Machine Learning Techniques for Nematode Microscopic Image Analysis: A Systematic Review
Abstract
1. Introduction
- What challenges are addressed by AI-based methods?
- What are the most effective ML/DL models? What are their strengths and limitations?
- What novel AI-driven techniques have been proposed to improve nematode inference?
- What trends and opportunities exist for improving AI-based nematode inference?
2. Methodology
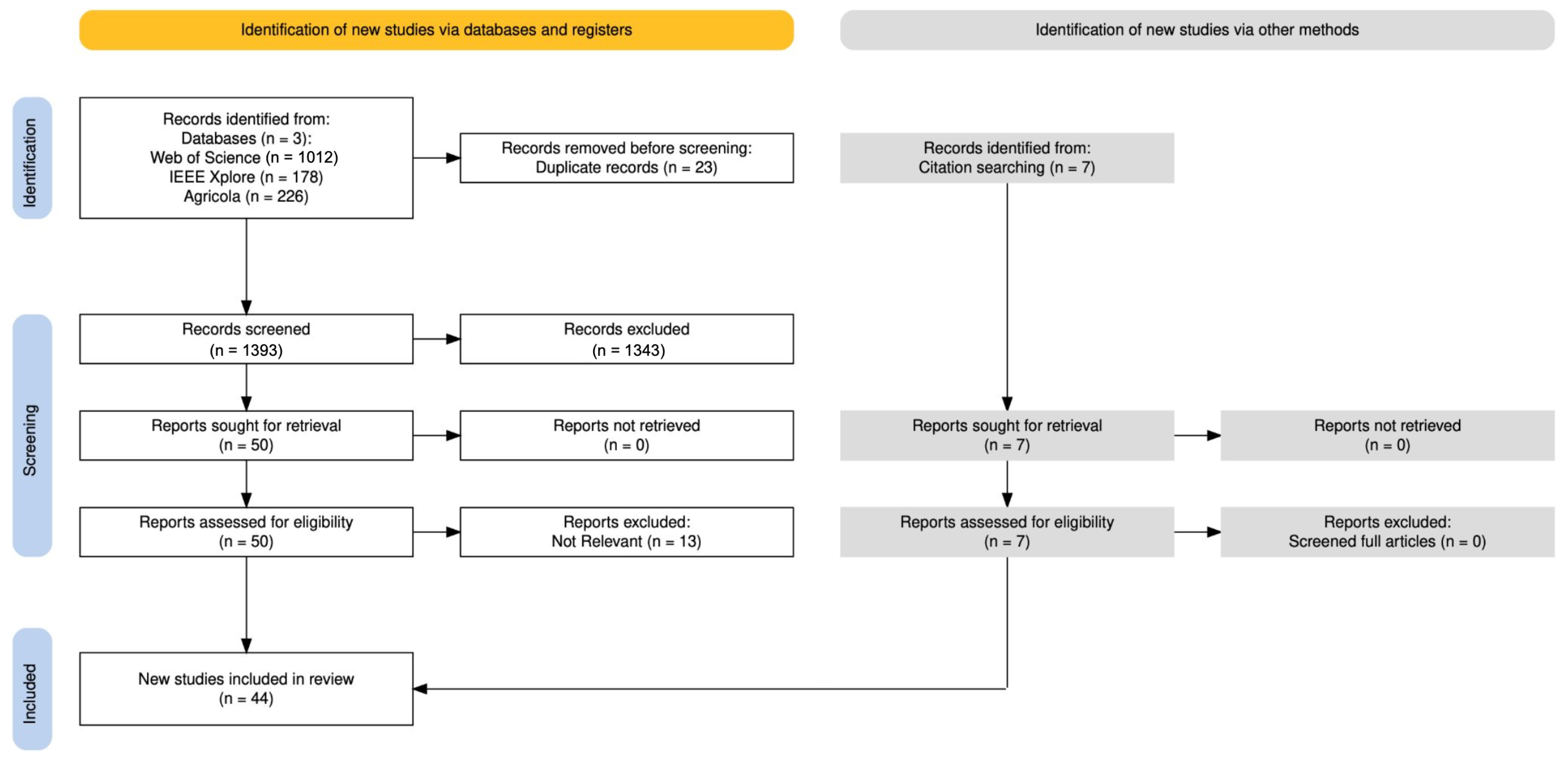
2.1. Review Process
2.2. Search Strategy
2.3. Selection Criteria
2.4. Selection Process
2.5. Data Extraction Process
2.6. Other Methods
2.7. Performance Metrics
2.7.1. Standard Classification Metrics
2.7.2. Additional Metrics
2.8. Machine Learning Models
3. Results
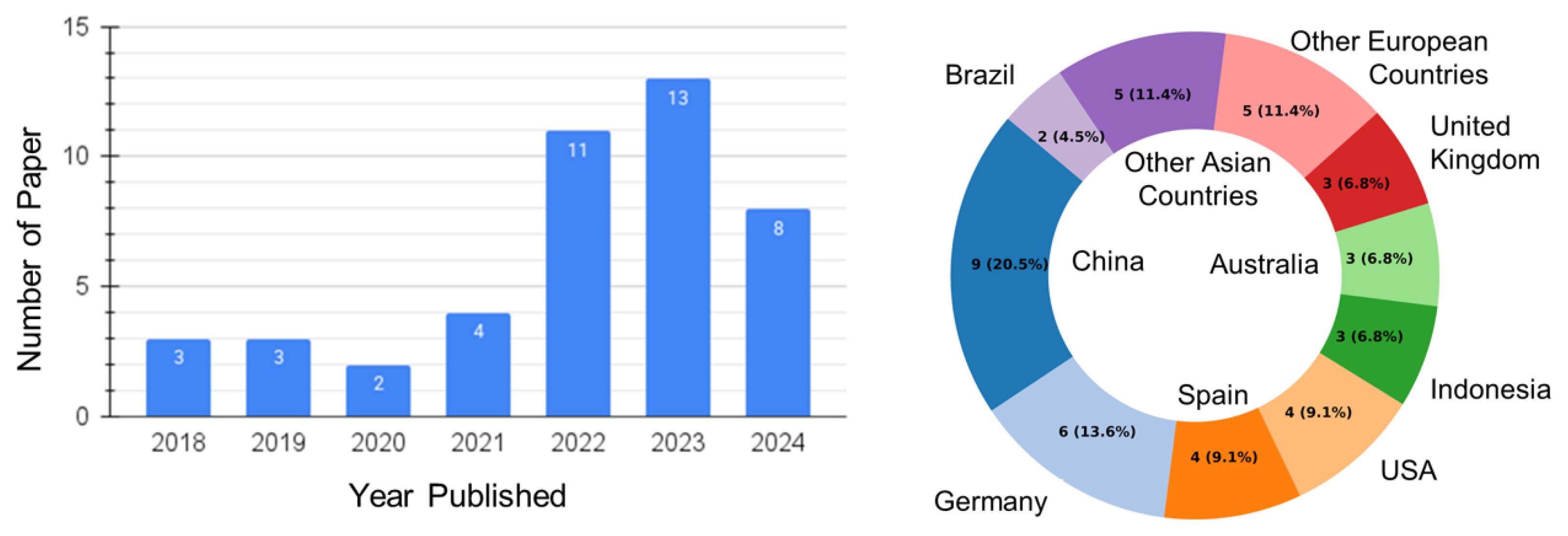
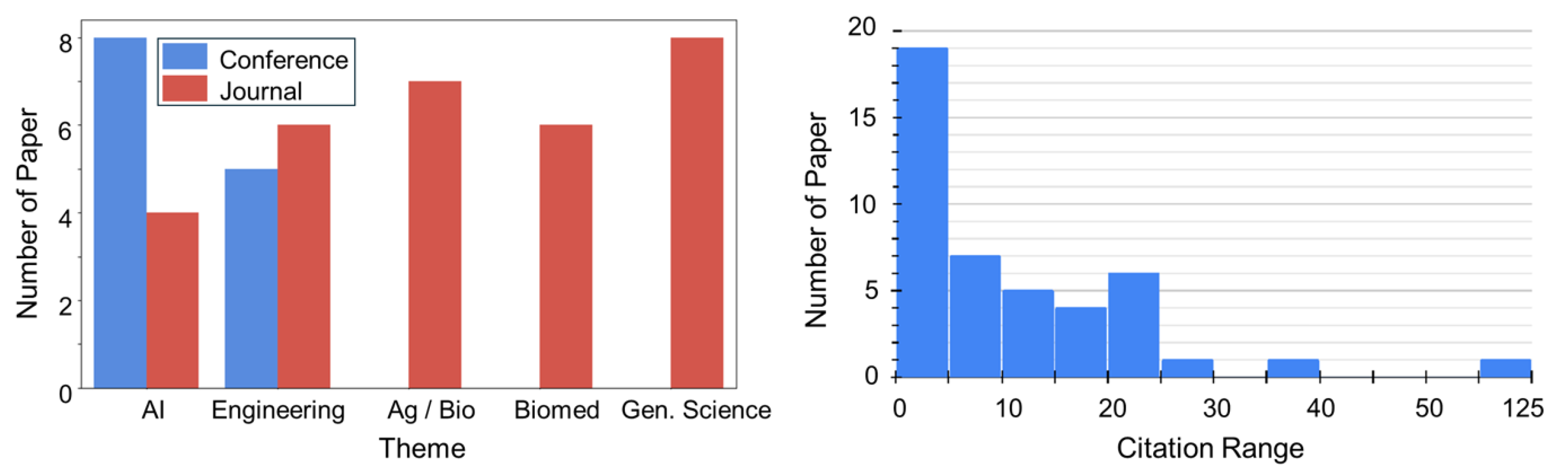
3.1. Publication Trends
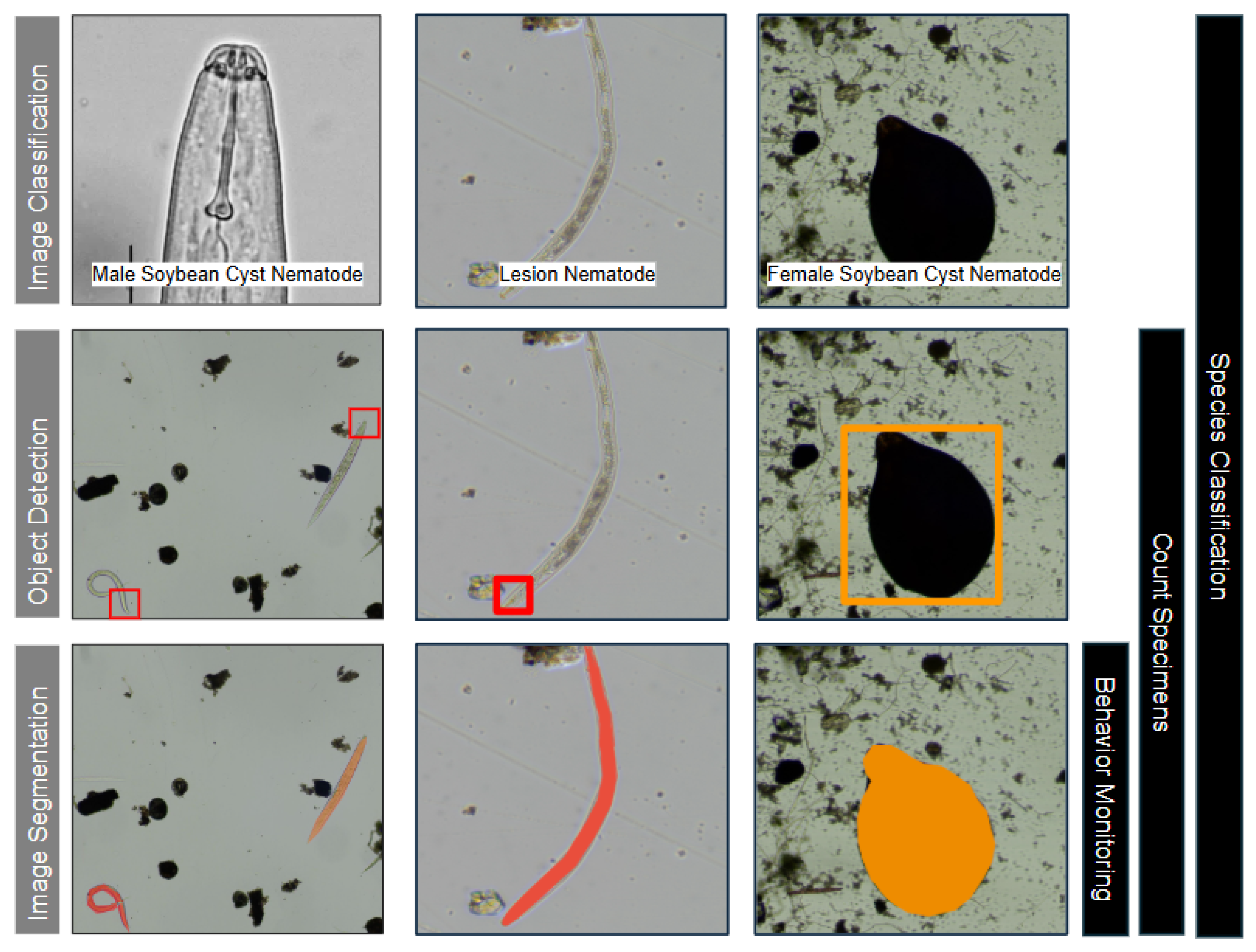
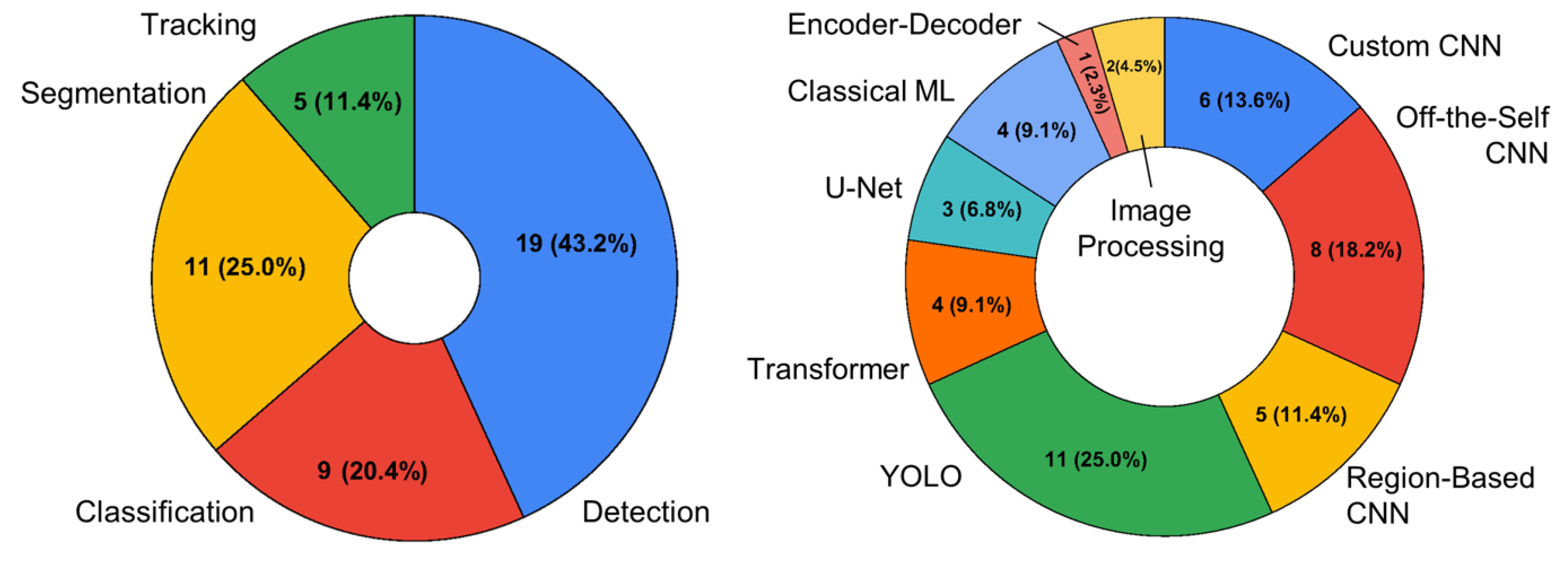
3.2. Image Analysis Trends
| Ref | Detection | ||||||
|---|---|---|---|---|---|---|---|
| and Year | Model Used | Dataset: Size | Train/Test | Acc | Prec | Rec | F1 |
| [44] 2018 | Conv. Selective AE | Cust: 3044 | 2435/609 | 0.95 | 0.94 | 0.94 | 0.94 |
| [57] 2018 | CNN | Cust: 11,820 | 10,047/1773 | 0.93 | |||
| [58] 2019 | HoG+SVM | Cust: 1200 | 996/204 | 0.87 | |||
| [45] 2019 | FCN | 0.99 | |||||
| [59] 2021 | CNN+LSTM | Cust: 23,584 | 17,923/5661 | 0.91 | |||
| [60] 2022 | YOLOv5s | Cust.: 1901 | 1520/380 | 0.93 | |||
| Faster R-CNN | 0.94 | ||||||
| [61] 2022 | YOLOv4-1 Class | 0.91 | 0.79 | 0.85 | |||
| YOLOv4-Multiclass | Cust.: 20,000 (augmented images) | 19,000/1000 | 0.93 | 0.93 | 0.92 | ||
| [62] 2022 | Image processing | N/A | 0.98 | 0.98 | 0.98 | ||
| [55] 2023 | YOLOv5s | BBBC010/11 [56]: 17,213 | 14,385/2828 | 0.88 | 0.76 | ||
| [63] 2023 | YOLOv7-640 | Cust: 415 | 332/83 | 0.99 | 0.99 | 0.99 | 0.99 |
| [64] 2023 | YOLOv5 | Cust:13k (augmented) | 10,400/2600 | 1.00 | 0.99 | 0.99 | |
| [65] 2023 | Faster R-CNN | Cust.: 182 | 163/19 | 0.86 | 0.94 | 0.83 | 0.88 |
| YOLOv4-CSP | 0.77 | 0.91 | 0.80 | 0.85 | |||
| [66] 2023 | YOLOv7 | Cust.: 3931 | 3537/394 | 0.99 | 0.97 | 0.96 | 0.97 |
| Faster R-CNN | 0.98 | 0.74 | 0.99 | 0.85 | |||
| [67] 2023 | YOLOv5x | Cust: 3503 | - | 0.85 | 0.75 | ||
| [68] 2023 | Faster R-CNN | Cust: 1625 | 1413/212 | 0.87 | 0.61 | 0.68 | |
| [69] 2024 | CNN and contour methods | Cust: 10,468 | 0.99 | ||||
| [70] 2024 | YOLOv8 | Cust: 19,929 | 15,677/4252 | 0.57 | 0.59 | 0.94 | 0.47 |
| [71] 2024 | YOLOv5 | Cust: 894 | 788/106 | 0.78 | 0.78 | ||
| [72] 2024 | YOLOv5 | Cust: 6000 | 0.97 | 0.94 | 0.95 | ||
| [73] 2024 | HoG+SVM | Cust: 20,647 | 0.91 | 0.92 | 0.97 | 0.95 | |
| Ref | Segmentation | ||||
|---|---|---|---|---|---|
| Data | and Year | Model Used | DC | AP50 | Prec/Rec @ 0.5 |
| C4 | [75] 2019 | Image proc. | 0.76 | ||
| C6 | [13] 2020 | U-Net | 0.90/0.87 | ||
| D1 | [76] 2021 | Mask R-CNN | 0.9 | ||
| C1 | [77] 2022 | U-Net | 0.46 | ||
| C2 | 0.47 | ||||
| C3 | [78] 2022 | Mask R-CNN | 0.89 | ||
| D2 | [18] 2023 | Transformer-WormSwin | 0.98 | ||
| D3 | [33] 2024 | Mask2Former | 0.62 | ||
| D4 | [13] 2020 | U-Net | 0.96/0.97 | ||
| [18] 2023 | Transformer-WormSwin | 0.95 | |||
| [33] 2024 | Mask2Former | 0.92 | |||
| [79] 2024 | Mask2Former | 0.71 | |||
| D5 | [18] 2023 | Transformer-WormSwin | 0.99 | ||
| [79] 2024 | Mask2former | 0.87 | |||
| D3 | [33] 2024 | Multiple Instance Learning | 0.65 | ||
| D4 | 0.92 | ||||
| ID | Name and Ref | No. of Images | Train/Test Split | Avail. |
|---|---|---|---|---|
| D1 | C. elegans [80] | 1908 | 1711/197 | Yes |
| D2 | Mating Dataset [18] | 450 patch images | 0/450 | Yes |
| D3 | Haemonchus Contortus [17] | 95 | 76/19 | Yes |
| D4 | BBBC010 [56] | 100 | N/A | Yes |
| D5 | CSB-1 [18] | 4631 (10 videos) | N/A | Yes |
| C1 | Custom Nematode Dataset [77] | 3967 | 3367/667 | No |
| C2 | Custom Cyst Dataset [77] | 487 | 387/100 | No |
| C3 | Custom Nematode Dataset [78] | 16k | 14,880/1120 | No |
| C4 | Custom Cyst Dataset [75] | 132 | N/A | No |
| C5 | Potato Cyst Nematode [13] | 1973 | 1606/367 | No |
Datasets
3.3. Biological Inference Trends
3.3.1. Species Classification
3.3.2. Counting
3.3.3. Behavioral Tracking
3.3.4. Lifespan Tracking
4. Discussion
4.1. Advantages
4.2. Challenges and Proposed Solutions
- Limited training data—Custom datasets are often small or inconsistently annotated. Another barrier identified in this review is the lack of standardized benchmark (SB) datasets for nematodes. In the context of ML, an SB is a common, objective framework that is used to evaluate and compare the performance of different models and algorithms. It refers to a curated and publicly available dataset with clearly defined tasks, consistent annotation standards, and well-established evaluation metrics. Solution: Wider use of data augmentation (e.g., rotation, scaling, and synthetic image generation), transfer learning, and collaborative dataset sharing can improve the generalizability of models. Many studies relied on private datasets and inconsistent metrics, making direct evaluation difficult. A prime example of the SB approach is the I-Nema dataset presented by Lu et al. [16], which provides images of 19 nematode species with a rigorous, repeatable experimental protocol. Adopting such benchmarks would directly address the challenges of reproducibility and inconsistent reporting.
- Occlusion and overlapping objects—Worms and eggs often overlap, complicating detection and counting. Solution: Advanced models such as transformer-based detectors and instance segmentation approaches can help to resolve occluded objects more reliably.
- Inconsistent metric reporting—Variation in reported evaluation metrics makes cross-study comparisons difficult. Solution: Adoption of standardized evaluation frameworks and transparent reporting of datasets and splits will facilitate reproducibility and fair benchmarking.
4.3. Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| ML | Machine Learning |
| DL | Deep Learning |
| AP | Average Precision |
| SLR | Systematic Literature Review |
| PRISMA | Pref. Reporting Items for Syst. Reviews and Meta-Analyses |
| C. elegans | Caenorhabditis elegans |
| MIA | Microscopy Image Analysis |
| CNN | Convolutional Neural Network |
| IoU | Intersection over Union |
| TP | True Positives |
| FP | False Positives |
| AP | Average Precision |
| mAP | mean Average Precision |
| SVM | Support Vector Machine |
| RF | Random Forest |
| ANN | Artificial Neural Network |
| DNN | Deep Neural Network |
| AE | Autoencoder |
| R-CNN | Region-Based Convolutional Neural Network |
| YOLO | You Only Look Once |
| HoG | Histogram of Gradients |
| SVM | Support Vector Machine |
| RKN | Root-knot nematode |
| CSAE | Convolutional Sparse Auto-Encoder |
| FLN | Free-living nematode |
| coefficient of determination | |
| RPN | Region Proposal Network |
| IJ | Infective Juvenile |
| LSTM | Long short-term memory |
| RoI | Region of Interest |
| SotA | State-of-the-Art |
Appendix A. Top-Performing DL and ML Models
Appendix A.1. Classical Machine Learning Models
Appendix A.2. Convolutional Neural Network (CNN) Models
Appendix A.3. Region-Based CNN Models
Appendix A.4. YOLO Object Detection Models
Appendix A.5. U-Net and Encoder–Decoder Models
Appendix A.6. Transformer Models
Appendix A.7. Segmentation Models
References
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M.L.; et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations (FAO). The State of the World’s Land and Water Resources for Food and Agriculture 2021 (SOLAW 2021): Systems at Breaking Point. 2021. Available online: https://openknowledge.fao.org/items/55def12b-2a81-41e5-91dc-ac6c42f1cd0f (accessed on 21 July 2025).
- Min, Y.Y.; Toyota, K.; Sato, E. A novel nematode diagnostic method using the direct quantification of major plant-parasitic nematodes in soil by real-time PCR. Nematology 2012, 14, 265–276. [Google Scholar] [CrossRef]
- Holladay, B.H.; Willett, D.S.; Stelinski, L.L. High throughput nematode counting with automated image processing. BioControl 2016, 61, 177–183. [Google Scholar] [CrossRef]
- Abdelsamea, M.M. A semi-automated system based on level sets and invariant spatial interrelation shape features for Caenorhabditis elegans phenotypes. J. Vis. Commun. Image Represent. 2016, 41, 314–323. [Google Scholar] [CrossRef]
- Uhlemann, J.; Cawley, O.; Kakouli-Duarte, T. Nematode Identification using Artificial Neural Networks. In Proceedings of the 1st International Conference on Deep Learning Theory and Applications—DeLTA. INSTICC, Virtual Event, 8–10 July 2020; SciTePress: Setubal, Portugal, 2020; pp. 13–22. [Google Scholar] [CrossRef]
- Shabrina, N.H.; Lika, R.A.; Indarti, S. The Effect of Data Augmentation and Optimization Technique on the Performance of EfficientNetV2 for Plant-Parasitic Nematode Identification. In Proceedings of the 2023 IEEE International Conference on Industry 4.0, Artificial Intelligence, and Communications Technology (IAICT), Bali, Indonesia, 13–15 July 2023; pp. 190–195. [Google Scholar] [CrossRef]
- Bates, K.; Le, K.N.; Lu, H. Deep learning for robust and flexible tracking in behavioral studies for C. elegans. PLoS Comput. Biol. 2022, 18, 1–24. [Google Scholar] [CrossRef]
- Banerjee, S.C.; Khan, K.A.; Sharma, R. Deep-worm-tracker: Deep learning methods for accurate detection and tracking for behavioral studies in C. elegans. Appl. Anim. Behav. Sci. 2023, 266, 106024. [Google Scholar] [CrossRef]
- Shabrina, N.H.; Lika, R.A.; Indarti, S. Deep learning models for automatic identification of plant-parasitic nematode. Artif. Intell. Agric. 2023, 7, 1–12. [Google Scholar] [CrossRef]
- Shabrina, N.H.; Indarti, S.; Lika, R.A.; Maharani, R. A comparative analysis of convolutional neural networks approaches for phytoparasitic nematode identification. Commun. Math. Biol. Neurosci. 2023, 2023, 65. [Google Scholar] [CrossRef]
- Chen, L.; Strauch, M.; Daub, M.; Jiang, X.; Jansen, M.; Luigs, H.G.; Schultz-Kuhlmann, S.; Krüssel, S.; Merhof, D. A CNN Framework Based on Line Annotations for Detecting Nematodes in Microscopic Images. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 508–512. [Google Scholar] [CrossRef]
- Wählby, C.; Kamentsky, L.; Liu, Z.H.; Riklin-Raviv, T.; Conery, A.L.; O’Rourke, E.J.; Sokolnicki, K.L.; Visvikis, O.; Ljosa, V.; Irazoqui, J.E.; et al. An image analysis toolbox for high-throughput C. elegans Assays. Nat. Methods 2012, 9, 714–716. [Google Scholar] [CrossRef]
- Abade, A.; Porto, L.F.; Ferreira, P.A.; de Barros Vidal, F. NemaNet: A convolutional neural network model for identification of soybean nematodes. Biosyst. Eng. 2022, 213, 39–62. [Google Scholar] [CrossRef]
- Lu, X.; Wang, Y.; Fung, S.; Qing, X. I-Nema: A Biological Image Dataset for Nematode Recognition. arXiv 2021, arXiv:2103.08335. [Google Scholar] [CrossRef]
- Žofka, M.; Thuy Nguyen, L.; Mašátová, E.; Matoušková, P. Image recognition based on deep learning in Haemonchus contortus motility assays. Comput. Struct. Biotechnol. J. 2022, 20, 2372–2380. [Google Scholar] [CrossRef] [PubMed]
- Deserno, M.; Bozek, K. WormSwin: Instance segmentation of C. elegans using vision transformer. Sci. Rep. 2023, 13, 11021. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Yang, L. Chapter 4-Machine Learning and Its Application in Microscopic Image Analysis. In Machine Learning and Medical Imaging; Wu, G., Shen, D., Sabuncu, M.R., Eds.; The Elsevier and MICCAI Society Book Series; Academic Press: Washington, DC, USA, 2016; pp. 97–127. [Google Scholar] [CrossRef]
- Wilson, M.; Kakouli-Duarte, T. Nematodes as Environmental Indicators; CABI: Wallingford, UK, 2009; pp. 1–326. [Google Scholar] [CrossRef]
- Walter, T.; Couzin, I.D. TRex, a fast multi-animal tracking system with markerless identification, and 2D estimation of posture and visual fields. eLife 2021, 10, e64000. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Shamseer, L.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A.; PRISMA-P Group. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015, 4, 1. [Google Scholar] [CrossRef]
- Kitchenham, B.A.; Charters, S. Guidelines for Performing Systematic Literature Reviews in Software Engineering; EBSE Technical Report EBSE-2007-001; Keele University: Keele, UK; Durham University: Durham, UK, 2007. [Google Scholar]
- Web of Science. Web of Science Digital Library. Available online: https://clarivate.com/webofsciencegroup/solutions/web-of-science (accessed on 12 August 2025).
- IEEE Xplore. IEEE Xplore Digital Library. Available online: https://ieeexplore.ieee.org (accessed on 12 August 2025).
- AGRICOLA. AGRICOLA Digital Library. Available online: https://www.nal.usda.gov/agricola (accessed on 12 August 2025).
- Haddaway, N.R.; Page, M.J.; Pritchard, C.C.; McGuinness, L.A. PRISMA2020: An R package and Shiny app for producing PRISMA 2020-compliant flow diagrams, with interactivity for optimised digital transparency and Open Synthesis. Campbell Syst. Rev. 2022, 18, e1230. [Google Scholar] [CrossRef]
- Gupta, M.; Mishra, A. A systematic review of deep learning based image segmentation to detect polyp. Artif. Intell. Rev. 2024, 57, 7. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; Available online: http://www.deeplearningbook.org (accessed on 15 August 2025).
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized Intersection Over Union: A Metric and a Loss for Bounding Box Regression. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 658–666. [Google Scholar] [CrossRef]
- Shamir, R.R.; Duchin, Y.; Kim, J.; Sapiro, G.; Harel, N. Continuous Dice Coefficient: A Method for Evaluating Probabilistic Segmentations. bioRxiv 2018. Available online: https://www.biorxiv.org/content/early/2018/04/25/306977.full.pdf (accessed on 15 August 2025). [CrossRef]
- Ma, T.; Zhang, K.; Xu, J.; Xu, L.; Zhang, C.; Guo, L.; Li, Y. Instance Segmentation of Nematode Image Based on Transformer with Skeleton Points. In Proceedings of the 2024 7th International Conference on Advanced Algorithms and Control Engineering (ICAACE), Shanghai, China, 1–3 March 2024; pp. 442–448. [Google Scholar] [CrossRef]
- Davis, J.; Goadrich, M. The relationship between Precision-Recall and ROC curves. In Proceedings of the 23rd International Conference on Machine Learning, ICML ’06, Pittsburgh, PA, USA, 25–29 June 2006; pp. 233–240. [Google Scholar] [CrossRef]
- Everingham, M.; Van Gool, L.; Williams, C.K.I.; Winn, J.; Zisserman, A. The Pascal Visual Object Classes (VOC) Challenge. Int. J. Comput. Vis. 2010, 88, 303–338. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context. In Proceedings of the Computer Vision—ECCV 2014, Zurich, Switzerland, 6–12 September 2014; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer: Cham, Switzerland, 2014; pp. 740–755. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J.H.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: Berlin/Heidelberg, Germany, 2009; Volume 2. [Google Scholar]
- Jiménez-Chavarría, J. SegNema: Nematode Segmentation strategy in Digital Microscopy Images Using Deep Learning and Shape Models. 2019. Available online: https://api.semanticscholar.org/CorpusID:204827619 (accessed on 15 August 2025).
- Haque, S.; Lobaton, E.; Nelson, N.; Yencho, G.C.; Pecota, K.V.; Mierop, R.; Kudenov, M.W.; Boyette, M.; Williams, C.M. Computer vision approach to characterize size and shape phenotypes of horticultural crops using high-throughput imagery. Comput. Electron. Agric. 2021, 182, 106011. [Google Scholar] [CrossRef]
- Ropelewska, E.; Skwiercz, A.; Sobczak, M. Distinguishing Cyst Nematode Species Using Image Textures and Artificial Neural Networks. Agronomy 2023, 13, 2277. [Google Scholar] [CrossRef]
- Yuan, Z.; Musa, N.; Dybal, K.; Back, M.; Leybourne, D.; Yang, P. Quantifying Nematodes through Images: Datasets, Models, and Baselines of Deep Learning. arXiv 2024, arXiv:2404.19748. [Google Scholar] [CrossRef]
- Chou, Y.; Dah-Jye, L.; Zhang, D. Edge Detection Using Convolutional Neural Networks for Nematode Development and Adaptation Analysis. In Computer Vision Systems. ICVS 2017; Springer: Cham, Switzerland, 2017; pp. 228–238. [Google Scholar] [CrossRef]
- Zhan, M.; Crane, M.M.; Entchev, E.V.; Caballero, A.; Fernandes de Abreu, D.A.; Ch’ng, Q.; Lu, H. Automated processing of imaging data through multi-tiered classification of biological structures illustrated using Caenorhabditis elegans. PLoS Comput. Biol. 2015, 11, e1004194. [Google Scholar] [CrossRef]
- Akintayo, A.; Tylka, G.L.; Singh, A.K.; Ganapathysubramanian, B.; Singh, A.; Sarkar, S. A deep learning framework to discern and count microscopic nematode eggs. Sci. Rep. 2018, 8, 9145. [Google Scholar] [CrossRef]
- Javer, A.; Brown, A.E.X.; Kokkinos, I.; Rittscher, J. Identification of C. elegans Strains Using a Fully Convolutional Neural Network on Behavioural Dynamics. In Proceedings of the Computer Vision—ECCV 2018 Workshops, Munich, Germany, 8–14 September 2018; Leal-Taixé, L., Roth, S., Eds.; Springer: Cham, Switzerland, 2019; pp. 455–464. [Google Scholar]
- Galimov, E.; Yakimovich, A. A tandem segmentation-classification approach for the localization of morphological predictors of C. elegans Lifesp. Motility. Aging 2022, 14, 1665–1677. [Google Scholar] [CrossRef]
- Qing, X.; Wang, Y.; Lu, X.; Li, H.; Wang, X.; Li, H.; Xie, X. NemaRec: A deep learning-based web application for nematode image identification and ecological indices calculation. Eur. J. Soil Biol. 2022, 110, 103408. [Google Scholar] [CrossRef]
- Liu, M.; Wang, X.; Zhang, H. Taxonomy of multi-focal nematode image stacks by a CNN based image fusion approach. Comput. Methods Programs Biomed. 2018, 156, 209–215. [Google Scholar] [CrossRef]
- Javer, A.; Currie, M.; Lee, C.W.; Hokanson, J.; Li, K.; Martineau, C.N.; Yemini, E.; Grundy, L.J.; Li, C.; Ch’ng, Q.; et al. An open source platform for analyzing and sharing worm behavior data. bioRxiv 2018. Available online: https://www.biorxiv.org/content/early/2018/07/26/377960.full.pdf (accessed on 15 August 2025). [CrossRef]
- Cook, D.E.; Zdraljevic, S.; Roberts, J.P.; Andersen, E.C. CeNDR, the Caenorhabditis elegans natural diversity resource. Nucleic Acids Res. 2017, 45, D650–D657. [Google Scholar] [CrossRef]
- Thevenoux, R.; LE, V.L.; Villessèche, H.; Buisson, A.; Beurton-Aimar, M.; Grenier, E.; Folcher, L.; Parisey, N. Image based species identification of Globodera quarantine nematodes using computer vision and deep learning. Comput. Electron. Agric. 2021, 186, 106058. [Google Scholar] [CrossRef]
- EMBL-EBI BioImage Archive. Raw images of Caenorhabditis elegans with Lifespan, Movement, and Segmentation Annotations (Accession: S-BIAD300). 2022. Available online: https://www.ebi.ac.uk/biostudies/bioimages/studies/S-BIAD300 (accessed on 15 August 2025).
- Qayyum, A.; Benzinou, A.; Nasreddine, K.; Foulon, V.; Martinel, A.; Borremans, C.; Hagemann, R.; Zeppilli, D. A Deep Learning Vision Transformer using Majority Voting Ensemble for Nematode Classification with Test Time Augmentation Technique. In Proceedings of the 4rd International Conference on Advances in Signal Processing and Artificial Intelligence (ASPAI’ 2022), Corfu, Greece, 19–21 October 2022. [Google Scholar]
- Verma, M.; Manhas, J.; Parihar, R.D.; Sharma, V. A Framework for Classification of Nematodes Species Using Deep Learning. In Emerging Trends in Expert Applications and Security; Rathore, V.S., Piuri, V., Babo, R., Ferreira, M.C., Eds.; Springer: Singapore, 2023; pp. 71–79. [Google Scholar]
- Escobar-Benavides, S.; García-Garví, A.; Layana-Castro, P.E.; Sánchez-Salmerón, A.J. Towards generalization for Caenorhabditis elegans detection. Comput. Struct. Biotechnol. J. 2023, 21, 4914–4922. [Google Scholar] [CrossRef] [PubMed]
- Ljosa, V.; Sokolnicki, K.L.; Carpenter, A.E. Annotated high-throughput microscopy image sets for validation. Nat. Methods 2012, 9, 637. [Google Scholar] [CrossRef] [PubMed]
- Hakim, A.; Mor, Y.; Toker, I.A.; Levine, A.; Neuhof, M.; Markovitz, Y.; Rechavi, O. WorMachine: Machine learning-based phenotypic analysis tool for worms. BMC Biol. 2018, 16, 8. [Google Scholar] [CrossRef] [PubMed]
- Bornhorst, J.; Nustede, E.J.; Fudickar, S. Mass Surveilance of C.elegans—Smartphone-Based DIY Microsc. Mach.-Learn. Approach Worm Detect. Sensors 2019, 19, 1468. [Google Scholar] [CrossRef]
- García Garví, A.; Puchalt, J.C.; Layana Castro, P.E.; Navarro Moya, F.; Sánchez-Salmerón, A.J. Towards Lifespan Automation for Caenorhabditis elegans Based on Deep Learning: Analysing Convolutional and Recurrent Neural Networks for Dead or Live Classification. Sensors 2021, 21, 4943. [Google Scholar] [CrossRef]
- Rico-Guardiola, E.J.; Layana-Castro, P.E.; García-Garví, A.; Sánchez-Salmerón, A.J. Caenorhabditis Elegans Detection Using YOLOv5 and Faster R-CNN Networks. In Optimization, Learning Algorithms and Applications; Pereira, A.I., Košir, A., Fernandes, F.P., Pacheco, M.F., Teixeira, J.P., Lopes, R.P., Eds.; Springer: Cham, Switzerland, 2022; pp. 776–787. [Google Scholar]
- Mori, S.; Tachibana, Y.; Suzuki, M.; Harada, Y. Automatic worm detection to solve overlapping problems using a convolutional neural network. Sci. Rep. 2022, 12, 8521. [Google Scholar] [CrossRef]
- Pun, T.B.; Neupane, A.; Koech, R.; Owen, K.J. Detection and Quantification of Root-Knot Nematode (Meloidogyne Spp.) Eggs from Tomato Plants Using Image Analysis. IEEE Access 2022, 10, 123190–123204. [Google Scholar] [CrossRef]
- Pun, T.B.; Neupane, A.; Koech, R. A Deep Learning-Based Decision Support Tool for Plant-Parasitic Nematode Management. J. Imaging 2023, 9, 240. [Google Scholar] [CrossRef]
- Pun, T.B.; Neupane, A.; Koech, R.; Walsh, K. Detection and counting of root-knot nematodes using YOLO models with mosaic augmentation. Biosens. Bioelectron. X 2023, 15, 100407. [Google Scholar] [CrossRef]
- Ribeiro, P.; Sobieranski, A.C.; Gonçalves, E.C.; Dutra, R.C.; von Wangenheim, A. Automated classification and tracking of microscopic holographic patterns of nematodes using machine learning methods. Nematology 2023, 26, 183–201. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, S.; Yuan, H.; Yong, R.; Duan, S.; Li, Y.; Spencer, J.; Lim, E.G.; Yu, L.; Song, P. Deep Learning for Microfluidic-Assisted Caenorhabditis elegans Multi-Parameter Identification Using YOLOv7. Micromachines 2023, 14, 1339. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Curran, Z.C.; Yu, G.; Mishra, S.; Baniya, A.; Bogale, M.; Hughes, K.; Salichs, O.; Zare, A.; Jiang, Z.; et al. Plant Parasitic Nematode Identification in Complex Samples with Deep Learning. J. Nematol. 2023, 55, 20230045. [Google Scholar] [CrossRef] [PubMed]
- Angeline, N.; Husna Shabrina, N.; Indarti, S. Faster region-based convolutional neural network for plant-parasitic and non-parasitic nematode detection. Indones. J. Electr. Eng. Comput. Sci. 2023, 30, 316. [Google Scholar] [CrossRef]
- Fraher, S.P.; Watson, M.; Nguyen, H.; Moore, S.; Lewis, R.S.; Kudenov, M.; Yencho, G.C.; Gorny, A.M. A Comparison of Three Automated Root-Knot Nematode Egg Counting Approaches Using Machine Learning, Image Analysis, and a Hybrid Model. Plant Dis. 2024, 108, 2625–2629. [Google Scholar] [CrossRef] [PubMed]
- Saikai, K.K.; Bresilla, T.; Kool, J.; de Ruijter, N.C.A.; van Schaik, C.; Teklu, M.G. Counting nematodes made easy: Leveraging AI-powered automation for enhanced efficiency and precision. Front. Plant Sci. 2024, 15, 1349209. [Google Scholar] [CrossRef]
- Phuyued, U.; Kangkachit, T.; Jitkongchuen, D. Detection of Infective Juvenile Stage of Entomopathogenic Nematodes Using Deep Learning. In Proceedings of the 2024 5th International Conference on Big Data Analytics and Practices (IBDAP), Bangkok, Thailand, 23–25 August 2024; pp. 50–55. [Google Scholar] [CrossRef]
- Weng, J.; Lin, Q.; Chen, S.; Chen, W. Method for Live Determination of Caenorhabditis Elegans Based on Deep Learning and Image Processing. In Proceedings of the 2024 IEEE International Conference on Advanced Information, Mechanical Engineering, Robotics and Automation (AIMERA), Wulumuqi, China, 18–19 May 2024; pp. 62–68. [Google Scholar] [CrossRef]
- Radvansky, M.; Kadlecova, A.; Kudelka, M. Utilizing Image Recognition Methods for the Identification and Counting of Live Nematodes in Petri Dishes. In Proceedings of the 2024 25th International Carpathian Control Conference (ICCC), Krynica Zdrój, Poland, 22–24 May 2024; pp. 1–6. [Google Scholar] [CrossRef]
- Lehner, T.; Pum, D.; Rollinger, J.M.; Kirchweger, B. Workflow for Segmentation of Caenorhabditis elegans from Fluorescence Images for the Quantitation of Lipids. Appl. Sci. 2021, 11, 1420. [Google Scholar] [CrossRef]
- Chen, L.; Strauch, M.; Daub, M.; Jansen, M.; Luigs, H.G.; Merhof, D. Instance Segmentation of Nematode Cysts in Microscopic Images of Soil Samples. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 5932–5936. [Google Scholar] [CrossRef]
- Fudickar, S.; Nustede, E.J.; Dreyer, E.; Bornhorst, J. Mask R-CNN Based C. Elegans Detection with a DIY Microscope. Biosensors 2021, 11, 257. [Google Scholar] [CrossRef]
- Chen, L.; Strauch, M.; Daub, M.; Luigs, H.G.; Jansen, M.; Merhof, D. Learning to Segment Fine Structures Under Image-Level Supervision With an Application to Nematode Segmentation. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; pp. 2128–2131. [Google Scholar] [CrossRef]
- Liu, T.; Liu, Y.; Long, R.; Song, Y. Case Segmentation of Nematode Microscopic Image Based on Improved Mask R-CNN. In Proceedings of the CAIBDA 2022: 2nd International Conference on Artificial Intelligence, Big Data and Algorithms, Nanjing, China, 17–19 June 2022; pp. 1–5. [Google Scholar]
- Cheng, Y.; Xu, J.; Xu, L.; Guo, L.; Li, Y. Semantic-guided Nematode Instance Segmentation. In Proceedings of the 2024 5th International Conference on Artificial Intelligence and Electromechanical Automation (AIEA), Shenzhen, China, 14–16 June 2024; pp. 282–286. [Google Scholar] [CrossRef]
- Fudickar, S.; Nustede, E.J.; Dreyer, E.; Bornhorst, J. C. elegans Detection with DIY Microscope Dataset; Zenodo. 2021. Available online: https://doi.org/10.5281/zenodo.5144802 (accessed on 15 August 2025).
- Zong, Z.; Song, G.; Liu, Y. DETRs with Collaborative Hybrid Assignments Training. arXiv 2023, arXiv:2211.12860. [Google Scholar] [CrossRef]
- Cheng, B.; Misra, I.; Schwing, A.G.; Kirillov, A.; Girdhar, R. Masked-attention Mask Transformer for Universal Image Segmentation. arXiv 2022, arXiv:2112.01527. [Google Scholar] [CrossRef]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and Efficient Design for Semantic Segmentation with Transformers. arXiv 2021, arXiv:2105.15203. [Google Scholar] [CrossRef]
- Layana Castro, P.E.; Garví, A.G.; Sánchez-Salmerón, A.J. Automatic segmentation of Caenorhabditis elegans skeletons in worm aggregations using improved U-Net in low-resolution image sequences. Heliyon 2023, 9, e14715. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, W. Automated recognition and analysis of body bending behavior in C. elegans. BMC Bioinform. 2023, 24, 175. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Moher, D. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich Feature Hierarchies for Accurate Object Detection and Semantic Segmentation. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.K.; Girshick, R.B.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. arXiv 2015, arXiv:1506.02640. [Google Scholar]
- Terven, J.; Córdova-Esparza, D.M.; Romero-González, J.A. A Comprehensive Review of YOLO Architectures in Computer Vision: From YOLOv1 to YOLOv8 and YOLO-NAS. Mach. Learn. Knowl. Extr. 2023, 5, 1680–1716. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Ding, M.; Liu, J.; Zhao, Z.; Luo, Y.; Tang, J. WSM-MIL: A weakly supervised segmentation method with multiple instance learning for C elegans image. Phys. Scr. 2024, 99, 065043. [Google Scholar] [CrossRef]
| Database Name | Search String | Results |
|---|---|---|
| Web of Science | TI = (Nematode OR Nematodes OR elegans OR C. elegans) AND TI = (detecting OR detect OR detection OR tracking OR track OR identifying OR identify OR identification OR classifying OR classify OR classification OR segmentation OR segment OR recognizing OR recognize OR recognition OR automating OR automate OR automation OR automated OR sorting OR sort OR counting OR count OR phenotypic OR phenotype OR phenotypes OR deciding OR decide OR decision) | 1012 |
| Agricola | 178 | |
| IEEE Xplore | 226 |
| Category | Data Collected |
|---|---|
| Metadata | Title, Authors, Year, Citations, Publication Type, Location (Country), Institution/University, Journal/Publisher |
| Methods | ML/DL Model Type, Architecture, Computational and Pre-processing Techniques |
| Inference Tasks | Counting, Lifespan, Tracking, Morphological Analysis, Segmentation, Classification |
| Performance Metrics | Accuracy, Precision, Recall, F1-Score, mAP, AP, mAP50, AP@0.5, Avg Mask, IoU, Dice Coefficient |
| Dataset Information | Dataset Size, Availability, Splits (Train, Validation, Test) |
| Nematode Species | Species studied in each paper |
| Code Availability | Publicly available source code |
| Challenges | Identified gaps, limitations, and proposed improvements |
| Ref | Classification | |||||||
|---|---|---|---|---|---|---|---|---|
| and Year | Model Used | Dataset: Size | Train/Test | Class | Acc | Prec | Rec | F1 |
| [48] 2018 | Cust. Siamese | Cust.: 50 k | - | 5 (S) | 0.93 | 0.94 | ||
| Neural Net | ||||||||
| [45] 2019 | Cust. Fully Conv. Nets (FCN) | Open Worm Mov. [49] (Single): 10,476 (V) | - | 5 (S) | 0.81 | 0.53 | ||
| Open Worm Mov. [50] (Multiple): 308 (V) | 95 / 5 | 12 (S) | 0.99 | 0.98 | ||||
| [51] 2021 | Custom CNN Model | Cust.: 390 | 330/60 | 2 | 0.83 | |||
| [7] 2020 | Xception | Cust.: 188 | - | 3 (S) | 0.88 | |||
| Cust.: 234 | - | 3 (S) | 0.69 | |||||
| [46] 2022 | Cust. CNN-WormNet | BioImage Archive (EMBL) [52]: 734 | - | 2 (L) | 0.72 | |||
| [47] 2022 | ResNet101 | Cust.: 257 | 190 / 67 | 19 (S) | 0.94 | |||
| [11] 2022 | EfficientNet | Cust.: 957 | 862 / 95 | 11 (S) | 0.97 | 0.98 | 0.97 | 0.97 |
| [15] 2022 | Cust. CNN-NemaNet | Cust.: 3063 | - | 5 (S) | 0.98 | 0.98 | 0.98 | 0.98 |
| [53] 2022 | Vision Transformer | Cust.: 2769 | 2215 / 554 | 19 (S) | 0.83 | 0.82 | 0.81 | 0.81 |
| [54] 2023 | Inception V3 | I-Nema Dataset [16]: 277 | 227 / 50 | 2 (S) | 0.9 | |||
| [8] 2023 | EfficientNet | Cust.: 957 | - | 11 (S) | 0.96 | 0.97 | 0.96 | 0.96 |
| Biological Inference Task | Proportion of Papers |
|---|---|
| Species Classification | |
| Counting Specimens | |
| Behavior Monitoring | |
| Lifespan Tracking |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jimenez, J.L.; Gandhi, P.; Ayyappan, D.; Gorny, A.; Ye, W.; Lobaton, E. Machine Learning Techniques for Nematode Microscopic Image Analysis: A Systematic Review. AgriEngineering 2025, 7, 356. https://doi.org/10.3390/agriengineering7110356
Jimenez JL, Gandhi P, Ayyappan D, Gorny A, Ye W, Lobaton E. Machine Learning Techniques for Nematode Microscopic Image Analysis: A Systematic Review. AgriEngineering. 2025; 7(11):356. https://doi.org/10.3390/agriengineering7110356
Chicago/Turabian StyleJimenez, Jose Luis, Prem Gandhi, Devadharshini Ayyappan, Adrienne Gorny, Weimin Ye, and Edgar Lobaton. 2025. "Machine Learning Techniques for Nematode Microscopic Image Analysis: A Systematic Review" AgriEngineering 7, no. 11: 356. https://doi.org/10.3390/agriengineering7110356
APA StyleJimenez, J. L., Gandhi, P., Ayyappan, D., Gorny, A., Ye, W., & Lobaton, E. (2025). Machine Learning Techniques for Nematode Microscopic Image Analysis: A Systematic Review. AgriEngineering, 7(11), 356. https://doi.org/10.3390/agriengineering7110356







