Understanding Corn Production Complexity: Causal Structure Learning and Variable Ranking from Agricultural Simulations
Abstract
1. Introduction
- Identify and rank the most influential variables impacting corn yield among N fertilizer application rates, irrigation management rules, weather conditions, climate change scenarios, planting date, soil characteristics, and cultivar types;
- Elucidate the underlying causal structure describing the interactions between yield and key G, E, and M variables to enable a deeper understanding of the corn production that can inform experimental design and support mechanistically grounded decision support systems.
2. Materials and Methods
2.1. APSIM Interface and Model Description
2.2. Experimental Setup
2.3. Site Description and Agrometeorological Data
2.4. Random Forest
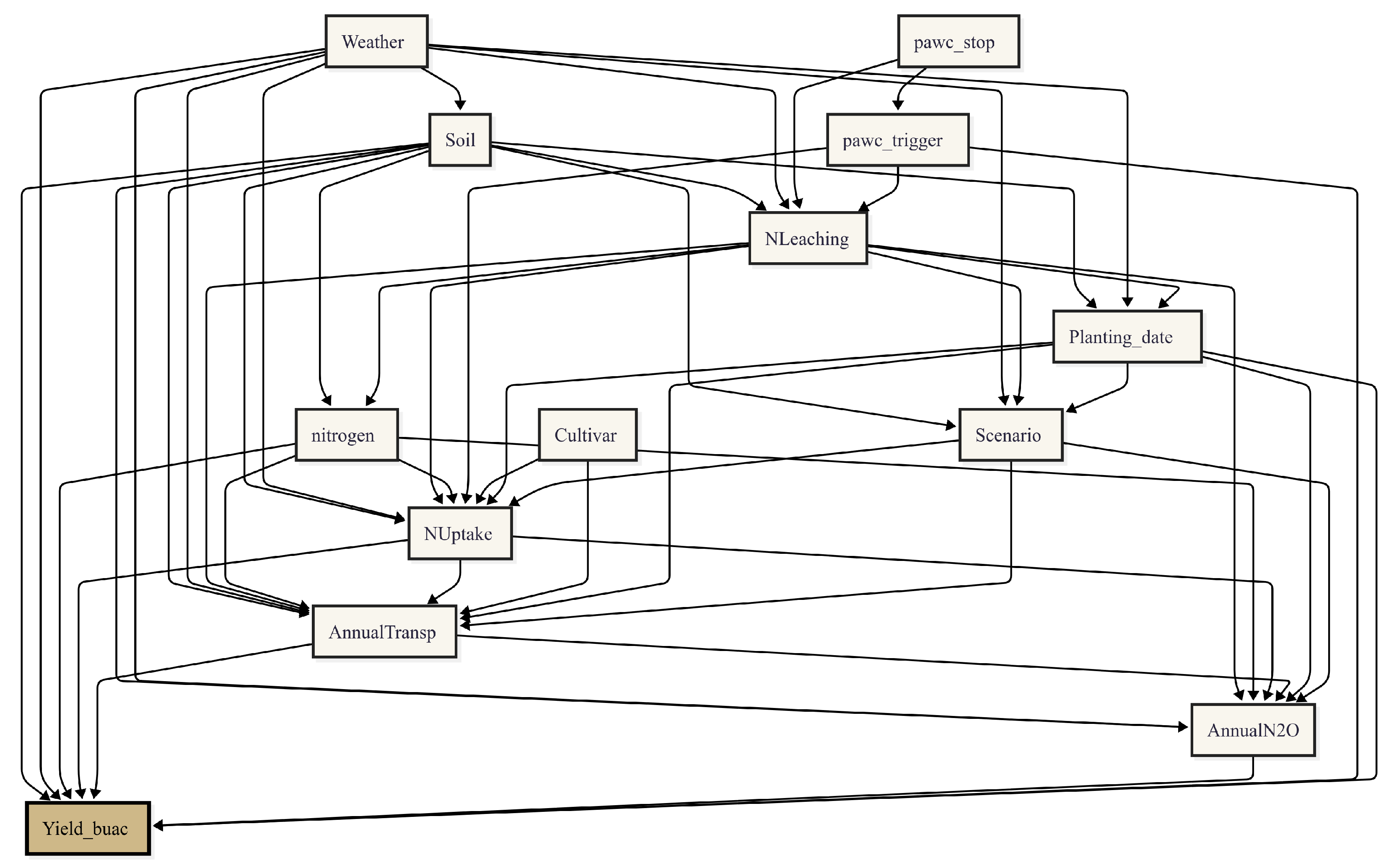
2.5. Causal Learning
3. Results and Discussions
3.1. Random Forest
3.2. Causal Learning
4. Limitations and Directions for Future Work
- This study employed a limited set of input variables and management scenarios that may not fully capture the heterogeneity of agricultural systems observed under field conditions. Future research could be expanded to larger-scale, regional simulations encompassing broader geographical regions, longer temporal periods, and more comprehensive treatment matrices. These expanded studies should include diverse cropping systems, management practices, soil types, and climatic conditions to better assist both policymakers in regional decision-making and farmers in making informed, farm-specific management decisions.
- This study employed a discretization approach using five bins with the HillClimb model. While the model struggled to map some edges correctly and occasionally reversed their direction, it successfully mapped around most edges in ways that were practical and intuitive, though not entirely identical to how those relationships were encoded in APSIM. Although these results are sensible and practically valuable, users interested in tracing causal relationships more accurately as they exist in APSIM or other crop growth models should consider several improvements. These include implementing temporal constraints (as we carried out specifically for yield, being the main focus of this study), tuning discretization hyperparameters, selecting the optimal number of bins, and exploring alternative causal discovery models.
- In most studies, including this one, spatial resolution mismatches exist between biophysical crop simulations and the weather datasets or climate change projections used to drive them [2]. Future research should quantify the effects of these scale disparities by downscaling climate projections and meteorological data to match the spatial resolution of field-scale crop models, with particular emphasis on precipitation-sensitive crops such as maize, where rainfall variability significantly affects yield outcomes [2,57].
- This study focused exclusively on the APSIM crop simulation model and aimed to elucidate the causal relationships among key variables within the model framework with yield. Future research should apply similar analytical approaches to other crop growth models to characterize their structural components, develop comprehensive model mappings, and establish evidence-based guidelines for selecting optimal models or model components for specific agricultural applications [17].
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| N | Nitrogen |
| PAWC | Plant Available Water Content |
| PAWCtrigger | Irrigation Triggering point |
| PAWCstop | Irrigation Stopping point |
| EPIC | Environmental Policy Integrated Climate |
| NUptake | Nitrogen Uptake |
| AnnualTrans | Annual Transpiration |
| Yield_buac | Yield bushels per acre |
| DUL | Drain Upper Limit |
| CLL | Crop Lower Limit |
| APSIM | Agricultural Production Systems sIMulator |
| DSSAT | ecision Support System for Agrotechnology Transfer |
| WOFOST | WOrld FOod STudies |
| RF | Random Forest |
| G | Genetics |
| GSCV | Grid Search CV |
| E | Environment |
| M | Management |
| GDD | Growing Degree Day |
| kg | Kilogram |
| ha | Hectare |
| SOILN | Soil Nitrogen |
| SOILWAT | Soil Water |
| SOILTEMP | Soil Temperature |
| C | Carbon |
| V | Vegetative Growth Stage |
| ppm | parts per million |
| C | Celsius |
| K | Kelvin |
| MDI | Mean Decrease in Impurity |
| DAG | Directed Acyclic Graph |
| HC | HillClimb |
| RM | Relative Maturity |
| PPAC | Pinney Purdue Agriculture Center |
| ACRE | Agronomy Center for Research and Education |
| SEPAC | Southeast Purdue Agriculture Center |
Appendix A
| Depth (cm) | BD | AD | LL15 | DUL | SAT | KS | LL | KL | XF | PAWC | pH | Carbon (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0–15 | 1.40 | 0.12 | 0.229 | 0.345 | 0.442 | 29.57 | 0.229 | 0.08 | 1 | 0.116 | 6.59 | 4.500 |
| 15–30 | 1.40 | 0.21 | 0.229 | 0.345 | 0.442 | 21.70 | 0.229 | 0.08 | 1 | 0.116 | 6.59 | 4.500 |
| 30–60 | 1.49 | 0.23 | 0.230 | 0.346 | 0.408 | 15.78 | 0.230 | 0.08 | 1 | 0.116 | 7.12 | 2.250 |
| 60–90 | 1.55 | 0.18 | 0.182 | 0.312 | 0.385 | 13.26 | 0.182 | 0.08 | 1 | 0.130 | 7.12 | 1.420 |
| 90–120 | 1.64 | 0.13 | 0.125 | 0.270 | 0.351 | 14.46 | 0.125 | 0.08 | 1 | 0.145 | 7.23 | 0.750 |
| 120–150 | 1.80 | 0.11 | 0.111 | 0.254 | 0.291 | 20.48 | 0.111 | 0.06 | 0.9 | 0.143 | 7.86 | 0.750 |
| 150–180 | 1.80 | 0.11 | 0.111 | 0.254 | 0.291 | 26.13 | 0.111 | 0.03 | 0.5 | 0.143 | 7.86 | 0.750 |
| Depth (cm) | BD | AD | LL15 | DUL | SAT | KS | LL | KL | XF | PAWC | pH | Carbon (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0–15 | 1.42 | 0.08 | 0.233 | 0.365 | 0.420 | 38.76 | 0.233 | 0.06 | 1.000 | 0.132 | 5.90 | 2.281 |
| 15–30 | 1.54 | 0.08 | 0.230 | 0.335 | 0.385 | 27.37 | 0.230 | 0.06 | 0.876 | 0.105 | 5.80 | 1.041 |
| 30–60 | 1.59 | 0.08 | 0.233 | 0.324 | 0.375 | 17.72 | 0.235 | 0.06 | 0.748 | 0.089 | 5.70 | 0.590 |
| 60–90 | 1.64 | 0.08 | 0.237 | 0.316 | 0.355 | 14.05 | 0.264 | 0.04 | 0.602 | 0.052 | 5.75 | 0.374 |
| 90–120 | 1.68 | 0.08 | 0.228 | 0.304 | 0.340 | 15.87 | 0.291 | 0.01 | 0.516 | 0.013 | 5.94 | 0.295 |
| 120–150 | 1.68 | 0.07 | 0.224 | 0.300 | 0.340 | 17.01 | 0.299 | 0.00 | 0.509 | 0.001 | 6.02 | 0.287 |
| 150–180 | 1.69 | 0.07 | 0.219 | 0.294 | 0.340 | 18.56 | 0.294 | 0.00 | 0.000 | 0.000 | 6.12 | 0.277 |
| Depth (cm) | BD | AD | LL15 | DUL | SAT | KS | LL | KL | XF | PAWC | pH | Carbon (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0–15 | 1.36 | 0.06 | 0.170 | 0.286 | 0.430 | 77.75 | 0.170 | 0.06 | 1.000 | 1.360 | 6.00 | 3.123 |
| 15–30 | 1.45 | 0.05 | 0.160 | 0.267 | 0.400 | 73.37 | 0.160 | 0.06 | 1.000 | 1.450 | 5.95 | 1.979 |
| 30–60 | 1.47 | 0.05 | 0.158 | 0.257 | 0.395 | 69.23 | 0.158 | 0.06 | 1.000 | 1.472 | 5.93 | 1.418 |
| 60–90 | 1.51 | 0.04 | 0.142 | 0.232 | 0.380 | 73.37 | 0.160 | 0.05 | 1.000 | 0.072 | 6.05 | 0.874 |
| 90–120 | 1.53 | 0.03 | 0.118 | 0.200 | 0.380 | 90.40 | 0.156 | 0.03 | 1.000 | 0.044 | 6.24 | 0.607 |
| 120–150 | 1.53 | 0.03 | 0.114 | 0.196 | 0.380 | 96.92 | 0.163 | 0.02 | 1.000 | 0.033 | 6.32 | 0.604 |
| 150–180 | 1.53 | 0.03 | 0.109 | 0.190 | 0.380 | 105.70 | 0.171 | 0.01 | 1.000 | 0.019 | 6.42 | 0.600 |
References
- U.S. Grains Council (USGC). 2024/2025 Corn Harvest Quality Report. Available online: https://grains.org/corn_report/corn-harvest-quality-report-2024-2025-2/ (accessed on 6 October 2025).
- Pathak, H.; Buckmaster, D.; Messina, C.; Wang, D. Crop growth model: Optimal application of nitrogen fertilizer in corn for economic returns and environmental sustainability. In Proceedings of the 2023 ASABE Annual International Meeting, Omaha, NE, USA, 9–12 July 2023; p. 1. [Google Scholar] [CrossRef]
- Pathak, H. Machine Vision Methods for Evaluating Plant Stand Count and Weed Classification Using Open-Source Platforms. Master’s Thesis, North Dakota State University, Fargo, ND, USA, 2021. [Google Scholar]
- Pathak, H.; Warren, C.J.; Buckmaster, D.R.; Wang, D.R. Advancing adaptive agricultural strategies: Unraveling impacts of climate change and soils on corn productivity using APSIM. In Proceedings of the 16th International Conference on Precision Agriculture, Manhattan, KS, USA, 21–24 July 2024. [Google Scholar]
- Boyer, J.S.; Byrne, P.; Cassman, K.G.; Cooper, M.; Delmer, D.; Greene, T.; Gruis, F.; Habben, J.; Hausmann, N.; Kenny, N.; et al. The US drought of 2012 in perspective: A call to action. Glob. Food Sec. 2013, 2, 139–143. [Google Scholar] [CrossRef]
- Singh, G.; Sharma, V.; Mulla, D.; Tahir, M.; Fernandez, F.G. Effect of irrigation scheduling methods on maize grain yield and nitrate leaching in central Minnesota. J. Nat. Resour. Agric. Ecosyst. 2023, 1, 13–31. [Google Scholar] [CrossRef]
- Bowling, L.C.; Cherkauer, K.A.; Lee, C.I.; Beckerman, J.L.; Brouder, S.; Buzan, J.R.; Doering, O.C.; Dukes, J.S.; Ebner, P.D.; Frankenberger, J.R.; et al. Agricultural impacts of climate change in Indiana and potential adaptations. Clim. Change 2020, 163, 2005–2027. [Google Scholar] [CrossRef]
- Deines, J.M.; Swatantran, A.; Ye, D.; Myers, B.; Archontoulis, S.; Lobell, D.B. Field-scale dynamics of planting dates in the US Corn Belt from 2000 to 2020. Remote Sens. Environ. 2023, 291, 113551. [Google Scholar] [CrossRef]
- Dong, Y.; Christenson, C.; Kelley, L.; Miller, S. Trends and future of agricultural irrigation in Michigan and Indiana. Irrig. Drain. 2024, 73, 346–358. [Google Scholar] [CrossRef]
- Rai, N. Weed Identification on Drone-Captured Images Using Edge Device for Spot Spraying Application. Ph.D. Thesis, North Dakota State University, Fargo, ND, USA, 2023. [Google Scholar]
- McCown, R.L.; Hammer, G.L.; Hargreaves, J.N.G.; Holzworth, D.P.; Freebairn, D.M. APSIM: A novel software system for model development, model testing and simulation in agricultural systems research. Agric. Syst. 1996, 50, 255–271. [Google Scholar] [CrossRef]
- Jones, J.W.; Hoogenboom, G.; Porter, C.H.; Boote, K.J.; Batchelor, W.D.; Hunt, L.A.; Wilkens, P.W.; Singh, U.; Gijsman, A.J.; Ritchie, J.T. The DSSAT cropping system model. Eur. J. Agron. 2003, 18, 235–265. [Google Scholar] [CrossRef]
- Van Diepen, C.A.; Van Wolf, J.; Van Keulen, H.; Rappoldt, C. WOFOST: A simulation model of crop production. Soil Use Manag. 1989, 5, 16–24. [Google Scholar] [CrossRef]
- Mandrini, G.; Bullock, D.S.; Martin, N.F. Modeling the economic and environmental effects of corn nitrogen management strategies in Illinois. Field Crops Res. 2021, 261, 108000. [Google Scholar] [CrossRef]
- Shahhosseini, M.; Hu, G.; Huber, I.; Archontoulis, S.V. Coupling machine learning and crop modeling improves crop yield prediction in the US Corn Belt. Sci. Rep. 2021, 11, 1606. [Google Scholar] [CrossRef]
- Kim, M.; Sung, K. Assessment of causality between climate variables and production for whole crop maize using structural equation modeling. J. Anim. Sci. Technol. 2021, 63, 339. [Google Scholar] [CrossRef]
- Sitokonstantinou, V.; Díaz Salas Porras, E.; Cerdà Bautista, J.; Piles, M.; Athanasiadis, I.; Kerner, H.; Martini, G.; Sweet, L.-b.; Tsoumas, I.; Zscheischler, J.; et al. Causal machine learning for sustainable agroecosystems. arXiv 2024, arXiv:2408.13155. [Google Scholar] [CrossRef]
- Holzworth, D.P.; Huth, N.I.; deVoil, P.G.; Zurcher, E.J.; Herrmann, N.I.; McLean, G.; Chenu, K.; van Oosterom, E.J.; Snow, V.; Murphy, C.; et al. APSIM—Evolution towards a new generation of agricultural systems simulation. Environ. Model. Softw. 2014, 62, 327–350. [Google Scholar] [CrossRef]
- Winn, C.A.; Archontoulis, S.; Edwards, J. Calibration of a crop growth model in APSIM for 15 publicly available corn hybrids in North America. Crop Sci. 2023, 63, 511–534. [Google Scholar] [CrossRef]
- Probert, M.E.; Dimes, J.P.; Keating, B.A.; Dalal, R.C.; Strong, W.M. APSIM’s water and nitrogen modules and simulation of the dynamics of water and nitrogen in fallow systems. Agric. Syst. 1998, 56, 1–28. [Google Scholar] [CrossRef]
- Soufizadeh, S.; Munaro, E.; McLean, G.; Massignam, A.; van Oosterom, E.J.; Chapman, S.C.; Messina, C.; Cooper, M.; Hammer, G.L. Modelling the nitrogen dynamics of maize crops—Enhancing the APSIM maize model. Eur. J. Agron. 2018, 100, 118–131. [Google Scholar] [CrossRef]
- Mandrini, G.; Archontoulis, S.V.; Pittelkow, C.M.; Mieno, T.; Martin, N.F. Simulated dataset of corn response to nitrogen over thousands of fields and multiple years in Illinois. Data Brief 2022, 40, 107753. [Google Scholar] [CrossRef]
- Mandrini, G.; Pittelkow, C.M.; Archontoulis, S.V.; Mieno, T.; Martin, N.F. Understanding differences between static and dynamic nitrogen fertilizer tools using simulation modeling. Agric. Syst. 2021, 194, 103275. [Google Scholar] [CrossRef]
- Mandrini, G.; Pittelkow, C.M.; Archontoulis, S.; Kanter, D.; Martin, N.F. Exploring trade-offs between profit, yield, and the environmental footprint of potential nitrogen fertilizer regulations in the US Midwest. Front. Plant Sci. 2022, 13, 852116. [Google Scholar] [CrossRef]
- Pimentel, J.; Quinn, D.; Bower, B. From South to North: Tracking Indiana’s Planting Progress, Issue #1–23 April 2025. Available online: https://ag.purdue.edu/news/department/agry/kernel-news/2025/04/_docs/indiana-corn-update-issue-1-april23.pdf (accessed on 6 October 2025).
- Nielsen, R.L.; Camberato, J.; Lee, J. Yield Response of Corn to Plant Population in Indiana. Available online: https://www.agry.purdue.edu/ext/corn/news/timeless/CornPopulations.pdf (accessed on 6 October 2025).
- Camberato, J.; Nielsen, R.L.; Quinn, D. Nitrogen Management Guidelines for Corn in Indiana. Available online: https://www.agry.purdue.edu/ext/corn/news/timeless/nitrogenmgmt.pdf (accessed on 6 October 2025).
- He, D.; Oliver, Y.; Rab, A.; Fisher, P.; Armstrong, R.; Kitching, M.; Wang, E. Plant available water capacity (PAWC) of soils predicted from crop yields better reflects within-field soil physicochemical variations. Geoderma 2022, 422, 115958. [Google Scholar] [CrossRef]
- Filippelli, G.M.; Freeman, J.L.; Gibson, J.; Jay, S.; Moreno-Madrinán, M.J.; Ogashawara, I.; Rosenthal, F.S.; Wang, Y.; Wells, E. Climate change impacts on human health at an actionable scale: A state-level assessment of Indiana, USA. Clim. Change 2020, 163, 1985–2004. [Google Scholar] [CrossRef]
- Stoner, E.R.; Baumgardner, M.F.; Weismiller, R.A.; Biehl, L.L.; Robinson, B.F. Extension of laboratory-measured soil spectra to field conditions. Soil Sci. Soc. Am. J. 1980, 44, 572–574. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Rai, N.; Pathak, H.; Mahecha, M.V.; Buckmaster, D.R.; Huang, Y.; Overby, P.; Sun, X. A case study on canola (Brassica napus L.) potential yield prediction using remote sensing imagery and advanced data analytics. Smart Agric. Technol. 2024, 9, 100698. [Google Scholar] [CrossRef]
- Louppe, G. An Introduction to Machine Learning with Scikit-Learn. LHCb Scikit-Learn Tutorial, 2015. Available online: https://github.com/glouppe/tutorials-scikit-learn/tree/master (accessed on 5 August 2025).
- Kakimoto, S.; Mieno, T.; Tanaka, T.S.T.; Bullock, D.S. Causal forest approach for site-specific input management via on-farm precision experimentation. Comput. Electron. Agric. 2022, 199, 107164. [Google Scholar] [CrossRef]
- Pearl, J. An introduction to causal inference. Int. J. Biostat. 2010, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- VanderWeele, T.J.; Hernán, M.A.; Robins, J.M. Causal directed acyclic graphs and the direction of unmeasured confounding bias. Epidemiology 2008, 19, 720–728. [Google Scholar] [CrossRef]
- Scutari, M.; Graafland, C.E.; Gutiérrez, J.M. Who learns better Bayesian network structures: Constraint-based, score-based or hybrid algorithms? In Proceedings of the International Conference on Probabilistic Graphical Models, Alicante, Spain, 13–15 September 2018; pp. 416–427. [Google Scholar]
- Tsamardinos, I.; Brown, L.E.; Aliferis, C.F. The max-min hill-climbing Bayesian network structure learning algorithm. Mach. Learn. 2006, 65, 31–78. [Google Scholar] [CrossRef]
- Valleggi, L.; Scutari, M.; Stefanini, F.M. Learning Bayesian networks with heterogeneous agronomic data sets via mixed-effect models and hierarchical clustering. Eng. Appl. Artif. Intell. 2024, 131, 107867. [Google Scholar] [CrossRef]
- Adhitama, R.P.; Saputro, D.R.S. Hill climbing algorithm for Bayesian network structure. AIP Conf. Proc. 2022, 2479, 020035. [Google Scholar] [CrossRef]
- Carvalho, A.M. Scoring Functions for Learning Bayesian Networks. Available online: https://www.lx.it.pt/~asmc/pub/talks/09-TA/ta_pres.pdf (accessed on 28 July 2025).
- Goswami, M.; Mohanty, S.; Pattnaik, P.K. Optimization of machine learning models through quantization and data bit reduction in healthcare datasets. Frankl. Open 2024, 8, 100136. [Google Scholar] [CrossRef]
- Ghazizadeh, A.; Ambroggi, F. Optimal Binning of Peri-Event Time Histograms Using Akaike Information Criterion. bioRxiv 2020. 2020-02. Available online: https://doi.org/10.1101/2020.02.06.937367 (accessed on 23 September 2025). [CrossRef]
- Boulle, M. Optimal Bin Number for Equal Frequency Discretizations in Supervised Learning. Intell. Data Anal. 2005, 9, 175–188. Available online: https://doi.org/10.3233/IDA-2005-9204 (accessed on 23 September 2025). [CrossRef]
- Gokmen, S.; Lyhagen, J. The Performance of Restricted AIC for Irregular Histogram Models. PLoS ONE 2024, 19, e0289822. [Google Scholar] [CrossRef]
- Apsim Info. The APSIM Maize Model. Available online: https://docs.apsim.info/validation/Maize (accessed on 6 October 2025).
- Licht, M.A. Corn Water Use and Evapotranspiration. Available online: https://crops.extension.iastate.edu/cropnews/2017/06/corn-water-use-and-evapotranspiration (accessed on 5 May 2025).
- Barideh, R.; Besharat, S.; Morteza, M.; Rezaverdinejad, V. Effects of partial root-zone irrigation on the water use efficiency and root water and nitrate uptake of corn. Water 2018, 10, 526. [Google Scholar] [CrossRef]
- Raza, S.; Farmaha, B.S. Contrasting corn yield responses to nitrogen fertilization in southeast coastal plain soils. Front. Environ. Sci. 2022, 10, 955142. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, W.; Wang, W.; Cui, T.; Chen, S.; Xu, C.; Engel, B. FACE-ing climate change: Propagation of risks and opportunities for cropping systems in mid-high-latitude regions: A case study between US and China corn belts. Agric. Syst. 2024, 220, 104087. [Google Scholar] [CrossRef]
- Apsim Info. SoilN—APSIM Soil Nitrogen Module Documentation. Available online: https://www.apsim.info/documentation/model-documentation/soil-modules-documentation/soiln/ (accessed on 28 June 2025).
- Williams, J.R.; Jones, C.A.; Dyke, P.T. The EPIC model and its application. In Proceedings of the ICRISAT-IBSNAT-SYSS Symposium on Minimum Data Sets for Agrotechnology Transfer, Hyderabad, India, 21–26 March 1984; pp. 111–121. [Google Scholar]
- Verburg, K.; Pasley, H.R.; Biggs, J.S.; Vogeler, I.; Wang, E.; Mielenz, H.; Snow, V.O.; Smith, C.J.; Pasut, C.; Basche, A.D.; et al. Review of APSIM’s soil nitrogen modelling capability for agricultural systems analyses. Agric. Syst. 2025, 224, 104213. [Google Scholar] [CrossRef]
- Tahir, N.; Li, J.; Ma, Y.; Ullah, A.; Zhu, P.; Peng, C.; Hussain, B.; Danish, S. 20 years nitrogen dynamics study by using APSIM nitrogen model simulation for sustainable management in Jilin, China. Sci. Rep. 2021, 11, 17505. [Google Scholar] [CrossRef]
- Snow, V.; Huth, N. The APSIM–MICROMET Module. March 2004. Available online: https://www.apsim.info/wp-content/uploads/2019/09/Micromet.pdf (accessed on 5 August 2025).
- Brown, H.E.; Huth, N.I.; Holzworth, D.P.; Teixeira, E.I.; Zyskowski, R.F.; Hargreaves, J.N.G.; Moot, D.J. Plant modelling framework: Software for building and running crop models on the APSIM platform. Environ. Model. Softw. 2014, 62, 385–398. [Google Scholar] [CrossRef]
- Tyagi, S.; Sahany, S.; Saraswat, D.; Mishra, S.K.; Dubey, A.; Niyogi, D. Assessing regional-scale heterogeneity in blue–green water availability under the 1.5 °C global warming scenario. J. Appl. Meteorol. Climatol. 2024, 63, 553–574. [Google Scholar] [CrossRef]


| Factor | Levels |
|---|---|
| Cultivars (Genetics) | 105 RM, 115 RM, 130 RM |
| Soil Types | North, Central, South Indiana |
| Weather Locations | North, Central, South Indiana |
| Planting Dates | 1 April, 30 April, 30 May |
| Nitrogen Rates | 0, 142, 190, 237 kg ha−1 |
| Irrigation management | No irrigation and Irrigation Triggers (0%, 50%, 75% PAWC), |
| which stops at 100%PAWC | |
| Climate Scenarios | Historical, Mid-century (precipitation increased by 6%, to |
| 550 ppm, and temperature by 5.5 K), End-century (precipitation | |
| increased by 10%, to 650 ppm, and temperature by 5.5 K) | |
| Years Simulated | 1984–2021 (38 years) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pathak, H.; Buckmaster, D.R.; Kaur, U.; Mandrini, G.; Poudel, P. Understanding Corn Production Complexity: Causal Structure Learning and Variable Ranking from Agricultural Simulations. AgriEngineering 2025, 7, 366. https://doi.org/10.3390/agriengineering7110366
Pathak H, Buckmaster DR, Kaur U, Mandrini G, Poudel P. Understanding Corn Production Complexity: Causal Structure Learning and Variable Ranking from Agricultural Simulations. AgriEngineering. 2025; 7(11):366. https://doi.org/10.3390/agriengineering7110366
Chicago/Turabian StylePathak, Harsh, Dennis R. Buckmaster, Upinder Kaur, German Mandrini, and Pratishtha Poudel. 2025. "Understanding Corn Production Complexity: Causal Structure Learning and Variable Ranking from Agricultural Simulations" AgriEngineering 7, no. 11: 366. https://doi.org/10.3390/agriengineering7110366
APA StylePathak, H., Buckmaster, D. R., Kaur, U., Mandrini, G., & Poudel, P. (2025). Understanding Corn Production Complexity: Causal Structure Learning and Variable Ranking from Agricultural Simulations. AgriEngineering, 7(11), 366. https://doi.org/10.3390/agriengineering7110366






