of zinc make it ideal for binding with histidine-containing motifs and coordinating with the oxygen center of water molecules, thus polarizing and activating it, facilitating rapid hydrolysis reaction [35].
Thus, our findings demonstrate the pivotal role of the peripheral moiety in governing the catalytic properties of a peptide amphiphile. Further, we also highlight a relationship between the coordination-driven supramolecular assemblies of the amphiphiles with their catalytic activity. The results from our study bear substantial implications in the endeavors towards a futuristic de novo metalloenzyme mimic.
4. Conclusions
In summary, we designed two amyloid-inspired histidine-appended peptide amphiphiles with differential potency to coordinate with metal ions by tweaking their C-terminal functionality. The amphiphiles, with structurally tuned degrees of supramolecular interactions, self-assembled to form nanofibers, were confirmed through a series of spectroscopic and microscopic studies. We rationally choose the metal-coordinated peripheral His moiety as the catalytically active site present in metalloenzymes and anchored it to a Zn(II) or Cu(II) ion. The stability of the peptide–metal co-assemblies being dictated by supramolecular interactions was evident from the gelation ability of the co-assemblies compared to the native peptide ones. Using a stable co-assembly of 2 and Zn as catalysts, we could achieve enhancement in the hydrolytic ability against p-NPA. The superiority of nanofibrous morphology over the nanoparticle one also suggested morphology-dependent catalytic activity. The metals failed to activate the catalyst where the terminal functionality is an amide, implying the significance of the carboxylic group in enzymatic activity. Such strategic fine-tuning of a catalyst’s activity is highly desirable in biotechnology to increase the production yields of robust biocatalysts.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/chemistry5020080/s1, Figure S1: AFM images of (a) 1+Cu, (b) 1+Zn, (c) 2+Cu and (d) 2+Zn in DMSO showing the presence of nanoparticles and lack of fibrillar assembly; Figure S2: ThT Fluorescence emission spectra of the peptide samples with and without metal ions. Control: ThT in water not bound to any peptide; Figure S3: UV-vis spectrophotometric titration of peptides 1 and 2 with different concentrations of Cu(II) and Zn(II). (c = 0.5 mM); Figure S4: Absorbance vs. time plot for p-NPA hydrolysis by co-assemblies of 1 and 2 with metals in DMSO, showing diminished catalytic activity due to lack of nanofibrous assembly in DMSO.
Author Contributions
The project was conceptualized by A.P. All the detailed methodologies and investigations were performed by S.P. under close supervision by N.A.M. and A.P. Rheological studies were conducted by L.R. and A.S. was consulted for detailed kinetic data analysis. All authors have read and agreed to the published version of the manuscript.
Funding
A. Pal thanks the Department of Science and Technology (DST, SERB project CRG/2020/004251) and the Department of Biotechnology (BIRAC-PACE, BT/AIR01159/PACE-21/20) for their grants of financial support.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank the Institute of Nano Science and Technology (INST), Mohali for providing infrastructure and characterization facilities. N.M. gratefully acknowledges the INST fellowship.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Benkovic, S.J.; Hammes-Schiffer, S. A Perspective on Enzyme Catalysis. Science 2003, 301, 1196–1202. [Google Scholar] [CrossRef] [PubMed]
- Changeux, J.-P.; Edelstein, S.J. Allosteric Mechanisms of Signal Transduction. Science 2005, 308, 1424–1428. [Google Scholar] [CrossRef] [PubMed]
- VanEsch, J.H.; Klajn, R.; Otto, S. Chemical systems out of equilibrium. Chem. Soc. Rev. 2017, 46, 5474–5475. [Google Scholar] [CrossRef]
- Sorrenti, A.; Leira-Iglesias, J.; Markvoort, A.J.; Greef, T.F.A.d.; Hermans, T.M. Non-equilibrium supramolecular polymerization. Chem. Soc. Rev. 2017, 46, 5476–5490. [Google Scholar] [CrossRef] [PubMed]
- McManus, J.J.; Charbonneau, P.; Zaccarelli, E.; Asherie, N. The physics of protein self-assembly. Curr. Opin. Colloid Interface Sci. 2016, 22, 73–79. [Google Scholar] [CrossRef]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Carny, O.; Gazit, E. A model for the role of short self-assembled peptides in the very early stages of the origin of life. FASEB J. 2005, 19, 1051–1055. [Google Scholar] [CrossRef]
- Greenwald, J.; Riek, R. On the Possible Amyloid Origin of Protein Folds. J. Mol. Biol. 2012, 421, 417–426. [Google Scholar] [CrossRef]
- Mattio, E.; Pal, A.; Leonetti, G.; Otto, S. Mechanism of building block exchange in stacks of self-replicating macrocycles. Synlett 2017, 28, 103–107. [Google Scholar] [CrossRef]
- Jaeger, K.; Eggert, T. Enantioselective biocatalysis optimized by directed evolution. Curr. Opin. Biotechnol. 2004, 15, 305–313. [Google Scholar] [CrossRef]
- Aldridge, S. Industry backs biocatalysis for greener manufacturing. Nat. Biotechnol. 2013, 31, 95–96. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xue, X.; Luo, Q.; Li, Y.; Yang, K.; Zhuang, X.; Jiang, Y.; Zhang, J.; Liu, J.; Zou, G.; et al. Self-Assembled Peptide Nanofibers Designed as Biological Enzymes for Catalyzing Ester Hydrolysis. ACS Nano 2014, 8, 11715–11723. [Google Scholar] [CrossRef] [PubMed]
- Rufo, C.M.; Moroz, Y.S.; Moroz, O.V.; Stöhr, J.; Smith, T.A.; Hu, X.; DeGrado, W.F.; Korendovych, I.V. Short peptides self-assemble to produce catalytic amyloids. Nat. Chem. 2014, 6, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.F.; Lopes, A.B.; Fernandes, P.A.; Ramos, M.J. The Zinc proteome: A tale of stability and functionality. Dalton Trans. 2009, 7946–7956. [Google Scholar] [CrossRef] [PubMed]
- Festa, R.A.; Thiele, D.J. Copper: An essential metal in biology. Curr. Biol. 2011, 21, R877–R883. [Google Scholar] [CrossRef]
- Frieden, E. Copper and iron metalloproteins. Trends Biochem. Sci. 1976, 1, 273–274. [Google Scholar] [CrossRef]
- Christianson, D.W.; Fierke, C.A. Carbonic Anhydrase: Evolution of the Zinc Binding Site by Nature and by Design. Acc. Chem. Res. 1996, 29, 331–339. [Google Scholar] [CrossRef]
- Zastrow, M.L.; Pecoraro, V.L. Designing Hydrolytic Zinc Metalloenzymes. Biochemistry 2014, 53, 957–978. [Google Scholar] [CrossRef]
- Singh, A.; Joo, J.-U.; Kim, D.-P. Microfluidic-driven ultrafast self-assembly of a dipeptide into stimuli-responsive 0D, 1D, and 2D nanostructures and as hydrolase mimic. Nanoscale 2022, 14, 15010–15020. [Google Scholar] [CrossRef]
- Makam, P.; Yamijala, S.S.R.K.C.; Tao, K.; Shimon, L.J.W.; Eisenberg, D.S.; Sawaya, M.R.; Wong, B.M.; Gazit, E. Non-proteinaceous hydrolase comprised of a phenylalanine metallo-supramolecular amyloid-like structure. Nat. Catal. 2019, 2, 977–985. [Google Scholar] [CrossRef]
- Lengyel, Z.; Rufo, C.M.; Moroz, Y.S.; Makhlynets, O.V.; Korendovych, I.V. Copper-Containing Catalytic Amyloids Promote Phosphoester Hydrolysis and Tandem Reactions. ACS Catal. 2018, 8, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.L.; Ulijn, R.V. Short Peptides in Minimalistic Biocatalyst Design. Biocatalysis 2015, 1, 67–81. [Google Scholar] [CrossRef]
- Gulseren, G.; Khalily, M.A.; Tekinay, A.B.; Guler, M.O. Catalytic supramolecular self-assembled peptide nanostructures for ester hydrolysis. J. Mater. Chem. B 2016, 4, 4605–4611. [Google Scholar] [CrossRef]
- Joseph, J.P.; Singh, A.; Gupta, D.; Miglani, C.; Pal, A. Tandem Interplay of the Host–Guest Interaction and Photoresponsive Supramolecular Polymerization to 1D and 2D Functional Peptide Materials. ACS Appl. Mater. Interfaces 2019, 11, 28213–28220. [Google Scholar] [CrossRef]
- Mavlankar, N.A.; Awasthi, A.K.; Ralhan, J.; Pal, A. Amyloid-inspired Peptide Self-assembly/Disassembly as Intervened by Gold Nanoparticles and Polydopamine Coating to Dictate Spatiotemporal Organization. ChemNanoMat 2022, 8, e202200368. [Google Scholar] [CrossRef]
- Singh, A.; Joseph, J.P.; Gupta, D.; Sarkar, I.; Pal, A. Pathway driven self-assembly and living supramolecular polymerization in an amyloid-inspired peptide amphiphile. Chem. Commun. 2018, 54, 10730–10733. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.P.; Gupta, N.; Miglani, C.; Nath, D.; Singh, A.; Gupta, D.; Pal, A. Unraveling On-demand Strain-Stiffening in Nanofibrous Peptide–Polymer Conjugates to Mimic Contractility in Actinomyosin Networks. Chem. Mater. 2022, 34, 4364–4374. [Google Scholar] [CrossRef]
- Gupta, D.; Bhatt, A.; Gupta, V.; Miglani, C.; Joseph, J.P.; Ralhan, J.; Mandal, D.; Ali, M.E.; Pal, A. Photochemically Sequestered Off-Pathway Dormant States of Peptide Amphiphiles for Predictive On-Demand Piezoresponsive Nanostructures. Chem. Mater. 2022, 34, 4456–4470. [Google Scholar] [CrossRef]
- Gupta, N.; Singh, A.; Dey, N.; Chattopadhyay, S.; Joseph, J.P.; Gupta, D.; Ganguli, M.; Pal, A. Pathway-Driven Peptide–Bioglass Nanocomposites as the Dynamic and Self-Healable Matrix. Chem. Mater. 2021, 33, 589–599. [Google Scholar] [CrossRef]
- Thomas, J.; Gupta, N.; Joseph, J.P.; Chopra, V.; Pal, A.; Ghosh, D. Mechanical Integrity in a Dynamic Interpenetrating Hydrogel Network of Supramolecular Peptide–Polysaccharide Supports Enhanced Chondrogenesis. ACS Biomater. Sci. Eng. 2021, 7, 5798–5809. [Google Scholar] [CrossRef]
- Gupta, D.; Sasmal, R.; Singh, A.; Joseph, J.P.; Miglani, C.; Agasti, S.S.; Pal, A. Enzyme-responsive chiral self-sorting in amyloid-inspired minimalistic peptide amphiphiles. Nanoscale 2020, 12, 18692–18700. [Google Scholar] [CrossRef]
- Singh, A.; Joseph, J.P.; Gupta, D.; Miglani, C.; Mavlankar, N.A.; Pal, A. Photothermally switchable peptide nanostructures towards modulating catalytic hydrolase activity. Nanoscale 2021, 13, 13401–13409. [Google Scholar] [CrossRef] [PubMed]
- Hartgerink, J.D.; Beniash, E.; Stupp, S.I. Self-Assembly and Mineralization of Peptide-Amphiphile Nanofibers. Science 2001, 294, 1684–1688. [Google Scholar] [CrossRef] [PubMed]
- Al-Garawi, Z.S.; McIntosh, B.A.; Neill-Hall, D.; Hatimy, A.A.; Sweet, S.M.; Bagley, M.C.; Serpell, L.C. The amyloid architecture provides a scaffold for enzyme-like catalysts. Nanoscale 2017, 9, 10773–10783. [Google Scholar] [CrossRef] [PubMed]
- Hernick, M.; Fierke, C.A. Zinc hydrolases: The mechanisms of zinc-dependent deacetylases. Arch. Biochem. Biophys. 2005, 433, 71–84. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).