Hydrogen and Halogen Bond Mediated Coordination Polymers of Chloro-Substituted Pyrazin-2-Amine Copper(I) Bromide Complexes
Abstract
1. Introduction
2. Materials and Methods
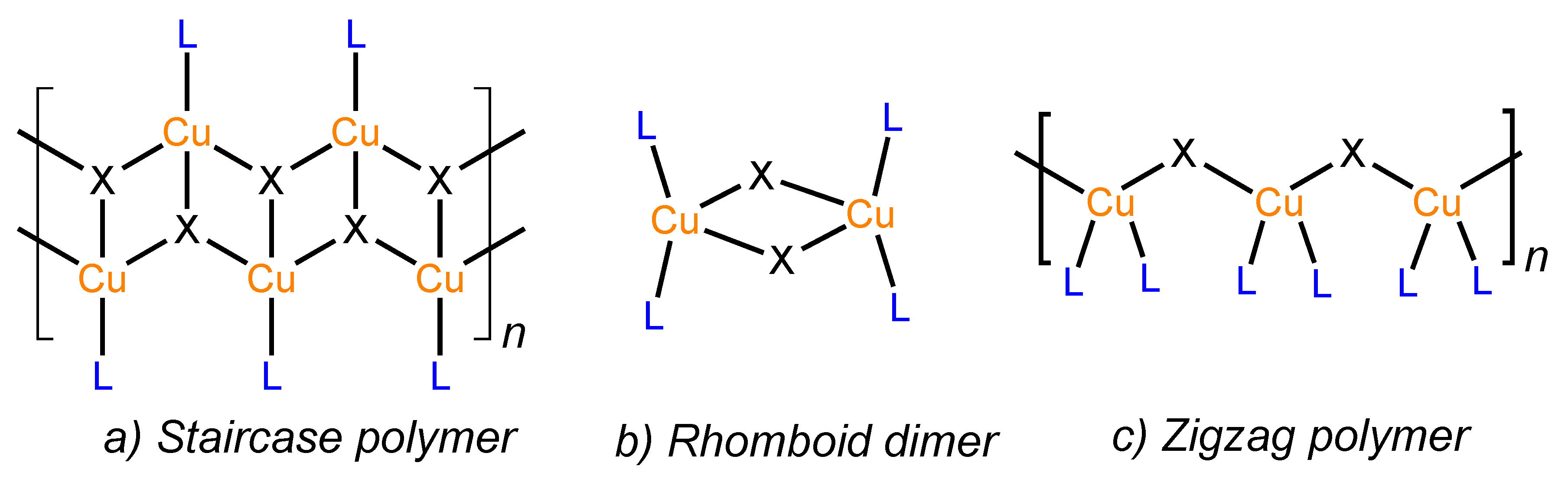
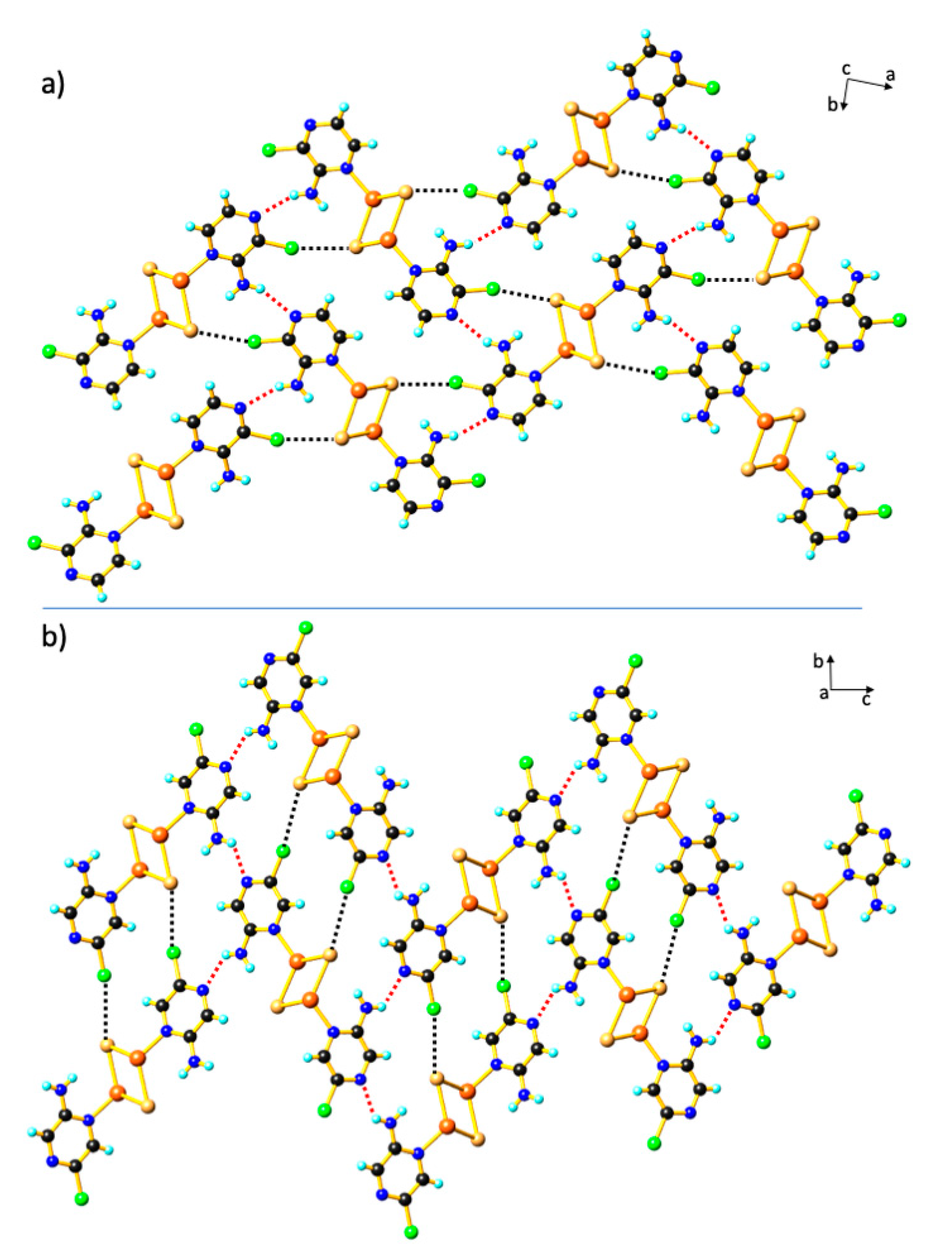
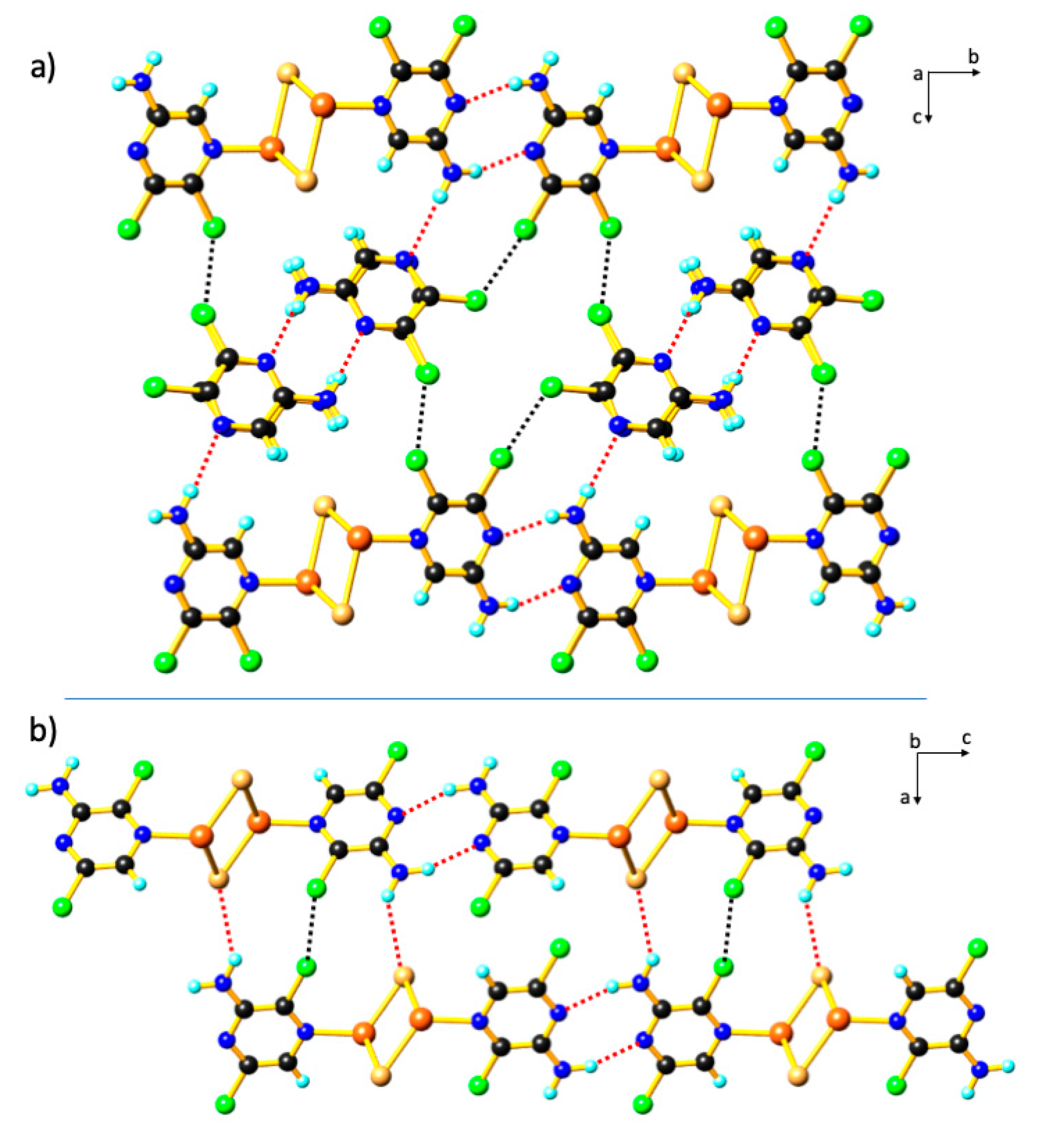
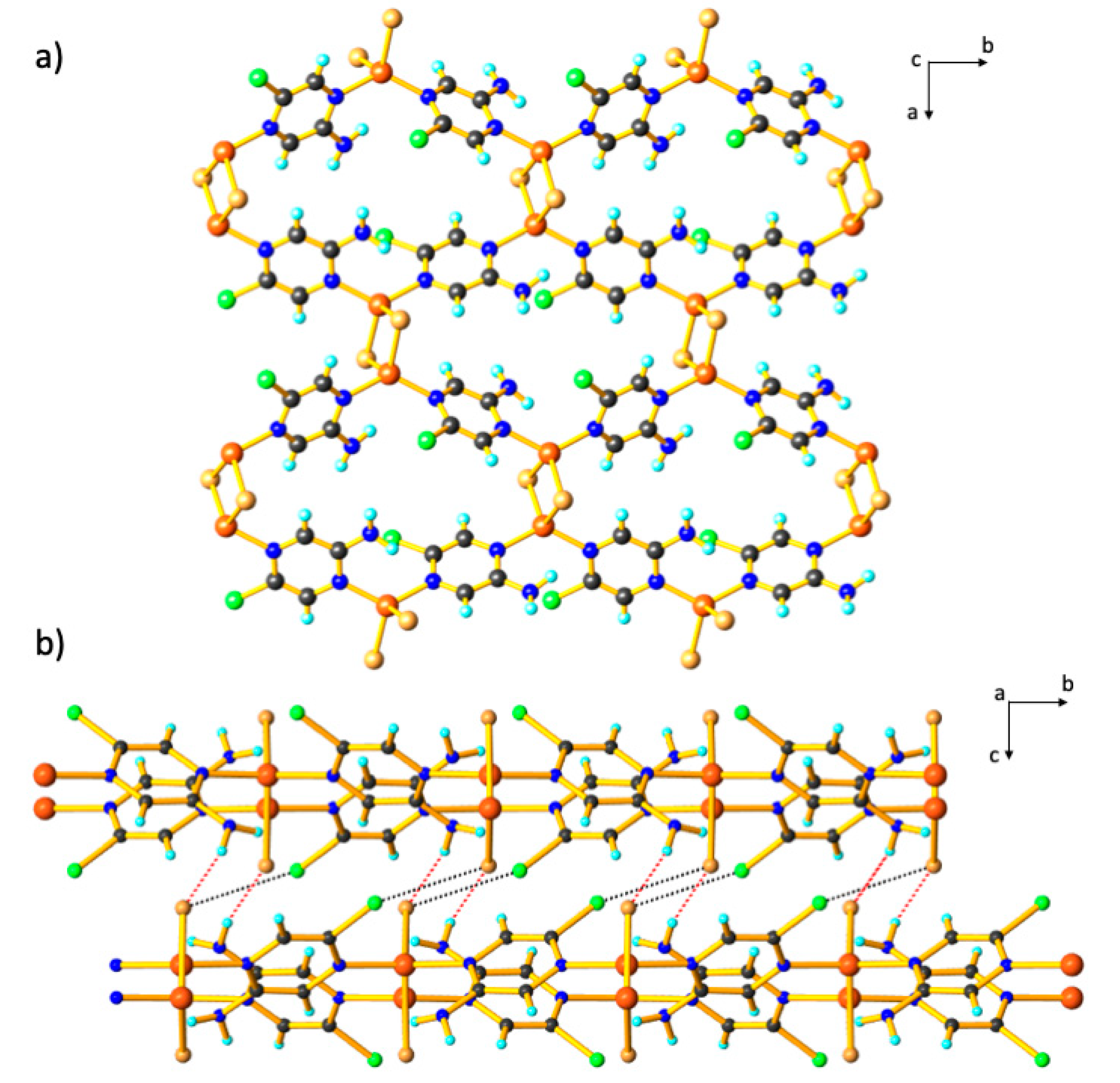
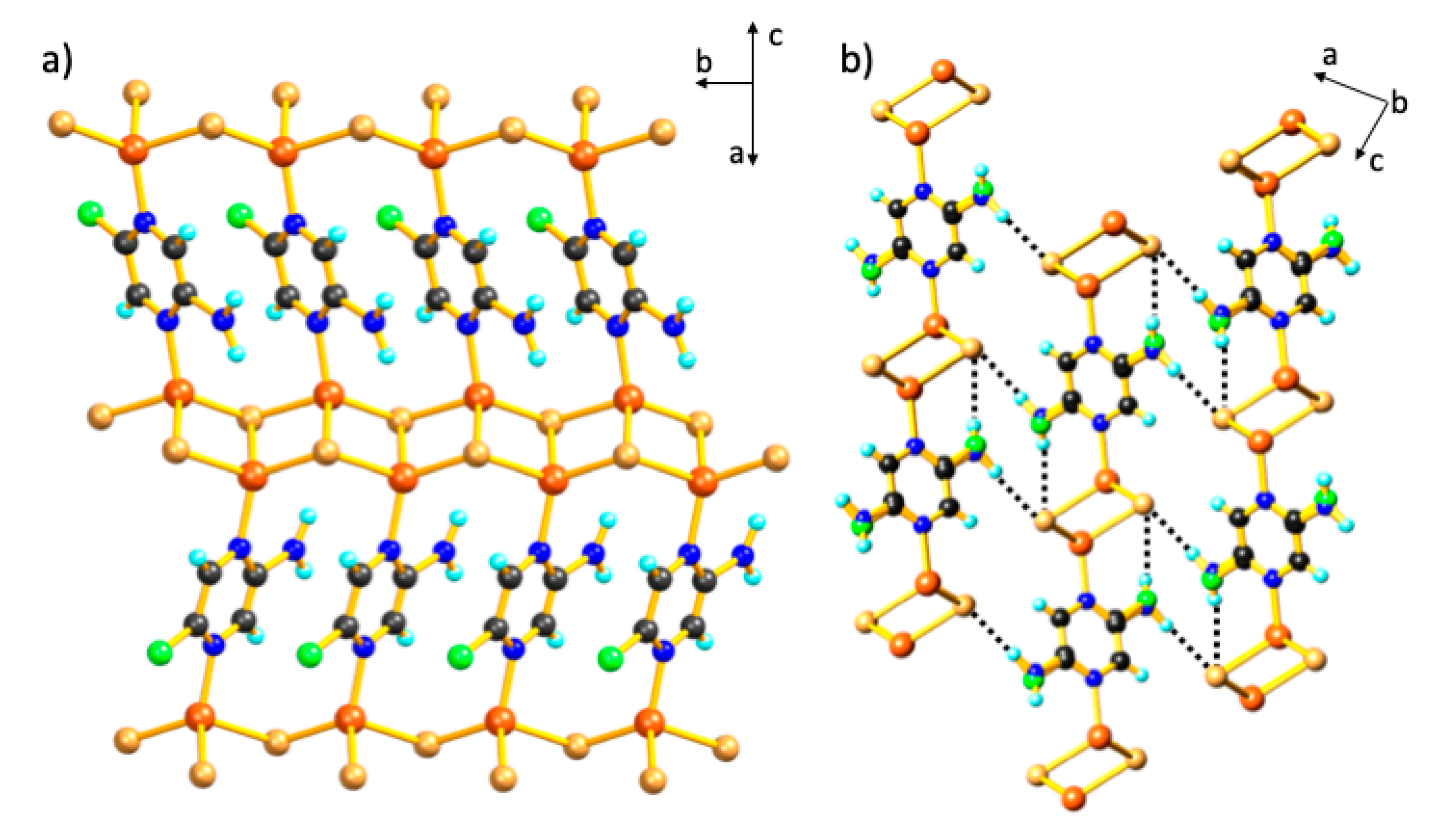
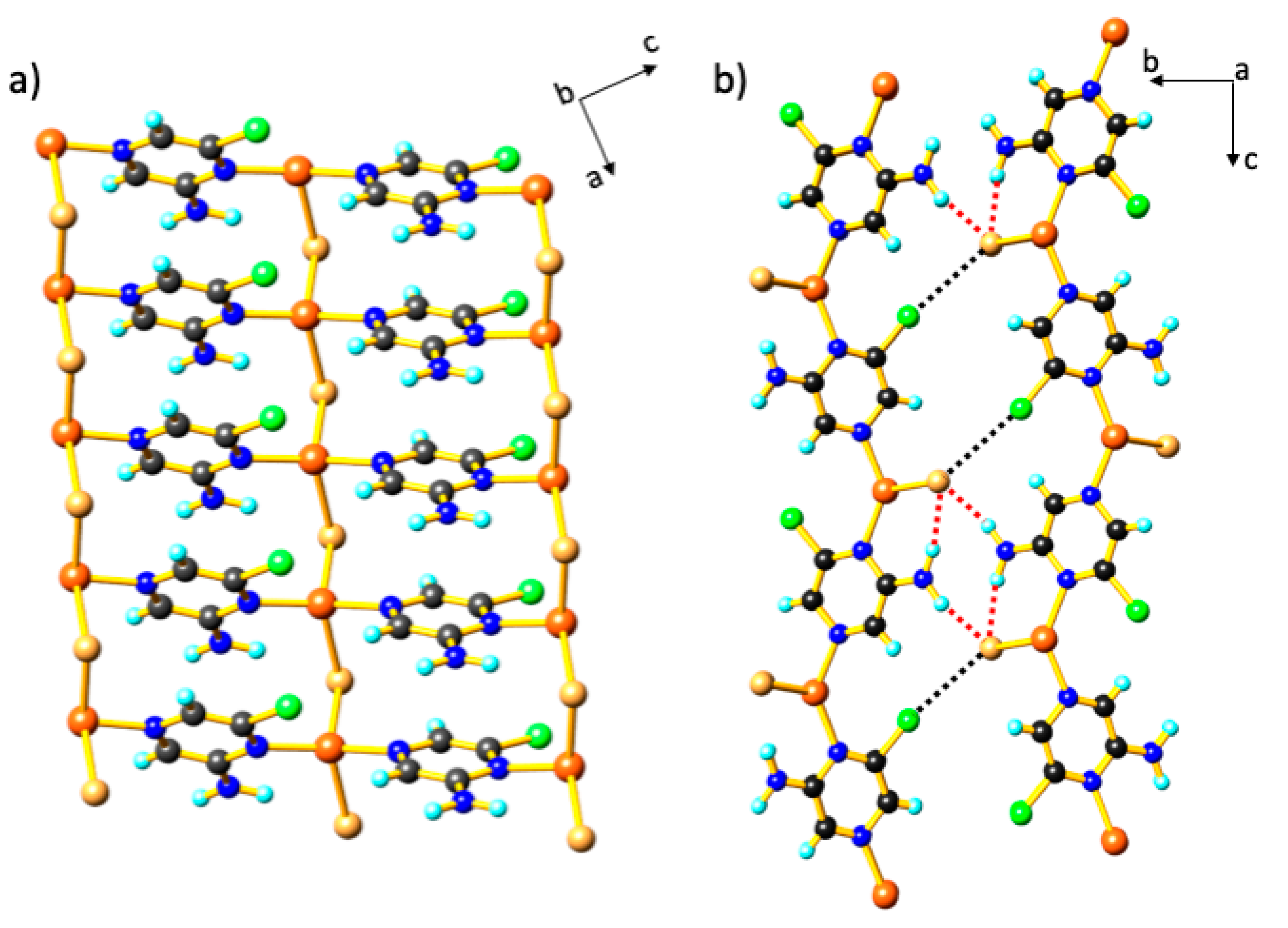
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Reference and Note
- Desiraju, G.R.; Vittal, J.J.; Ramanan, A. Crystal Engineering: A Textbook; World Scientific: Singapore, 2011. [Google Scholar]
- Jeffrey, G.A.; Saenger, W. Hydrogen Bonding in Biological Structures; Springer: Berlin/Heidelberg, Germany, 1991. [Google Scholar]
- Amabilino, D.B.; Smith, D.K.; Steed, J.W. Supramolecular materials. Chem. Soc. Rev. 2017, 46, 2404–2420. [Google Scholar] [CrossRef] [PubMed]
- Biradha, K.; Desiraju, G.R.; Braga, D.; Grepioni, F. Hydrogen Bonding in Organometallic Crystals. 3.1Transition-Metal Complexes Containing Amido Groups. Organometallics 1996, 15, 1284–1295. [Google Scholar] [CrossRef]
- Brammer, L.; Rivas, J.C.M.; Atencio, R.; Fang, S.; Pigge, F.C. Combining hydrogen bonds with coordination chemistry or organometallic π-arene chemistry: Strategies for inorganic crystal engineering. J. Chem. Soc. Dalton Trans. 2000, 3855–3867. [Google Scholar] [CrossRef]
- Beatty, A.M. Hydrogen bonded networks of coordination complexes. CrystEngComm 2001, 3, 243–255. [Google Scholar] [CrossRef]
- Epstein, L.M.; Shubina, E.S. New types of hydrogen bonding in organometallic chemistry. Coord. Chem. Rev. 2002, 231, 165–181. [Google Scholar] [CrossRef]
- Nair, K.P.; Pollino, J.M.; Weck, M. Noncovalently Functionalized Block Copolymers Possessing Both Hydrogen Bonding and Metal Coordination Centers. Macromolecules 2006, 39, 931–940. [Google Scholar] [CrossRef]
- Chandrasekhar, P.; Mukhopadhyay, A.; Savitha, G.; Moorthy, J.N. Orthogonal self-assembly of a trigonal triptycene triacid: Signaling of exfoliation of porous 2D metal–organic layers by fluorescence and selective CO2 capture by the hydrogen-bonded MOF. J. Mater. Chem. A 2017, 5, 5402–5412. [Google Scholar] [CrossRef]
- Aakeroy, C.B.; Beatty, A.M. Solid State, Crystal Engineering and Hydrogen Bonds. ChemInform 2004, 35. [Google Scholar] [CrossRef]
- Cook, S.A.; Borovik, A.S. Molecular Designs for Controlling the Local Environments around Metal Ions. Acc. Chem. Res. 2015, 48, 2407–2414. [Google Scholar] [CrossRef]
- Friese, V.A.; Kurth, D.G. Soluble dynamic coordination polymers as a paradigm for materials science. Coord. Chem. Rev. 2008, 252, 199–211. [Google Scholar] [CrossRef]
- Seetharaj, R.; Vandana, P.V.; Arya, P.; Mathew, S. Dependence of solvents, pH, molar ratio and temperature in tuning metal organic framework architecture. Arab. J. Chem. 2019, 12, 295–315. [Google Scholar] [CrossRef]
- Zhu, R.; Lübben, J.; Dittrich, B.; Clever, G.H. Stepwise Halide-Triggered Double and Triple Catenation of Self-Assembled Coordination Cages. Angew. Chem. Int. Ed. 2015, 54, 2796–2800. [Google Scholar] [CrossRef]
- Sun, Q.-F.; Iwasa, J.; Ogawa, D.; Ishido, Y.; Sato, S.; Ozeki, T.; Sei, Y.; Yamaguchi, K.; Fujita, M. Self-Assembled M24L48 Polyhedra and Their Sharp Structural Switch upon Subtle Ligand Variation. Science 2010, 328, 1144–1147. [Google Scholar] [CrossRef]
- Schultz, A.; Li, X.; Barkakaty, B.; Moorefield, C.N.; Wesdemiotis, C.; Newkome, G.R. Stoichiometric Self-Assembly of Isomeric, Shape-Persistent, Supramacromolecular Bowtie and Butterfly Structures. J. Am. Chem. Soc. 2012, 134, 7672–7675. [Google Scholar] [CrossRef]
- Li, S.; Moorefield, C.N.; Wang, P.; Shreiner, C.D.; Newkome, G.R. Self-Assembly of Shape-Persistent Hexagonal Macrocycles with Trimeric Bis(terpyridine)–FeII Connectivity. Eur. J. Org. Chem. 2008, 2008, 3328–3334. [Google Scholar] [CrossRef]
- Tominaga, M.; Suzuki, K.; Kawano, M.; Kusukawa, T.; Ozeki, T.; Sakamoto, S.; Yamaguchi, K.; Fujita, M. Finite, Spherical Coordination Networks that Self-Organize from 36 Small Components. Angew. Chem. Int. Ed. 2004, 43, 5621–5625. [Google Scholar] [CrossRef]
- Ronson, T.K.; Fisher, J.; Harding, L.P.; Rizkallah, P.J.; Warren, J.E.; Hardie, M.J. Stellated polyhedral assembly of a topologically complicated Pd4L4 ‘Solomon cube’. Nat. Chem. 2009, 1, 212–216. [Google Scholar] [CrossRef]
- Olenyuk, B.; Whiteford, J.A.; Fechtenkötter, A.; Stang, P.J. Self-assembly of nanoscale cuboctahedra by coordination chemistry. Nature 1999, 398, 796–799. [Google Scholar] [CrossRef]
- Song, B.; Kandapal, S.; Gu, J.; Zhang, K.; Reese, A.; Ying, Y.; Wang, L.; Wang, H.; Li, Y.; Wang, M.; et al. Self-assembly of polycyclic supramolecules using linear metal-organic ligands. Nat. Commun. 2018, 9, 4575. [Google Scholar] [CrossRef]
- Zhang, J.-W.; Kan, X.-M.; Li, X.-L.; Luan, J.; Wang, X.-L. Transition metal carboxylate coordination polymers with amide-bridged polypyridine co-ligands: Assemblies and properties. CrystEngComm 2015, 17, 3887–3907. [Google Scholar] [CrossRef]
- Ali Akbar Razavi, S.; Morsali, A. Linker functionalized metal-organic frameworks. Coord. Chem. Rev. 2019, 399, 213023. [Google Scholar] [CrossRef]
- Goura, J.; Chandrasekhar, V. Molecular Metal Phosphonates. Chem. Rev. 2015, 115, 6854–6965. [Google Scholar] [CrossRef]
- Lawrance, G.A. Mixed Donor Ligands Based in part on the article Mixed Donor Ligands by Nikolay N. Gerasimchuk & Kristin Bowman-James which appeared in the Encyclopedia of Inorganic Chemistry, First Edition. Available online: https://doi.org/10.1002/9781119951438.eibc0132 (accessed on 15 December 2011).
- Taghipour, F.; Mirzaei, M. A survey of interactions in crystal structures of pyrazine-based compounds. Acta Crystallogr. Sect. C 2019, 75, 231–247. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Stoeckli-Evans, H. The inner-salt zwitterion, the dihydrochloride dihydrate and the dimethyl sulfoxide disolvate of 3,6-bis(pyridin-2-yl)pyrazine-2,5-dicarboxylic acid. Acta Crystallogr. Sect. C 2012, 68, o431–o435. [Google Scholar] [CrossRef]
- Dobson, A.J.; Gerkin, R.E. 3-Aminopyrazine-2-carboxylic Acid. Acta Crystallogr. Sect. C 1996, 52, 1512–1514. [Google Scholar] [CrossRef] [PubMed]
- Berrah, F.; Bouacida, S.; Roisnel, T. 2-Amino-3-carboxypyrazin-1-ium dihydrogen phosphate. Acta Crystallogr. Sect. E 2011, 67, o1409–o1410. [Google Scholar] [CrossRef] [PubMed]
- Berrah, F.; Ouakkaf, A.; Bouacida, S.; Roisnel, T. Bis(2-amino-3-carboxypyrazin-1-ium) sulfate dihydrate. Acta Crystallogr. Sect. E 2011, 67, o677–o678. [Google Scholar] [CrossRef] [PubMed]
- Berrah, F.; Bouacida, S.; Bouhraoua, A.; Roisnel, T. 2-Amino-3-carboxypyrazin-1-ium perchlorate bis(2-aminopyrazin-1-ium-3-carboxylate) monohydrate. Acta Crystallogr. Sect. E 2012, 68, o1714–o1715. [Google Scholar] [CrossRef]
- Barszcz, B.; Masternak, J.; Hodorowicz, M.; Jabłońska-Wawrzycka, A. Cadmium(II) and calcium(II) complexes with N,O-bidentate ligands derived from pyrazinecarboxylic acid. J. Therm. Anal. Calorim. 2012, 108, 971–978. [Google Scholar] [CrossRef]
- Goher, M.A.S.; Al-Salem, N.A.; Mautner, F.A.; Klepp, K.O. A copper(II) azide compound of pyrazinic acid containing a new dinuclear complex anion [Cu2(N3)6]2−. Synthesis, spectral and study of KCu2(pyrazinato) (N3)4. Polyhedron 1997, 16, 825–831. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, Y.; Ran, J.; Li, L. Synthesis, crystal structure, photoluminescence and catalytic properties of a novel cuprous complex with 2,3-pyrazinedicarboxylic acid ligands. Sci. Rep. 2020, 10, 6273. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-W.; Tao, Y.; Hu, T.-L. Synthesis, structure, and photoluminescence of ZnII and CdII coordination complexes constructed by structurally related 5,6-substituted pyrazine-2,3-dicarboxylate ligands. Solid State Sci. 2012, 14, 1117–1125. [Google Scholar] [CrossRef]
- Bouchene, R.; Khadri, A.; Bouacida, S.; Berrah, F.; Merazig, H. Bis(3-amino-pyrazine-2-carboxyl-ato-κ2 N(1) O)di-aqua-nickel(II) dihydrate. Acta Crystallogr. Sect. E 2013, 69, m309–m310. [Google Scholar] [CrossRef] [PubMed]
- Koleša-Dobravc, T.; Maejima, K.; Yoshikawa, Y.; Meden, A.; Yasui, H.; Perdih, F. Vanadium and zinc complexes of 5-cyanopicolinate and pyrazine derivatives: Synthesis, structural elucidation and in vitro insulino-mimetic activity study. New J. Chem. 2017, 41, 735–746. [Google Scholar] [CrossRef]
- Benhamada, N.; Bouchene, R.; Bouacida, S.; Zouchoune, B. Molecular structure, bonding analysis and redox properties of transition metal–Hapca [bis(3-aminopyrazine-2-carboxylic acid)] complexes: A theoretical study. Polyhedron 2015, 91, 59–67. [Google Scholar] [CrossRef]
- Dehghanpour, S.; Jahani, K.; Mahmoudi, A.; Babakhodaverdi, M.; Notash, B. In situ hydrothermal synthesis of 2D mercury(I)–organic framework from 3-aminopyrazine-2-carboxylic acid and mercury(II) acetate. Inorg. Chem. Commun. 2012, 25, 79–82. [Google Scholar] [CrossRef]
- Desiraju, G.R.; Ho, P.S.; Kloo, L.; Legon, A.C.; Marquardt, R.; Metrangolo, P.; Politzer, P.; Resnati, G.; Rissanen, K. Definition of the halogen bond (IUPAC Recommendations 2013). Pure Appl. Chem. 2013, 85, 1711–1713. [Google Scholar] [CrossRef]
- Cavallo, G.; Metrangolo, P.; Milani, R.; Pilati, T.; Priimagi, A.; Resnati, G.; Terraneo, G. The Halogen Bond. Chem. Rev. 2016, 116, 2478–2601. [Google Scholar] [CrossRef]
- Gilday, L.C.; Robinson, S.W.; Barendt, T.A.; Langton, M.J.; Mullaney, B.R.; Beer, P.D. Halogen Bonding in Supramolecular Chemistry. Chem. Rev. 2015, 115, 7118–7195. [Google Scholar] [CrossRef]
- Troff, R.W.; Mäkelä, T.; Topić, F.; Valkonen, A.; Raatikainen, K.; Rissanen, K. Alternative Motifs for Halogen Bonding. Eur. J. Org. Chem. 2013, 2013, 1617–1637. [Google Scholar] [CrossRef]
- Rissanen, K. Halogen bonded supramolecular complexes and networks. CrystEngComm 2008, 10, 1107–1113. [Google Scholar] [CrossRef]
- Bui, T.T.T.; Dahaoui, S.; Lecomte, C.; Desiraju, G.R.; Espinosa, E. The Nature of Halogen⋅⋅⋅Halogen Interactions: A Model Derived from Experimental Charge-Density Analysis. Angew. Chem. Int. Ed. 2009, 48, 3838–3841. [Google Scholar] [CrossRef] [PubMed]
- Metrangolo, P.; Resnati, G. Type II halogen⋅⋅⋅halogen contacts are halogen bonds. IUCrJ 2014, 1, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Awwadi, F.F.; Willett, R.D.; Peterson, K.A.; Twamley, B. The Nature of Halogen⋅⋅⋅Halogen Synthons: Crystallographic and Theoretical Studies. Chem. A Eur. J. 2006, 12, 8952–8960. [Google Scholar] [CrossRef] [PubMed]
- Brammer, L.; Minguez Espallargas, G.; Adams, H. Involving metals in halogen-halogen interactions: Second-sphere Lewis acid ligands for perhalometallate ions (M-X⋅⋅⋅X’-C). CrystEngComm 2003, 5, 343–345. [Google Scholar] [CrossRef]
- Von Essen, C.; Rissanen, K.; Puttreddy, R. Halogen Bonds in 2,5-Dihalopyridine-Copper(I) Halide Coordination Polymers. Materials 2019, 12, 3305. [Google Scholar] [CrossRef]
- Puttreddy, R.; von Essen, C.; Peuronen, A.; Lahtinen, M.; Rissanen, K. Halogen bonds in 2,5-dihalopyridine-copper(ii) chloride complexes. CrystEngComm 2018, 20, 1954–1959. [Google Scholar] [CrossRef]
- Puttreddy, R.; von Essen, C.; Rissanen, K. Halogen Bonds in Square Planar 2,5-Dihalopyridine–Copper(II) Bromide Complexes. Eur. J. Inorg. Chem. 2018, 2018, 2393–2398. [Google Scholar] [CrossRef]
- Puttreddy, R.; Peuronen, A.; Lahtinen, M.; Rissanen, K. Metal-Bound Nitrate Anion as an Acceptor for Halogen Bonds in Mono-Halopyridine-Copper(II) Nitrate Complexes. Cryst. Growth Des. 2019, 19, 3815–3824. [Google Scholar] [CrossRef]
- Kwak, S.H.; Lee, G.-H.; Gong, Y.-D. Synthesis of N-Substituted-2-Aminothiazolo[4,5-b]pyrazines by Tandem Reaction of o-Aminohalopyrazines with Isothiocyanates. Bull. Korean Chem. Soc. 2012, 33. [Google Scholar] [CrossRef]
- Caldwell, J.J.; Veillard, N.; Collins, I. Design and synthesis of 2(1H)-pyrazinones as inhibitors of protein kinases. Tetrahedron 2012, 68, 9713–9728. [Google Scholar] [CrossRef]
- Bartolomé-Nebreda, J.M.; Delgado, F.; Martín-Martín, M.L.; Martínez-Viturro, C.M.; Pastor, J.; Tong, H.M.; Iturrino, L.; Macdonald, G.J.; Sanderson, W.; Megens, A.; et al. Discovery of a Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitor for the Potential Treatment of Schizophrenia. J. Med. Chem. 2014, 57, 4196–4212. [Google Scholar] [CrossRef] [PubMed]
- Reader, J.C.; Matthews, T.P.; Klair, S.; Cheung, K.-M.J.; Scanlon, J.; Proisy, N.; Addison, G.; Ellard, J.; Piton, N.; Taylor, S.; et al. Structure-Guided Evolution of Potent and Selective CHK1 Inhibitors through Scaffold Morphing. J. Med. Chem. 2011, 54, 8328–8342. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.G.; Meyrick, B.H.; Nelson, A.J. Colour and constitution of azo compounds derived from diaminoazines. Tetrahedron 1984, 40, 5081–5088. [Google Scholar] [CrossRef]
- Miesel, J.L. Novel 1-(mono-o-substituted benzoyl)-3-(substituted pyrazinyl) ureas. US Patent No. 4,293,552, 31 July 1979. [Google Scholar]
- Massaux, P.M.; Bernard, M.J.; Le Bihan, M.-T. Etude Structurale de CuBr.CH3CN. Acta Cryst. 1972, B27, 2419–2424. [Google Scholar] [CrossRef]
- Rigaku Oxford Diffraction. CrysAlisPro Version 38.46; Rigaku: Tokyo, Japan, 2018. [Google Scholar]
- Bruker AXS Inc. COLLECT. In Bruker AXS BV, 1997-2004; Bruker AXS Inc.: Madison, WI, USA.
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SADABS. Program for Empirical Absorption Correction; University of Göttingen: Göttingen, Lower Saxony, Germany, 1996. [Google Scholar]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Cryst. 2015, C71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H.J.J. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Gruene, T.; Hahn, H.W.; Luebben, A.V.; Meilleur, F.; Sheldrick, G.M.J. Refinement of macromolecular structures against neutron data with SHELXL2013. Appl. Cryst. 2014, 47, 462–466. [Google Scholar] [CrossRef]
- Degen, T.; Sadki, M.; Bron, E.; König, U.; Nénert, G. The HighScore suite. Powder Diffraction (Supplement S2). 20 December 2014; 29, S13–S18. [Google Scholar]
- Pawley, G.S. Unit-cell refinement from powder diffraction scans. J. Appl. Cryst. 1981, 14, 357–361. [Google Scholar] [CrossRef]
- Mailman, A.; unpublished results: A preliminary structure of the CuBr2 complex of 2-amino-5-chloropyrazine (2), i.e. 2∙CuBr2 has been determined by single crystal X-ray diffraction methods: Triclinic, P-1, a = 6.002(5), 7.078 (5), 8.467 (7), α = 94.225 (6), β =97.238(7) γ = 102.105(7), V = 347.03 Å3.
- Kochi, J.K. The Reduction of Cupric Chloride by Carbonyl Compounds. J. Am. Chem. Soc. 1955, 77, 5274–5278. [Google Scholar] [CrossRef]
- Yamada, Y.; Ohba, H.; Noboru, Y.; Daicho, S.; Nibu, Y. Solvation Effect on the NH Stretching Vibrations of Solvated Aminopyrazine, 2-Aminopyridine, and 3-Aminopyridine Clusters. J. Phys. Chem. A 2012, 116, 9271–9278. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Li, M.; Li, D. Copper(I) halides: A versatile family in coordination chemistry and crystal engineering. Coord. Chem. Rev. 2010, 254, 1–18. [Google Scholar] [CrossRef]
- Knorr, M.; Bonnot, A.; Lapprand, A.; Khatyr, A.; Strohmann, C.; Kubicki, M.M.; Rousselin, Y.; Harvey, P.D. Reactivity of CuI and CuBr toward Dialkyl Sulfides RSR: From Discrete Molecular Cu4I4S4 and Cu8I8S6 Clusters to Luminescent Copper(I) Coordination Polymers. Inorg. Chem. 2015, 54, 4076–4093. [Google Scholar] [CrossRef] [PubMed]
- Harisomayajula, N.V.S.; Makovetskyi, S.; Tsai, Y.-C. Cuprophilic Interactions in and between Molecular Entities. Chem. A Eur. J. 2019, 25, 8936–8954. [Google Scholar] [CrossRef] [PubMed]
- Barclay, T.M.; Cordes, A.W.; Oakley, R.T.; Preuss, K.E.; Zhang, H. 2,5-Diamino-3,6-dichloropyrazine. Acta Crystallogr. Sect. C 1998, 54, 1018–1019. [Google Scholar] [CrossRef]
- Chao, M.; Schempp, E.; Rosenstein, R.D. Aminopyrazine. Acta Crystallogr. Sect. B 1976, 32, 288–290. [Google Scholar] [CrossRef]
- Näther, C.; Greve, J.; Jeß, I. Synthesis, crystal structures and thermal properties of new copper(I) halide coordination polymers. Solid State Sci. 2002, 4, 813–820. [Google Scholar] [CrossRef]
- Conesa-Egea, J.; Gallardo-Martínez, J.; Delgado, S.; Martínez, J.I.; Gonzalez-Platas, J.; Fernández-Moreira, V.; Rodríguez-Mendoza, U.R.; Ocón, P.; Zamora, F.; Amo-Ochoa, P. Multistimuli Response Micro- and Nanolayers of a Coordination Polymer Based on Cu2I2 Chains Linked by 2-Aminopyrazine. Small 2017, 13, 1700965. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mailman, A.; Puttreddy, R.; Lahtinen, M.; Svahn, N.; Rissanen, K. Hydrogen and Halogen Bond Mediated Coordination Polymers of Chloro-Substituted Pyrazin-2-Amine Copper(I) Bromide Complexes. Chemistry 2020, 2, 700-713. https://doi.org/10.3390/chemistry2030045
Mailman A, Puttreddy R, Lahtinen M, Svahn N, Rissanen K. Hydrogen and Halogen Bond Mediated Coordination Polymers of Chloro-Substituted Pyrazin-2-Amine Copper(I) Bromide Complexes. Chemistry. 2020; 2(3):700-713. https://doi.org/10.3390/chemistry2030045
Chicago/Turabian StyleMailman, Aaron, Rakesh Puttreddy, Manu Lahtinen, Noora Svahn, and Kari Rissanen. 2020. "Hydrogen and Halogen Bond Mediated Coordination Polymers of Chloro-Substituted Pyrazin-2-Amine Copper(I) Bromide Complexes" Chemistry 2, no. 3: 700-713. https://doi.org/10.3390/chemistry2030045
APA StyleMailman, A., Puttreddy, R., Lahtinen, M., Svahn, N., & Rissanen, K. (2020). Hydrogen and Halogen Bond Mediated Coordination Polymers of Chloro-Substituted Pyrazin-2-Amine Copper(I) Bromide Complexes. Chemistry, 2(3), 700-713. https://doi.org/10.3390/chemistry2030045






