Abstract
Halogenated carboxylic acids have been important compounds in chemical synthesis and indispensable research tools in biochemical studies for decades. Nevertheless, the number of structurally characterized simple α-brominated monocarboxylic acids is still limited. We herein report the crystallization and structural elucidation of (R)- and rac-2-bromo-3-methylbutyric acid (2-bromo-3-methylbutanoic acid, 1) to shed light on intermolecular interactions, in particular hydrogen bonding motifs, packing modes and preferred conformations in the solid-state. The crystal structures of (R)- and rac-1 are revealed by X-ray crystallography. Both compounds crystallize in the triclinic crystal system with Z = 2; (R)-1 exhibits two crystallographically distinct molecules. In the crystal, (R)-1 forms homochiral O–H···O hydrogen-bonded carboxylic acid dimers with approximate non-crystallographic C2 symmetry. In contrast, rac-1 features centrosymmetric heterochiral dimers with the same carboxy syn···syn homosynthon. The crystal packing of centrosymmetric rac-1 is denser than that of its enantiopure counterpart (R)-1. The molecules in both crystal structures adopt a virtually identical staggered conformation, despite different crystal environments, which indicates a preferred molecular structure of 1. Intermolecular interactions apart from classical O–H···O hydrogen bonds do not appear to have a crucial bearing on the solid-state structures of (R)- and rac-1.
1. Introduction
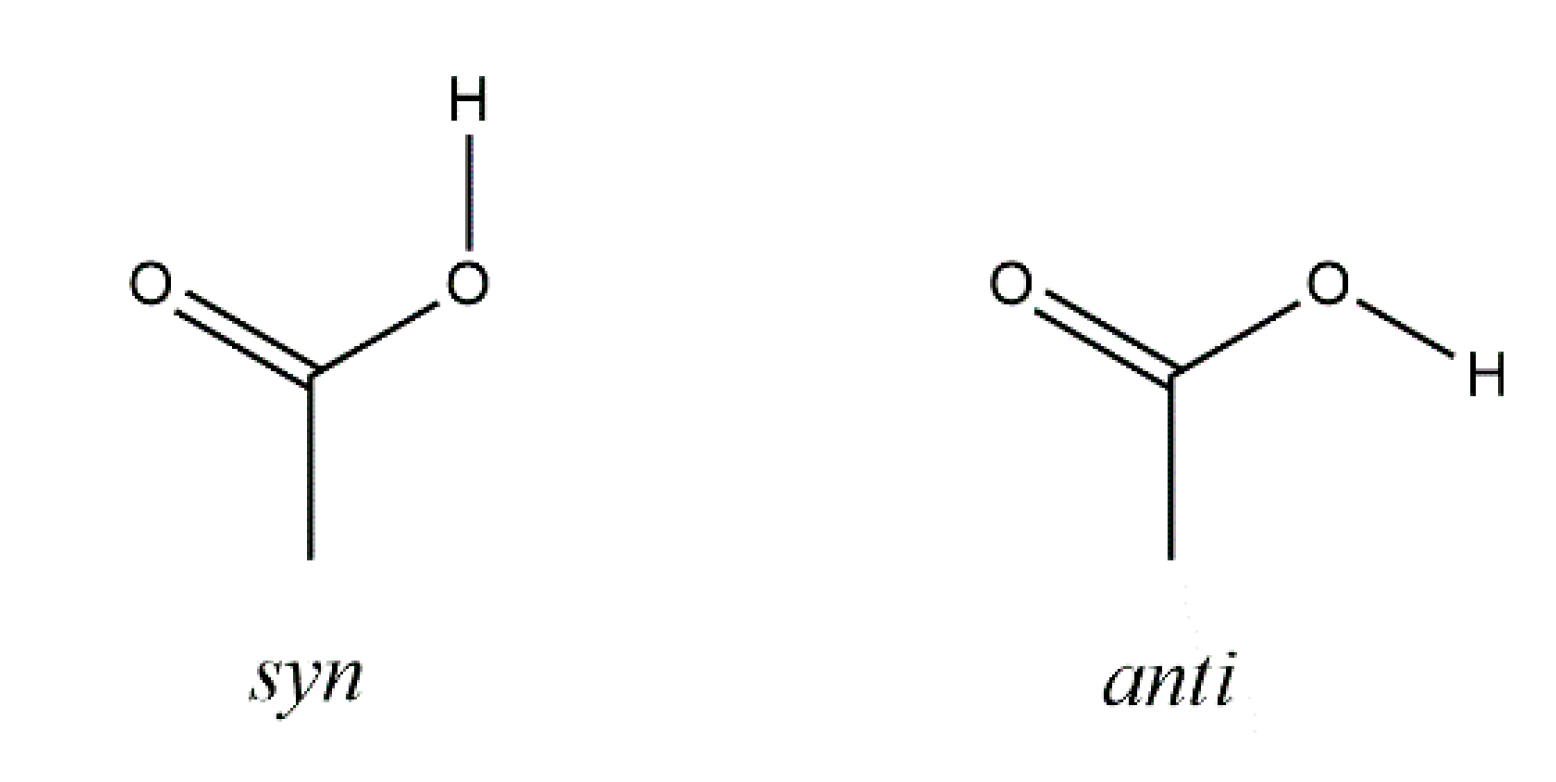
Halogenated organic compounds have received considerable research interest for decades, not only in the field of chemical synthesis [1,2,3] but also because of their biological properties [4,5]. In particular, a vast number of halogenated carboxylic acids have been synthesized and biochemically studied. Since mono-, di- and tricarboxylic acids are important intermediates in many biochemical pathways, their halogenated analogues have become an important research tool for the study of a wide range of biological processes owing to their ability to imitate the properties of the respective carboxylic acids or to inhibit crucial enzymes [6,7]. Despite tremendous research interest in halogenated carboxylic acids, the number of crystal structures of simple α-brominated monocarboxylic acids in the Cambridge Structural Database (CSD) is limited (13 as of June 2020) [8]. An example is bromoacetic acid, which forms a common syn···syn hydrogen-bonded carboxy dimer (Scheme 1) in the crystal (CSD refcode: BRMACA) [9]. Others are (−)-2-bromosuccinamic acid (BRSCAM) [10] and two crystal forms of 2,3-dibromo-3-phenylpropionic acid (CSD refcodes: ROFNOQ and ROFNOQ01) [11,12].
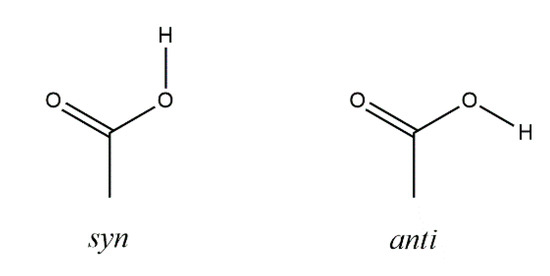
Scheme 1.
Carboxy group syn and anti conformations [13].
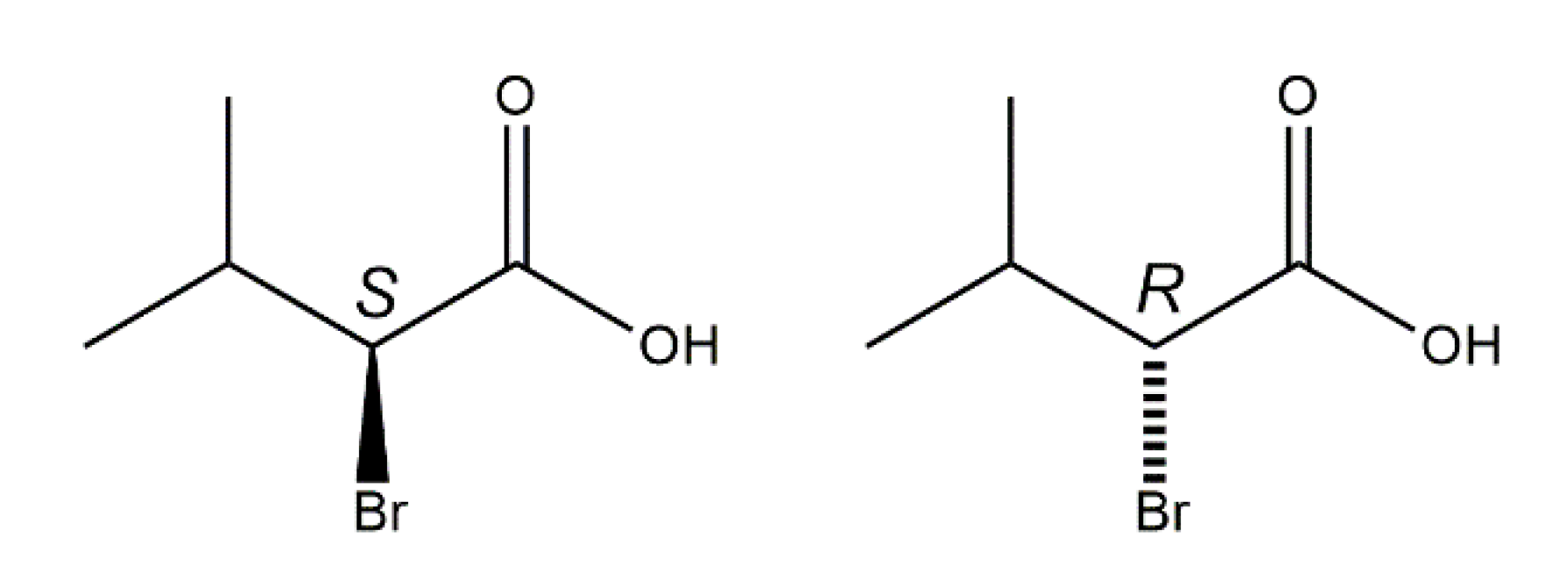
2-Bromo-3-methylbutyric acid (2-bromo-3-methylbutanoic acid, 1), commonly known as 2-bromoisovaleric or α-bromoisovaleric acid is a chiral α-halogenated monocarboxylic acid. Scheme 2 depicts the two enantiomers, (S)-1 and (R)-1. Their resolution by fractional crystallization was reported almost 100 years ago [14]. Auterhoff and Lang, for example, used this approach to prepare both enantiomers of the hypnotic and sedative agent bromisoval (2-bromo-3-methylbutyrylurea or commonly bromovalerylurea) from (S)-1 and (R)-1 by reaction of the respective acid chlorides with urea [15]. Despite the fact that 1 has long been known and is commercially available, to the best of our knowledge and based on a WebCSD search in May 2020 [16], a crystal structure of 1 has not been reported so far.
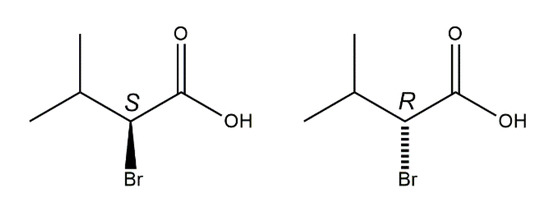
Scheme 2.
Chemical diagrams of the enantiomers of the title compound, (S)-1 (left) and (R)-1 (right) with stereodescriptors.
Chiral carboxylic acids have also attracted research interest in the fields of structural chemistry and crystal engineering, owing to phenomena such as frustration between molecular chirality and centrosymmetric hydrogen bond homosynthon formation in their crystal packing [17]. Enantiomeric mixtures can essentially crystallize as racemic crystals, racemic conglomerates (physical mixture of resolved crystals), inversion twins, disordered solid solutions [18] or, rarely, as kryptoracemates [19]. In this context, 1 attracted our attention. We have crystallized and investigated solvent-free 1 by X-ray crystallography in order to reveal preferred molecular conformations, crystal packing, intermolecular interactions and the outcome of crystallization of an enantiomeric mixture. Herein we report the crystal and molecular structures of (R)-1 and rac-1.
2. Materials and Methods
(S)- and (R)-1 were purchased from Sigma-Aldrich (purity 96%) and used as received. Solvents were of analytical grade and used without further purification. Crystals of (R)-1 suitable for single-crystal X-ray diffraction were obtained from an ethanolic solution by slow evaporation of the solvent at ambient conditions. To obtain rac-1, equimolar amounts of (S)- and (R)-1 were melted together on a Reichert hot-stage (Mikroheiztisch) mounted on a Nikon SMZ 1500 binocular microscope and cooled to room temperature [20]. The material so obtained was dissolved in ethyl acetate. Single-crystals of rac-1 suitable for X-ray analysis appeared when the solvent was allowed to evaporate slowly at ambient conditions.
The X-ray intensity data were collected at 100(2) K on an Enraf–Nonius Kappa CCD for (R)-1 and on a Bruker AXS Apex II for rac-1, using Mo Kα radiation in both cases. The data were scaled and corrected for absorption effects with SADABS [21]. The crystal structures were solved with SHELXT [22] and refined with SHELXL-2018/3 [23]. The highest residual difference electron density peak each for (R)-1 and rac-1 is ca. 0.7 Å from a bromine atom and can be ascribed to absorption effects. Carbon-bound hydrogen atoms were placed at geometrically calculated positions with Cmethine–H = 1.00 Å, Cmethyl–H = 0.98 Å and refined using a riding model with Uiso(H) = 1.2 Ueq(C) (1.5 for methyl groups). Torsion angles of the methyl groups were initially determined via difference Fourier syntheses and subsequently refined while maintaining tetrahedral angles at the carbon atoms. The carboxy hydrogen atoms in (R)-1 were located in difference electron density maps. In subsequent refinements, the O–H distances were restrained to a target value of 0.84(2) Å. In rac-1, the carboxy hydrogen atom was placed in an idealized hydrogen bonding position with O–H = 0.84 Å and refined using a riding model. Uiso(H) = 1.2 Ueq(O) was used for all carboxy hydrogen atoms. Refined and post-refinement values of the Flack x parameter [24] were obtained with SHELXL using TWIN/BASF instructions and Parsons’s method [25], respectively. The Hooft parameter [26,27,28] was calculated with PLATON [29]. Crystal data and refinement details for (R)-1 and rac-1 are listed in Table 1. Representations of the crystal and molecular structures were drawn with DIAMOND [30]. The structure overlay diagram and r.m.s. deviations of molecular structures from one another were obtained with MERCURY [31]. Packing indices were calculated with PLATON.

Table 1.
Crystal data and refinement details for (R)-1 and rac-1.
3. Results
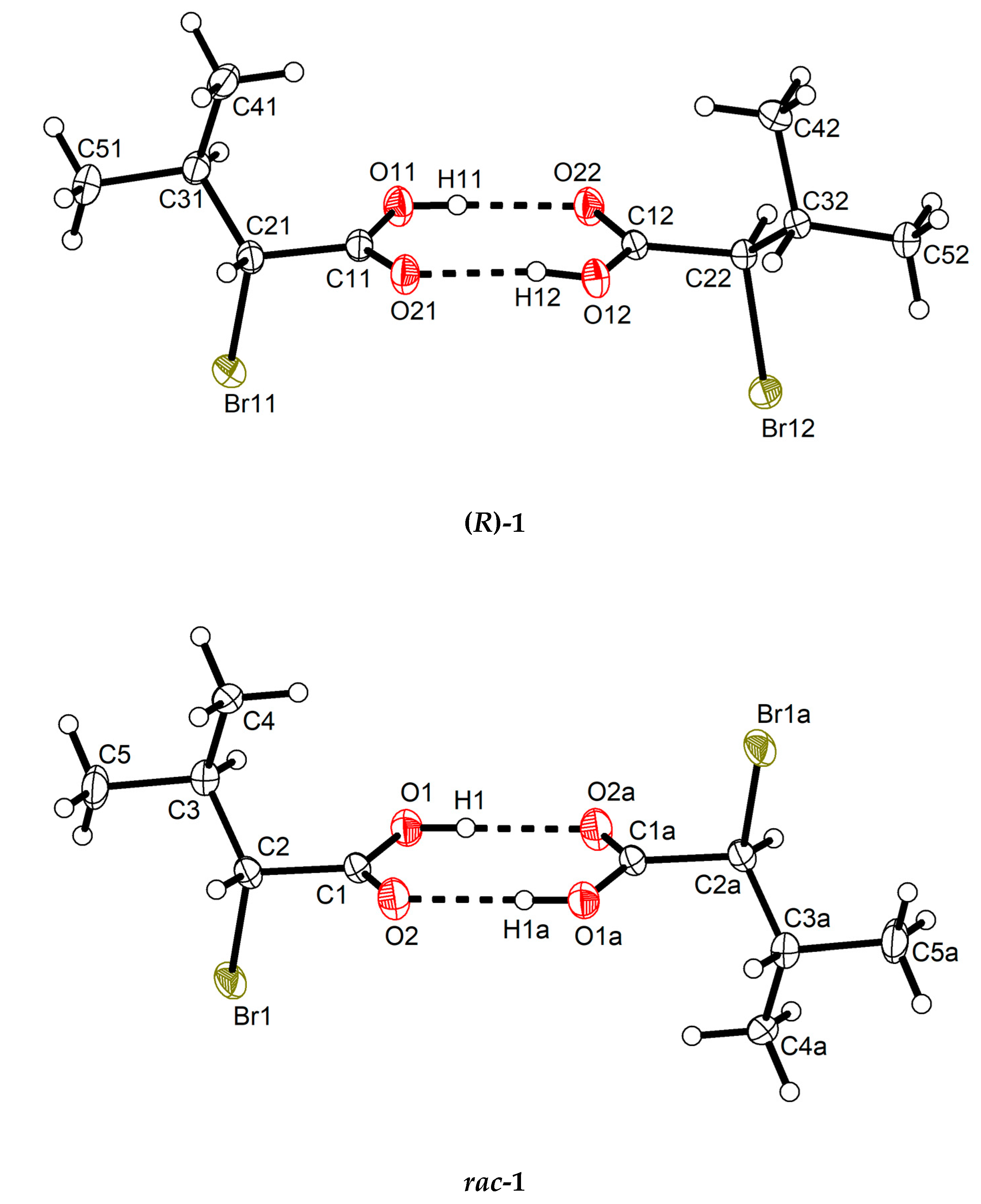
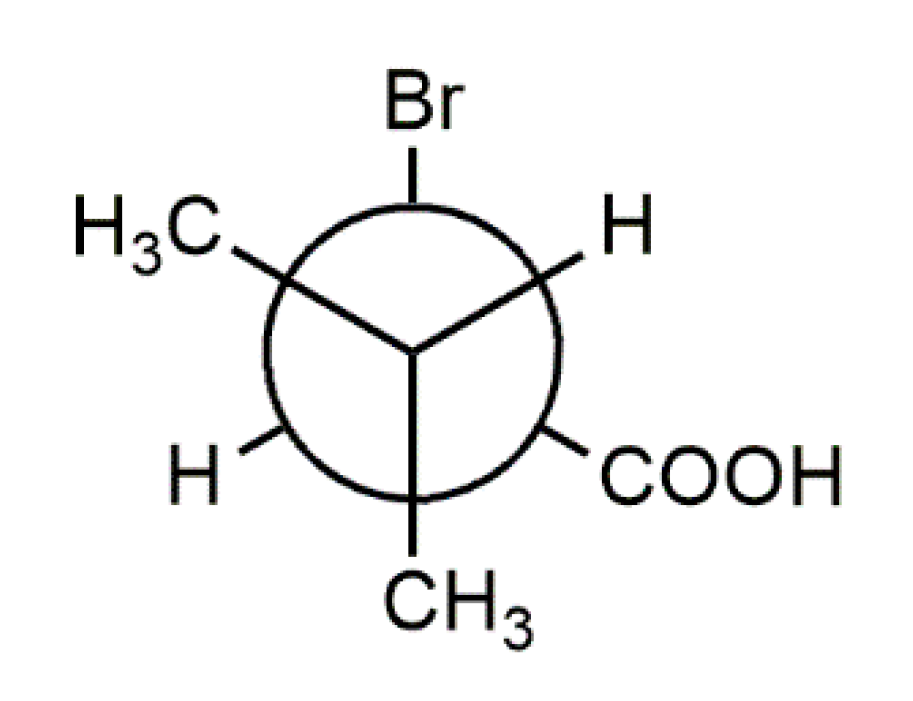
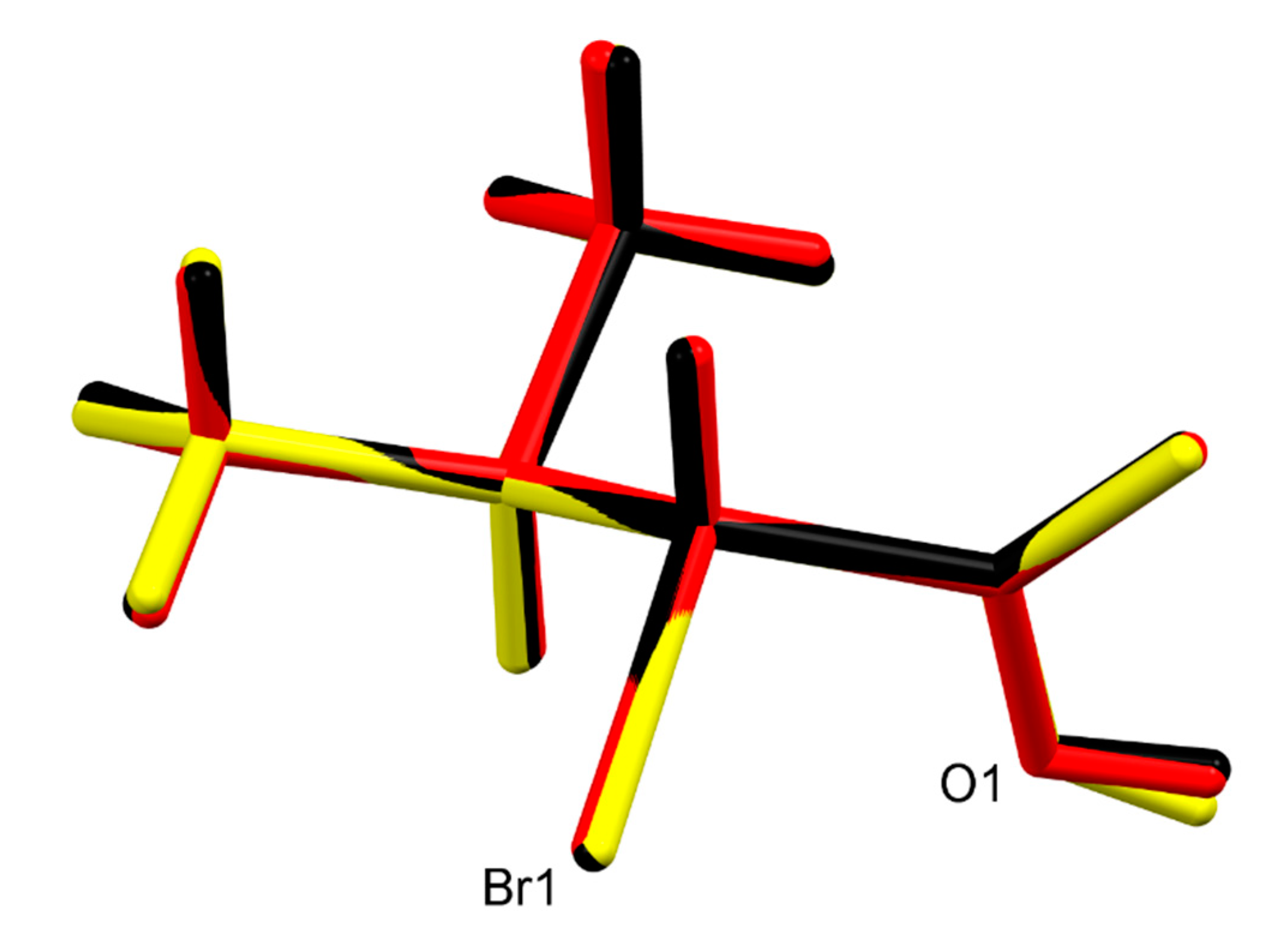
Both (R)-1 and rac-1 were found to crystallize in the triclinic crystal system with two molecules in the unit cell. As shown in Figure 1, the molecules form O–H···O hydrogen bonded dimers through the carboxy groups in the syn conformation with a motif [32] in both crystal structures. Geometric parameters of the hydrogen bonds in both (R)-1 and rac-1 are given in Table 2, and selected bond lengths, bond angles and torsion angles are listed in Table 3. The encountered homochiral hydrogen-bonded dimer in (R)-1 comprises two crystallographically unique molecules (Z′ = 2) and features approximate non-crystallographic C2 symmetry with the twofold rotation axis passing through the center of the hydrogen-bonded set and perpendicular to the mean plane of the two carboxy groups. In contrast, the hydrogen-bonded dimer in rac-1 is heterochiral and lies across a crystallographic inversion center and, thus, is centrosymmetric. The molecule in the chosen asymmetric unit of rac-1 (Z’ = 1) exhibits R configuration (see Figure 1, bottom). The two distinct molecules in (R)-1 and the R enantiomer in rac-1 adopt the same staggered conformation, as illustrated by a Newman projection in Scheme 3. A structure overlay diagram for the three molecular structures is depicted in Figure 2. The r.m.s. deviation of the respective non-hydrogen atoms in the two distinct molecules in (R)-1 is 0.0322 Å. Between the respective non-hydrogen atoms and those of the R enantiomer in rac-1, the r.m.s. deviations are 0.0234 and 0.0243 Å.
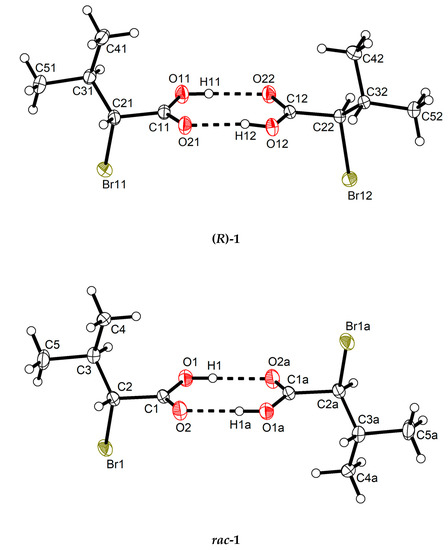
Figure 1.
Homochiral and heterochiral hydrogen-bonded dimers respectively in (R)-1 (top) and rac-1 (bottom) in their crystal structures. Displacement ellipsoids are drawn at the 50% probability level. Hydrogen atoms are represented by small spheres of arbitrary radii. Dashed lines represent hydrogen bonds. Symmetry code: (a) −x + 2, −y + 1, −z + 1.

Table 2.
Hydrogen bond geometry for (R)-1 and rac-1 (Å, °) 1.

Table 3.
Selected bond lengths, bond angles and torsion angles (°) for (R)-1 and rac-1 (Å, °) 1.
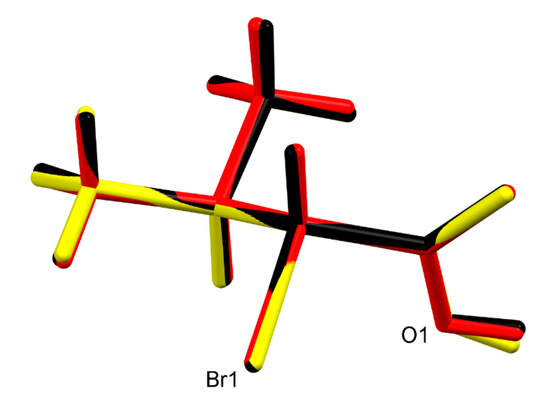
Figure 2.
Structure overlay of the two crystallographic distinct molecules in (R)-1 (red and yellow) and the R enantiomer in rac-1 (black).
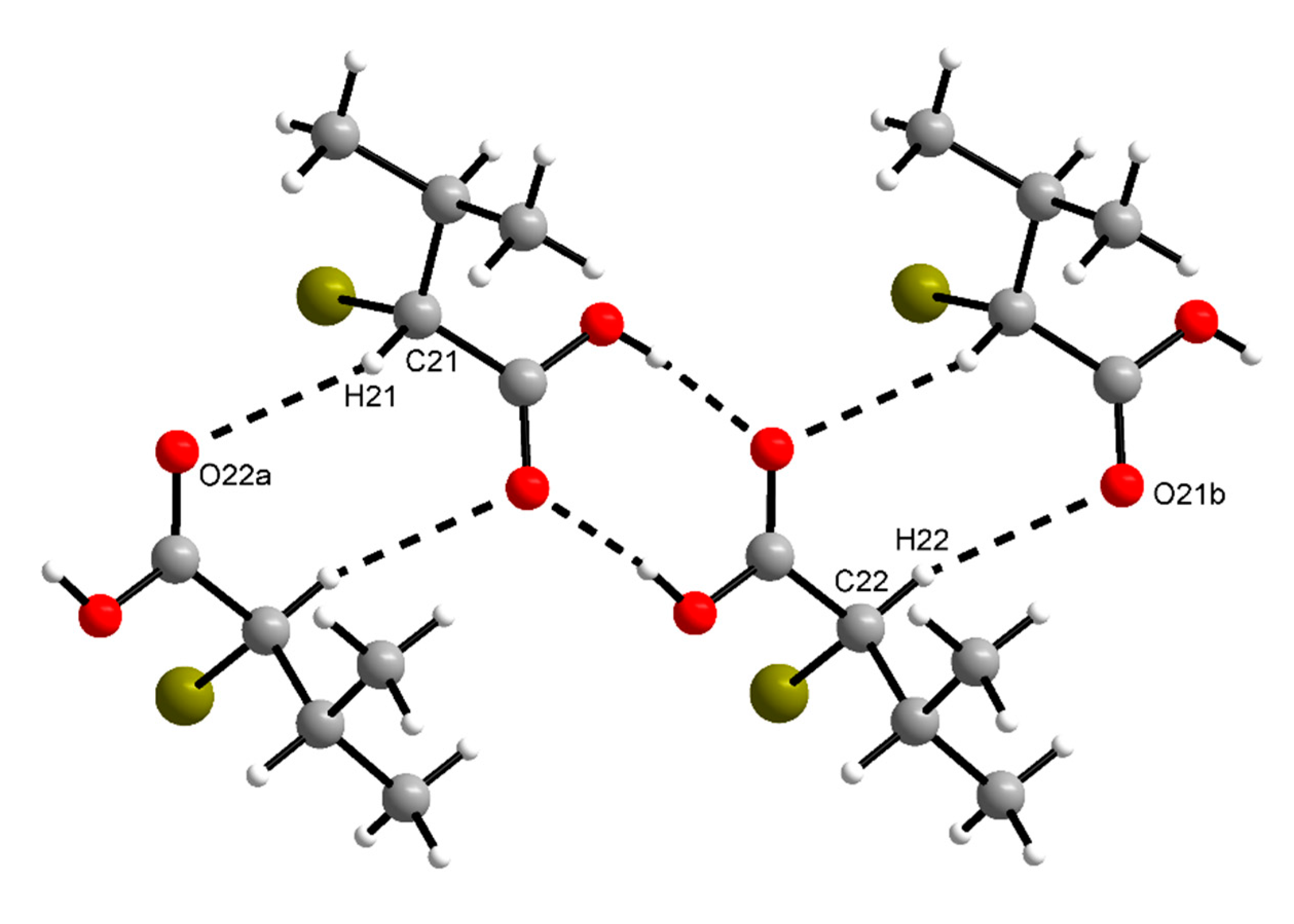
The supramolecular structure of (R)-1 in the crystal features short C–H···O contacts between the α-carbon atom of the carboxylic acid and the (formal) carboxy C=O moiety of an adjacent molecule (Figure 3). The hydrogen bonding motif descriptor is likewise . In rac-1, the methine group of the α-carbon atom does not form a similar short C–H···O contact. The (formal) carboxy C=O moiety, however, is approached by a methyl hydrogen atom of the isopropyl group of an adjacent molecule (C···O = 3.58 Å, see Figure S1). The crystal packing in rac-1 is denser than in (R)-1, which is evident from the volumes of the triclinic unit cells of both Z = 2 structures (Table 1). Thus, each molecule in rac-1 occupies 5.2 Å3 less space in the crystal than in (R)-1. The calculated densities (Table 1) and Kitaigorodskij packing indices [33] of 68.2% for rac-1 and 66.2% for (R)-1 further indicate a denser crystal packing in rac-1 than in (R)-1. Short contacts (with respect to the sum of the corresponding van der Waals radii) of the Br···Br or C–H···Br type are neither observed in (R)-1 nor in rac-1.
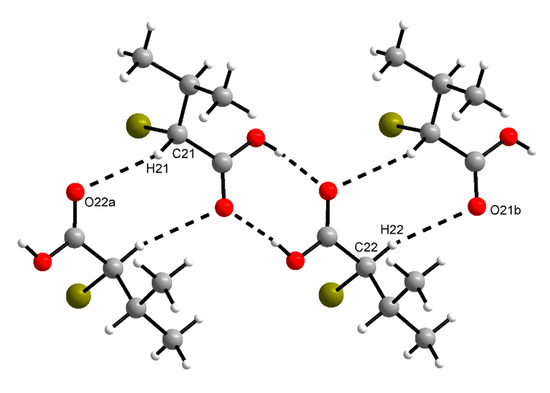
Figure 3.
Section of the crystal structure of (R)-1, showing
motifs of intermolecular C–H···O and O–H···O contacts (cf. Figure 1). C21–H21···O22a: d(D···A) = 3.384(3) Å, ˂(DHA) = 145°; C22–H22···O21b: d(D···A) = 3.519(3) Å, ˂(DHA) = 162°. Symmetry codes: (a) x − 1, y − 1, z; (b) x + 1, y + 1, z.
4. Discussion
The absolute structure of a single-crystal of (R)-1 grown from solution was established by anomalous-dispersion effects in the diffraction intensity measurements [34], thereby confirming the absolute configuration reported for the purchased bulk material [35,36]. Absolute structure parameters are listed in Table 1. The Flack x parameter estimated post-refinement based on quotients [25] and the Hooft parameter based on Bayesian statistics [26,27,28] are close to zero with adequately small standard uncertainties [18]. By way of comparison, the standard uncertainty of the refined Flack x parameter is larger than that of the former two parameters by a factor of ca. two [37].
The X-ray analysis of rac-1 clearly revealed that an equimolar mixture of both enantiomers (Scheme 2) forms a racemic crystal upon crystallization from ethyl acetate and not a racemic conglomerate, which is observed in only ca. 10% of cases [38]. The higher crystallographic density of rac-1 is in accord with Wallach’s rule from 1895, which states that racemic crystals tend to be denser than the chiral counterparts [39]. This phenomenon can essentially be explained by the fact that enantiopure compounds can only crystallize in a Sohncke space group, devoid of inversion symmetry. To enable densest packing with Z′ = 1 in rac-1, the molecular dimer must be placed across a crystallographic inversion center, which would be impossible for homochiral R···R and S···S dimers. (R)-1 crystallizes with Z′ = 2 in the space group P1, which is not among those available for densest packing of molecules of arbitrary shape [33]. Crystallization with Z′ > 1 is a common phenomenon for chiral carboxylic acids and has been described as frustration between chirality (referring to the whole molecule) and centrosymmetric dimer formation (referring to the hydrogen bond synthon) [17].
According to theoretical studies, the syn conformation of a carboxy group, as observed in (R)-1 and rac-1, is energetically more stable than the anti conformation by ca. 21.4–28.9 kJ mol−1 [13]. A syn···syn dimer (homosynthon) with a motif is a hydrogen bonding pattern commonly observed for carboxylic acids [40,41]. Its occurrence in the crystal structures of (R)-1 and rac-1 is thus as expected and also in accord with Etter’s rules for hydrogen bonding, whereby all acidic hydrogen atoms and all good hydrogen bond acceptors are involved in hydrogen bonds, and the best donor and the best acceptor are hydrogen-bonded to one another [42]. The short C–H···O contacts observed in (R)-1 (Figure 3) could be interpreted geometrically as weak hydrogen bonds [43]. It reasonable to assume that the hydrogen atom at the α-carbon atom here is prone to weak hydrogen bonds, since the carboxy group as well as the bromine atom should exert an electron-withdrawing effect. Since such contacts are not present in rac-1, their impact on the overall supramolecular structure in the crystal is probably minor.
The molecular conformations found in the crystal structures of (R)-1 and rac-1 are virtually identical, as evidenced by the calculated r.m.s. deviations of the corresponding heavy atom skeletons and visualized by a structure overlay diagram (Figure 2). It is reasonable to assume that a staggered conformation corresponds to a minimum energy structure, since not only the carbon chains but also the carboxy groups adopt the same orientation in the three molecular structures (Table 3) despite different crystal environments. This suggests that the observed conformation represents a preferred molecular structure of 1.
5. Conclusions
We have revealed the crystal and molecular structures of (R)-1 and rac-1 by single-crystal X-ray analysis. The absolute configuration of (R)-1 was confirmed by means of anomalous dispersion effects in the diffraction intensity measurements. Not unexpectedly, the ubiquitous carboxy syn···syn homosynthon was encountered in both structures. Clearly, O–H···O hydrogen bonds are the dominant intermolecular interaction in both structures. As compared with rac-1, the more open structure of (R)-1 and the existence of two molecules in its asymmetric unit can be ascribed to frustration between chirality and centrosymmetric homosynthon formation. The observed denser crystal packing of centrosymmetric rac-1 than of its enantiopure counterpart (R)-1 is in accord with Wallach’s rule. Short C–H···O contacts, as formed by the α-methine group in (R)-1, are not encountered in rac-1. This suggests that these weak intermolecular interactions may not have a crucial bearing on the packing of the hydrogen-bonded carboxylic acid dimers in the solid-state here, which appears to be essentially governed by close packing. A virtually identical molecular conformation in all in total three crystallographically distinct molecules in (R)-1 and rac-1, despite different crystal environments, suggests that the observed geometry represents the preferred low energy structure.
Supplementary Materials
The following are available online at https://www.mdpi.com/2624-8549/2/3/44/s1, Figure S1: Section of the crystal structure of rac-1, showing short contacts between methyl hydrogen atoms of the isopropyl groups and the (formal) carboxy C=O moieties of adjacent molecules (C···O = 3.58 Å). CCDC 200603 [(R)-1] and 2006031 (rac-1) contain the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/structures.
Author Contributions
Conceptualization, N.N., R.G. and R.W.S.; methodology, N.N. and R.G.; validation, R.W.S. and R.G.; formal analysis, R.W.S. and R.G.; investigation, N.N. and R.G.; resources, C.W.L.; data curation, R.W.S. and R.G.; writing—original draft preparation, R.W.S.; writing—review and editing, R.G.; visualization, R.W.S. and R.G.; supervision, C.W.L.; project administration, R.W.S.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors would like to thank Alois Fürstner for providing laboratory resources for this project. R.W.S. is grateful to Peter Imming for his support.
Conflicts of Interest
The authors declare no conflict of interest.
References and Note
- Czekelius, C.; Tzschucke, C.C. Synthesis of Halogenated Carboxylic Acids and Amino Acids. Synth. Stuttg. 2010, 543–566. [Google Scholar] [CrossRef]
- Kubitschke, J.; Lange, H.; Strutz, H. Carboxylic Acids, Aliphatic. Ullmann’s Encycl. Ind. Chem. 2014, 1–18. [Google Scholar] [CrossRef]
- Ullmann’s Fine Chemicals; Wiley: New York, NY, USA, 2014.
- Kirk, K.L. Biochemistry of halogenated organic compounds. In PATAI’S Chemistry of Functional Groups; Rappoport, Z., Ed.; Wiley: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Yue, Y.; Chen, J.; Bao, L.; Wang, J.; Li, Y.; Zhang, Q. Fluoroacetate dehalogenase catalyzed dehalogenation of halogenated carboxylic acids: A QM/MM approach. Chemosphere 2020, 254, 126803. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, S.; Cooper, R.H.; Randle, P.J. Mechanism of activation of pyruvate dehydrogenase by dichloroacetate and other halogenated carboxylic acids. Biochem. J. 1974, 141, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Kirk, K.L. Biochemistry of Halogenated Carboxylic Acids. In Biochemistry of Halogenated Organic Compounds; Kirk, K.L., Ed.; Springer: Boston, MA, USA, 1991. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. B Struct. Sci. Cryst. Eng. Mater. 2016, 72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Vor der Bruck, O.; Leiserowitz, L. Molecular packing modes. Two crystalline modifications of bromoacetic acid, C2H3BrO2. Cryst. Struct. Commun. 1975, 647–651. [Google Scholar]
- Murakami, Y.; Iitaka, Y. Determination of the Absolute Configuration of (-)-2-Bromosuccinamic Acid by X-Ray Diffraction Method. Chem. Pharm. Bull. 1969, 17, 2397–2404. [Google Scholar] [CrossRef]
- Thong, P.Y.; Lo, K.M.; Ng, S.W. 2,3-Dibromo-3-phenylpropionic acid. Acta Crystallogr. Sect. E 2008, 64, o1946. [Google Scholar] [CrossRef]
- Howard, T.R.; Mendez-deMello, K.A.; Cardenas, A.J.P. 2,3-Dibromo-3-phenylpropanoic acid: A monoclinic polymorph. IUCrData 2016, 1, x161885. [Google Scholar] [CrossRef]
- D’Ascenzo, L.; Auffinger, P. A comprehensive classification and nomenclature of carboxyl-carboxyl(ate) supramolecular motifs and related catemers: Implications for biomolecular systems. Acta Crystallogr. Sect. B 2015, 71, 164–175. [Google Scholar] [CrossRef]
- Levene, P.A.; Mori, T.; Mikeska, L.A. On Walden inversion: X. On the oxidation of 2-thiolcarboxylic acids to the corresponding sulfonic acids and on the Walden inversion in the series of 2-hydroxycarboxylic acids. J. Biol. Chem. 1927, 75, 337–365. [Google Scholar]
- Auterhoff, H.; Lang, W. Darstellung und Eigenschaften der optisch aktiven Bromisovale. Arch. Pharm. 1970, 303, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Thomas, I.R.; Bruno, I.J.; Cole, J.C.; Macrae, C.F.; Pidcock, E.; Wood, P.A. WebCSD: The online portal to the Cambridge Structural Database. J. Appl. Crystallogr. 2010, 43, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Steed, K.M.; Steed, J.W. Packing problems: High Z’ crystal structures and their relationship to cocrystals, inclusion compounds, and polymorphism. Chem. Rev. 2015, 115, 2895–2933. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.L.; Watkin, D.J. X-ray crystallography and chirality: Understanding the limitations. Tetrahedron Asymmetry 2009, 20, 712–717. [Google Scholar] [CrossRef]
- Rekis, T. Crystallization of chiral molecular compounds: What can be learned from the Cambridge Structural Database? Acta Crystallogr. Sect. B 2020, 76, 307–315. [Google Scholar] [CrossRef]
- A preliminary X-ray analysis of the crystals obtained from the melt revealed a severely disordered structure with unit cell parameters similar to those of rac-1 crystallized from ethyl acetate.
- SADABS; Bruker AXS Inc.: Madison, WI, USA, 2012.
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Flack, H. On enantiomorph-polarity estimation. Acta Crystallogr. Sect. A 1983, 39, 876–881. [Google Scholar] [CrossRef]
- Parsons, S.; Flack, H.D.; Wagner, T. Use of intensity quotients and differences in absolute structure refinement. Acta Crystallogr. B Struct. Sci. Cryst. Eng. Mater. 2013, 69, 249–259. [Google Scholar] [CrossRef]
- Hooft, R.W.W.; Straver, L.H.; Spek, A.L. Determination of absolute structure using Bayesian statistics on Bijvoet differences. J. Appl. Crystallogr. 2008, 41, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Hooft, R.W.W.; Straver, L.H.; Spek, A.L. Probability plots based on Student’s t-distribution. Acta Crystallogr. Sect. A 2009, 65, 319–321. [Google Scholar] [CrossRef] [PubMed]
- Hooft, R.W.W.; Straver, L.H.; Spek, A.L. Using the t-distribution to improve the absolute structure assignment with likelihood calculations. J. Appl. Crystallogr. 2010, 43, 665–668. [Google Scholar] [CrossRef]
- Spek, A.L. checkCIF validation ALERTS: What they mean and how to respond. Acta Crystallogr. E Crystallogr. Commun. 2020, 76, 1–11. [Google Scholar] [CrossRef]
- Brandenburg, K. DIAMOND, 3.2k3; Crystal Impact GbR: Bonn, Germany, 2018. [Google Scholar]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.L. Patterns in hydrogen bonding—Functionality and graph set analysis in crystals. Angew. Chem. Int. Ed. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- Kitajgorodskij, A.I. Molecular Crystals and Molecules; Academic Press: New York, NY, USA, 1973. [Google Scholar]
- Parsons, S. Determination of absolute configuration using X-ray diffraction. Tetrahedron Asymmetry 2017, 28, 1304–1313. [Google Scholar] [CrossRef]
- Flack, H.D.; Bernardinelli, G. Absolute structure and absolute configuration. Acta Crystallogr. Sect. A 1999, 55, 908–915. [Google Scholar] [CrossRef]
- Flack, H.D.; Bernardinelli, G. Reporting and evaluating absolute-structure and absolute-configuration determinations. J. Appl. Crystallogr. 2000, 33, 1143–1148. [Google Scholar] [CrossRef]
- Spek, A. Absolute structure determination: Pushing the limits. Acta Crystallogr. Sect. B 2016, 72, 659–660. [Google Scholar] [CrossRef]
- Collet, A.; Ziminski, L.; Garcia, C.; Vigné-Maeder, F. Chiral Discrimination in Crystalline Enantiomer Systems: Facts, Interpretations, and Speculations. In Supramolecular Stereochemistry; Siegel, J.S., Ed.; Springer: Dordrecht, The Netherlands, 1995; pp. 91–110. [Google Scholar] [CrossRef]
- Brock, C.P.; Schweizer, W.B.; Dunitz, J.D. On the validity of Wallach’s rule: On the density and stability of racemic crystals compared with their chiral counterparts. J. Am. Chem. Soc. 1991, 113, 9811–9820. [Google Scholar] [CrossRef]
- Leiserowitz, L. Molecular packing modes. Carboxylic acids. Acta Crystallogr. Sect. B 1976, 32, 775–802. [Google Scholar] [CrossRef]
- Corpinot, M.K.; Bučar, D.-K. A Practical Guide to the Design of Molecular Crystals. Cryst. Growth Des. 2018, 19, 1426–1453. [Google Scholar] [CrossRef]
- Etter, M.C. Encoding and decoding hydrogen-bond patterns of organic-compounds. Acc. Chem. Res. 1990, 23, 120–126. [Google Scholar] [CrossRef]
- Thakuria, R.; Sarma, B.; Nangia, A. 7.03—Hydrogen Bonding in Molecular Crystals. In Comprehensive Supramolecular Chemistry II; Atwood, J.L., Ed.; Elsevier: Oxford, UK, 2017; pp. 25–48. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).