Lightweight and Accurate Deep Learning for Strawberry Leaf Disease Recognition: An Interpretable Approach
Abstract
1. Introduction
- Development of a novel lightweight deep learning architecture, inspired by MobileNetV3Small, integrating Inverted Residual blocks with depthwise separable convolutions, Squeeze-and-Excitation modules, and explicit Swish activation to enhance feature extraction efficiency while maintaining low computational cost.
- Significant reduction in model complexity, achieving a smaller number of parameters and reduced model size compared to baseline architectures such as MobileNetV3Small, MobileNetV3Large, and EfficientNetB0, enabling deployment on low-cost mobile and edge devices.
- Interpretability through visual attention mechanisms (e.g., Grad-CAM) to highlight the region’s most influential in the model’s decision-making process.
2. Related Work
3. Materials and Methods
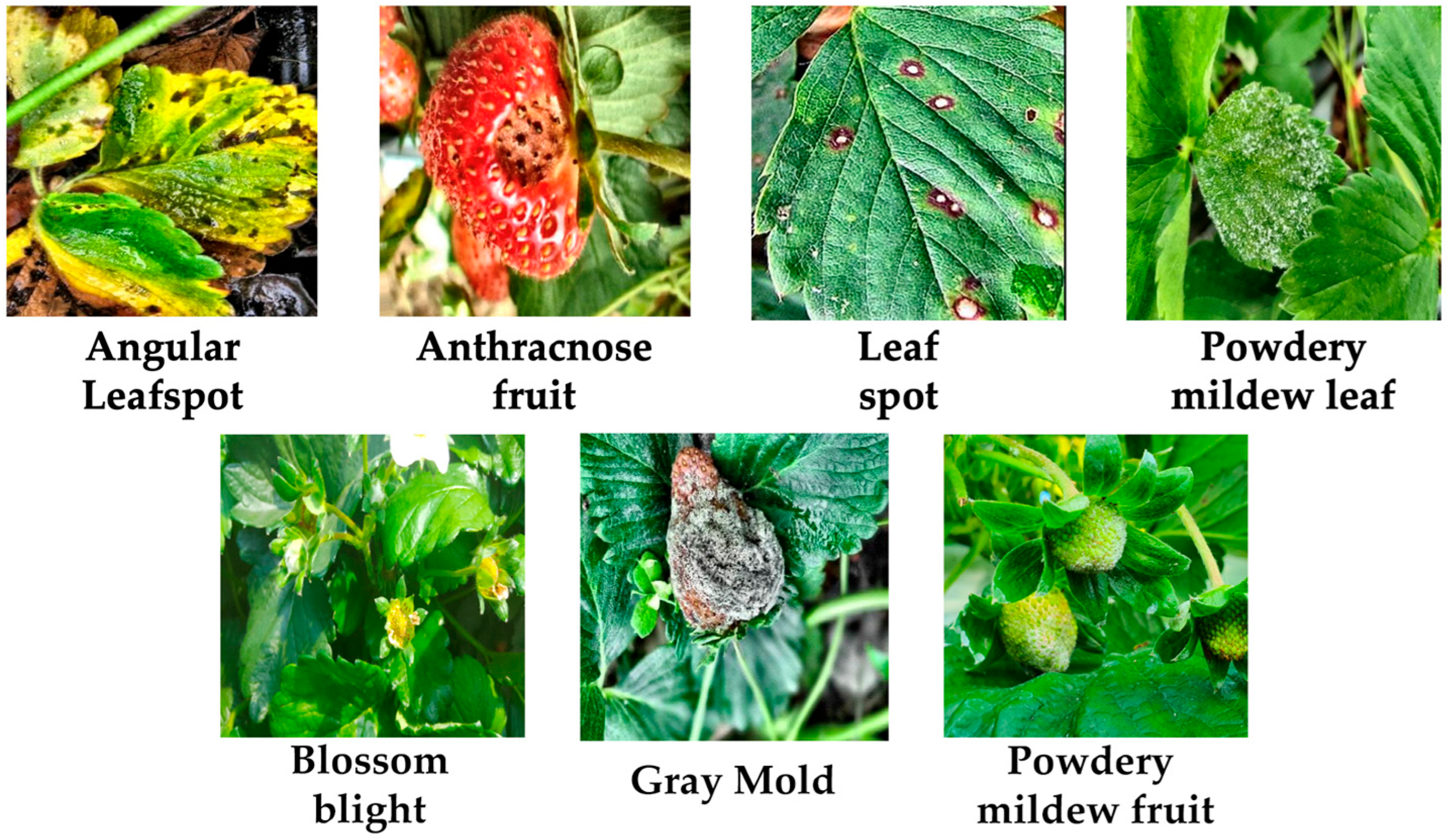
3.1. Image Datasets
3.2. Preprocessing
- Random rotations of up to 25° to simulate different camera angles;
- Zoom scaling within a range of 0.8 to 1.2 to account for variable shooting distances;
- Horizontal and vertical flips to replicate different leaf and fruit orientations in the field;
- Brightness adjustments within the range [0.5, 1.5] to handle variations in natural lighting;
- Nearest-neighbor filling for pixels introduced during geometric transformations.
3.3. Proposed Lightweight CNN Model
- A key efficiency lever, which involves is the use of depthwise separable convolutions in inverted-residual (IR) blocks;
- The ability to minimize architectural redundancy by carefully selecting expansion factors, strides, and block depths to preserve accuracy while reducing parameters and multiply–accumulate operations (MACs) [39].
3.3.1. Architecture of Light-MobileBerryNet
3.3.2. Layers of Light-MobileBerryNet
3.4. Visual Saliency Maps with Grad-CAM
4. Results
4.1. Performance Metrics
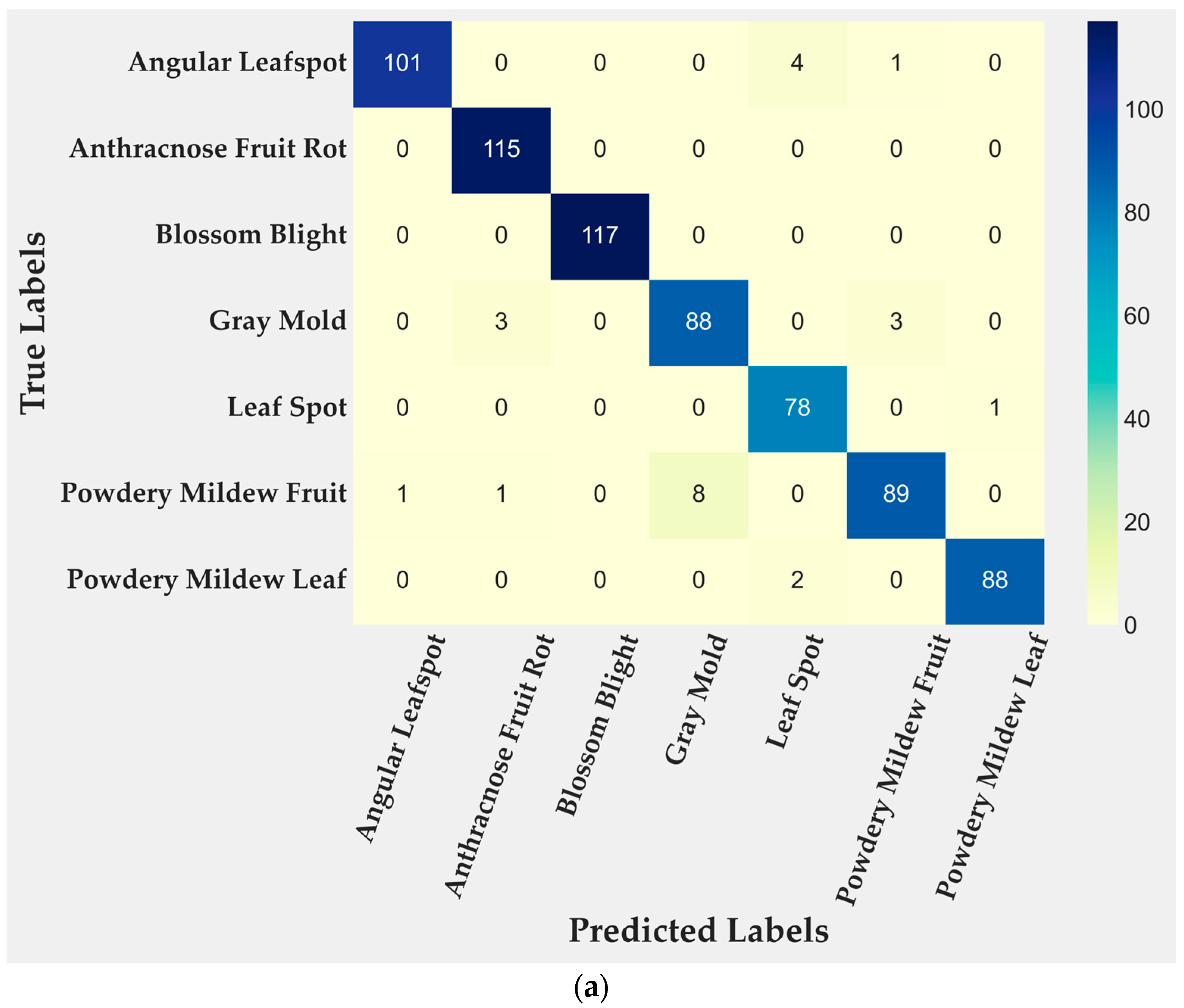
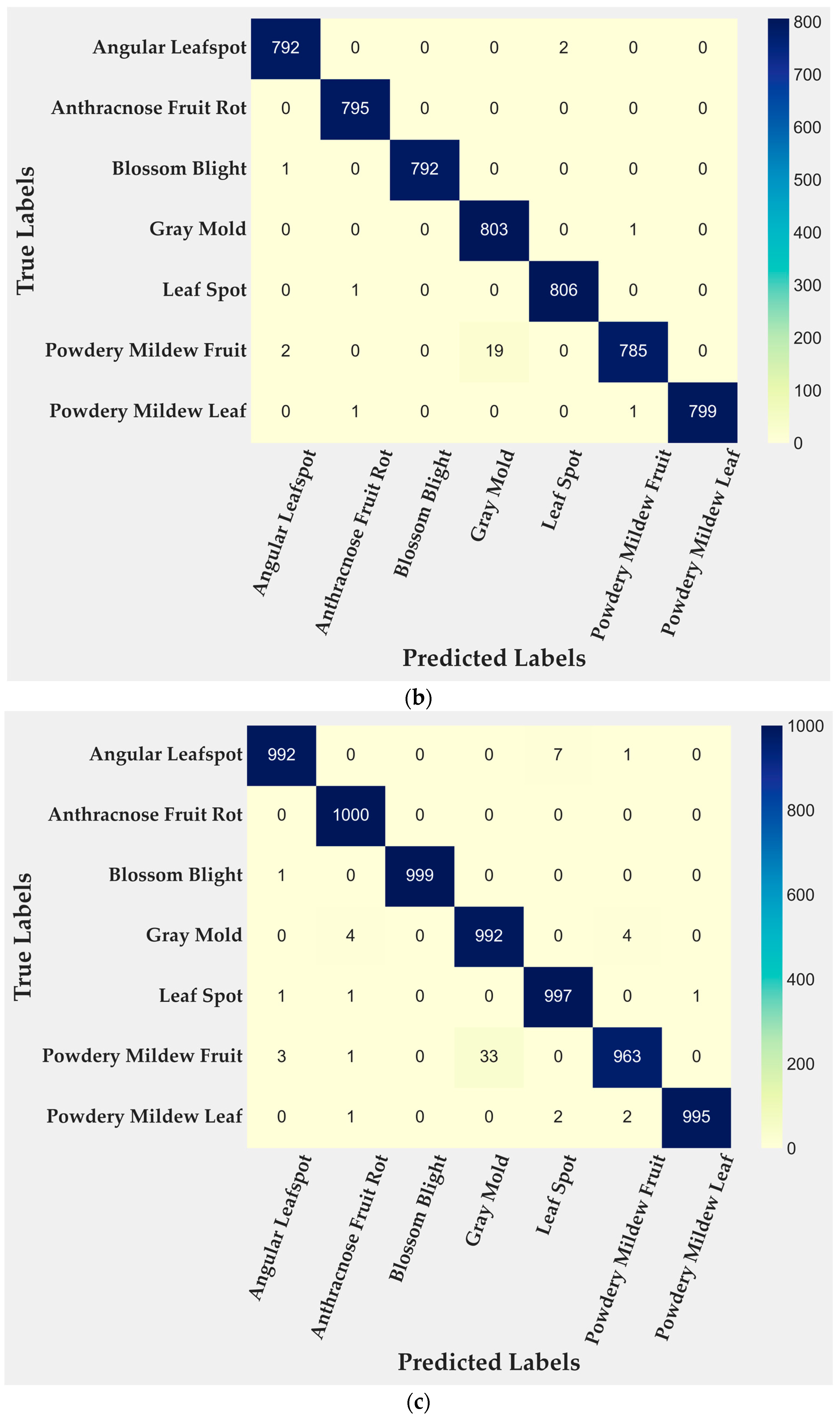
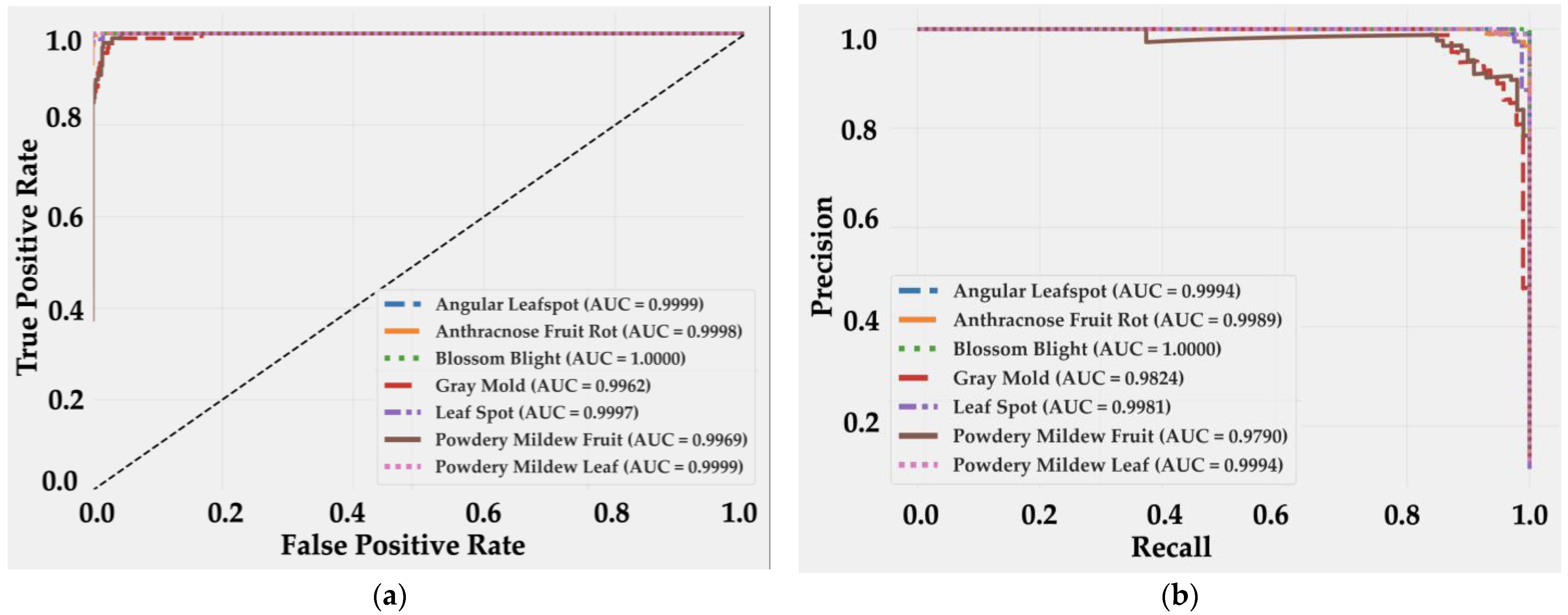
4.2. Model Evaluation
4.3. Ablation Study
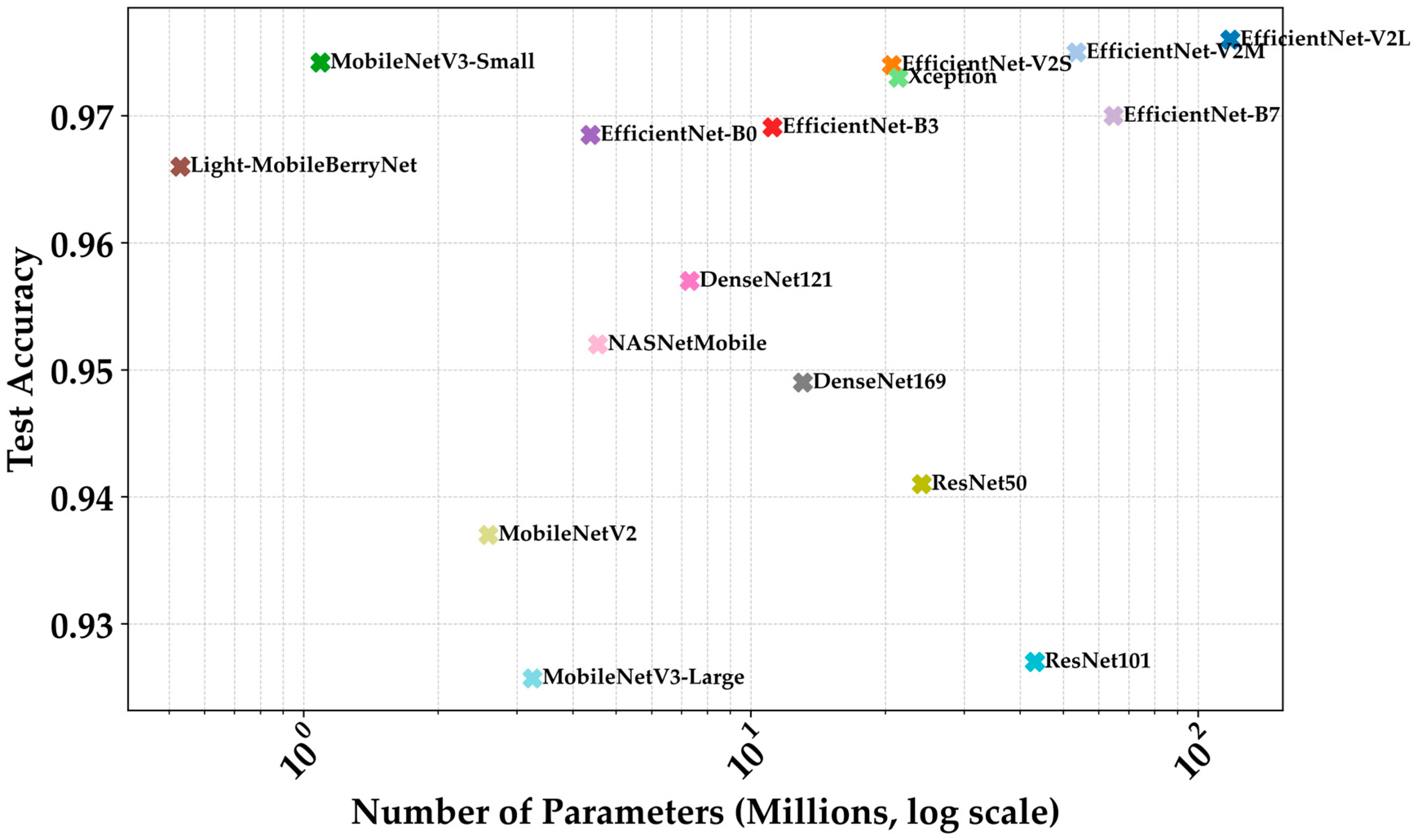
4.4. Comparative Analysis with State-of-the-Art Models
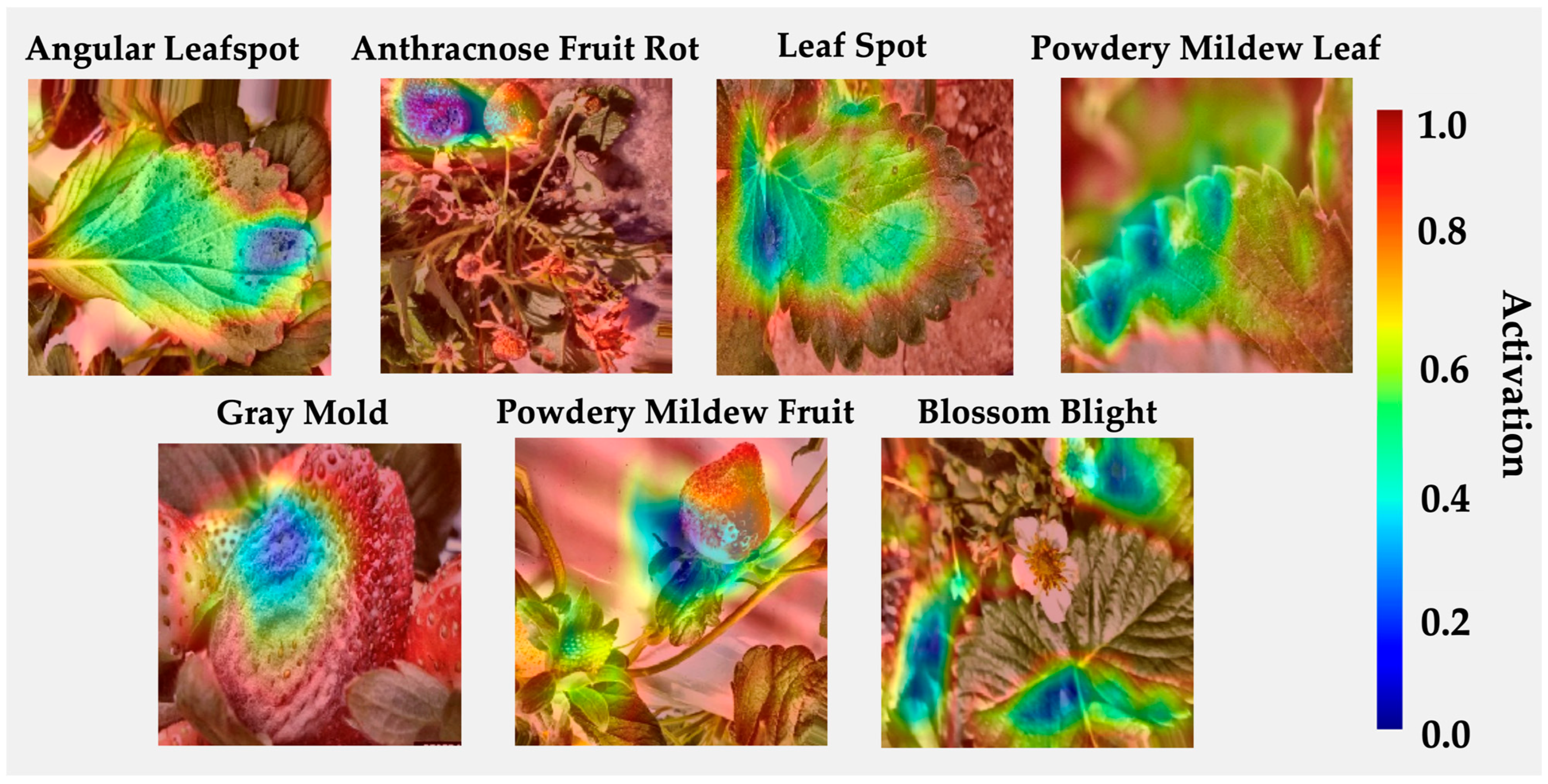
4.5. Visual Saliency Maps for Model Explainability
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| σ | Sigmoid activation function |
| β | Scaling parameter of the Swish activation |
| δ | ReLU activation function |
| λ | Regularization coefficient in the loss function |
| ARS | Agricultural Research Service |
| BN | Batch Normalization |
| CLAHE | Contrast Limited Adaptive Histogram Equalization |
| CNNs | Convolutional Neural Networks |
| ECA | Efficient Channel Attention |
| FN | False negatives |
| FP | False positives |
| FLOPs | Floating–point operations |
| FPS | Frames per second |
| GAP | Global Average Pooling |
| GPU | Graphics Processing Unit |
| H, W | Spatial height and width of the feature map |
| IoU | Intersection-over-Union |
| IR | Inverted Residual |
| K | Kernel size of convolution (e.g., 3 × 3) |
| M | Number of input channels in a convolutional layer |
| MACs | Multiply–Accumulate Operations |
| MCC | Matthews Correlation Coefficient |
| MLP | Multilayer Perceptron |
| N | Number of output channels in a convolutional layer |
| PR | Precision–Recall |
| r | Reduction ratio in the SE (Squeeze-and-Excitation) bottleneck |
| RGB | Red–Green–Blue |
| ROC-AUC | Receiver Operating Characteristic–Area Under the Curve |
| SE | Squeeze-and-Excitation |
| t | Expansion factor in inverted-residual block |
| tM | Expanded number of channels after applying the factor t |
| TB | Terabyte |
| TN | True negatives |
| TP | True positives |
| W1, W2 | Weight matrices in the SE attention mechanism |
| y, ŷ | Ground truth and predicted class probability vectors |
| x | Input activation or feature map |
| YOLO | You Only Look Once |
References
- Karki, S.; Basak, J.K.; Tamrakar, N.; Deb, N.C.; Paudel, B.; Kook, J.H.; Kim, H.T. Strawberry disease detection using transfer learning of deep convolutional neural networks. Sci. Hortic. 2024, 332, 113241. [Google Scholar] [CrossRef]
- Saha, R.; Shaikh, A.; Tarafdar, A.; Majumder, P.; Baidya, A.; Bera, U.K. Deep learning-based comparative study on multi-disease identification in strawberries. In Proceedings of the 2024 IEEE Silchar Subsection Conference (SILCON), Silchar, India, 15–17 November 2024; IEEE: Piscataway, NJ, USA, 2024; pp. 1–6. [Google Scholar]
- Venkatesh, R.; Vijayalakshmi, K.; Geetha, M.; Bhuvanesh, A. Optimized deep belief network for multi-disease classification and severity assessment in strawberries. J. Anim. Plant Sci. 2025, 35, 482–497. [Google Scholar] [CrossRef]
- Jiang, H.; Xue, Z.P.; Guo, Y. Research on plant leaf disease identification based on transfer learning algorithm. J. Phys. Conf. Ser. 2020, 1576, 012023. [Google Scholar] [CrossRef]
- Parameshwari, V.; Brundha, A.; Gomathi, P.; Gopika, R. An intelligent plant leaf syndrome identification derived from pathogen-based deep learning algorithm by interfacing IoT in smart irrigation system. In Proceedings of the 2nd Int. Conf. on Artificial Intelligence and Machine Learning Applications (AIMLA), Tiruchengode, India, 15–16 March 2024; IEEE: Piscataway, NJ, USA, 2024; pp. 1–7. [Google Scholar]
- Kaushik, A.; Attri, S.H.; Chauhan, S.S. Elucidating deep transfer learning approach for early plant disease detection through spot and lesion analysis. In Proceedings of the 3rd International Conference on Disruptive Technologies (ICDT), Greater Noida, India, 7–8 March 2025; IEEE: Piscataway, NJ, USA, 2025; pp. 1145–1150. [Google Scholar]
- Adiga, A.; Gagandeep, N.K.; Prabhu, A.A.; Pai, H.; Kumar, R.A. Comparative analysis on deep learning models for plant disease detection. In Proceedings of the International Conference on Emerging Technologies in Computer Science for Interdisciplinary Applications (ICETCS), Bengaluru, India, 22–23 April 2024; IEEE: Piscataway, NJ, USA, 2024; pp. 1–6. [Google Scholar]
- Wang, W.; Chen, W.; Xu, D.; An, Y. Diseased plant leaves identification by deep transfer learning. In Artificial Intelligence Technologies and Applications; Chen, C., Ed.; IOS Press: Amsterdam, The Netherlands, 2024. [Google Scholar]
- Aybergüler, A.; Arslan, E.; Kayaarma, S.Y. Deep learning-based growth analysis and disease detection in strawberry cultivation. In Proceedings of the 2023 Innovations in Intelligent Systems and Applications Conference (ASYU), Sivas, Turkey, 11–13 October 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–5. [Google Scholar]
- Wang, J.; Li, Z.; Gao, G.; Wang, Y.; Zhao, C.; Bai, H.; Li, Q. BerryNet-lite: A lightweight convolutional neural network for strawberry disease identification. Agriculture 2024, 14, 665. [Google Scholar] [CrossRef]
- Shang, C.; Wu, F.; Wang, M.; Gao, Q. Cattle behavior recognition based on feature fusion under a dual attention mechanism. J. Vis. Commun. Image Represent. 2022, 85, 103524. [Google Scholar] [CrossRef]
- Singh, R.; Sharma, N.; Gupta, R. Strawberry leaf disease detection using transfer learning models. In Proceedings of the IEEE 2nd Int. Conf. on Industrial Electronics: Developments & Applications (ICIDeA), Imphal, India, 29–30 September 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 341–346. [Google Scholar]
- Hu, X.; Xu, T.; Wang, C.; Zhu, H.; Gan, L. Domain generalization method of strawberry disease recognition based on instance whitening and restitution. Smart Agric. 2025, 7, 124–135. [Google Scholar]
- Jiang, Q.; Wu, G.; Tian, C.; Li, N.; Yang, H.; Bai, Y.; Zhang, B. Hyperspectral imaging for early identification of strawberry leaves diseases with machine learning and spectral fingerprint features. Infrared Phys. Technol. 2021, 118, 103898. [Google Scholar] [CrossRef]
- Tumpa, S.B.; Halder, K.K. A comparative study on different transfer learning approaches for identification of plant diseases. In Proceedings of the Int. Conf. on Next-Generation Computing, IoT and Machine Learning (NCIM), Gazipur, Bangladesh, 16–17 June 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–6. [Google Scholar]
- Das, S.; Karna, H.B.; Das, S.; Hazra, R. Disease detection in plant leaves using transfer learning. In Proceedings of the First Int. Conf. on Electronics, Communication and Signal Processing (ICECSP), New Delhi, India, 8–10 August 2024; IEEE: Piscataway, NJ, USA, 2024; pp. 1–6. [Google Scholar]
- Wang, J.; Li, J.; Meng, F. Recognition of strawberry powdery mildew in complex backgrounds: A comparative study of deep learning models. AgriEngineering 2025, 7, 182. [Google Scholar] [CrossRef]
- Pertiwi, S.; Wibowo, D.H.; Widodo, S. Deep learning model for identification of diseases on strawberry (Fragaria sp.) plants. Int. J. Adv. Sci. Eng. Inf. Technol. 2023, 13, 1342–1348. [Google Scholar] [CrossRef]
- Dong, C.; Zhang, Z.; Yue, J.; Zhou, L. Automatic recognition of strawberry diseases and pests using convolutional neural network. Smart Agric. Technol. 2021, 1, 100009. [Google Scholar] [CrossRef]
- Shin, J.; Chang, Y.K.; Heung, B.; Nguyen-Quang, T.; Price, G.W.; Al-Mallahi, A. A deep learning approach for RGB image-based powdery mildew disease detection on strawberry leaves. Comput. Electron. Agric. 2021, 183, 106042. [Google Scholar] [CrossRef]
- Li, G.; Jiao, L.; Chen, P.; Liu, K.; Wang, R.; Dong, S.; Kang, C. Spatial Convolutional Self-Attention-Based Transformer Module for Strawberry Disease Identification under Complex Background. Comput. Electron. Agric. 2023, 212, 108121. [Google Scholar] [CrossRef]
- Aghamohammadesmaeilketabforoosh, K.; Nikan, S.; Antonini, G.; Pearce, J.M. Optimizing Strawberry Disease and Quality Detection with Vision Transformers and Attention-Based Convolutional Neural Networks. Foods 2024, 13, 1869. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Tran, T.D.; Nguyen, T.T.; Pham, N.M.; Nguyen Ly, P.H.; Luong, H.H. Strawberry Disease Identification with Vision Transformer-Based Models. Multimed. Tools Appl. 2024, 83, 73101–73126. [Google Scholar] [CrossRef]
- Chen, M.; Zou, W.; Niu, X.; Fan, P.; Liu, H.; Li, C.; Zhai, C. Improved YOLOv8-based segmentation method for strawberry leaf and powdery mildew lesions in natural backgrounds. Agronomy 2025, 15, 525. [Google Scholar] [CrossRef]
- Alhwaiti, Y.; Khan, M.; Asim, M.; Siddiqi, M.H.; Ishaq, M.; Alruwaili, M. Leveraging YOLO deep learning models to enhance plant disease identification. Sci. Rep. 2025, 15, 7969. [Google Scholar] [CrossRef]
- Kim, H.; Kim, D. Deep-learning-based strawberry leaf pest classification for sustainable smart farms. Sustainability 2023, 15, 7931. [Google Scholar] [CrossRef]
- Ou, Y.; Yan, J.; Liang, Z.; Zhang, B. Hyperspectral imaging combined with deep learning for the early detection of strawberry leaf gray mold disease. Agronomy 2024, 14, 2694. [Google Scholar] [CrossRef]
- Xu, M.; Yoon, S.; Jeong, Y.; Park, D.S. Transfer learning for versatile plant disease recognition with limited data. Front. Plant Sci. 2022, 13, 1010981. [Google Scholar] [CrossRef]
- Ochoa-Ornelas, R.; Gudiño-Ochoa, A.; García-Rodríguez, J.A.; Uribe-Toscano, S. A robust transfer learning approach with histopathological images for lung and colon cancer detection using EfficientNetB3. Healthc. Anal. 2025, 7, 100391. [Google Scholar] [CrossRef]
- Ochoa-Ornelas, R.; Gudiño-Ochoa, A.; García-Rodríguez, J.A.; Uribe-Toscano, S. Enhancing early lung cancer detection with MobileNet: A comprehensive transfer learning approach. Franklin Open 2025, 10, 100222. [Google Scholar] [CrossRef]
- Ochoa-Ornelas, R.; Gudiño-Ochoa, A.; García-Rodríguez, J.A. A hybrid deep learning and machine learning approach with Mobile-EfficientNet and Grey Wolf Optimizer for lung and colon cancer histopathology classification. Cancers 2024, 16, 3791. [Google Scholar] [CrossRef]
- Afzaal, U.; Bhattarai, B.; Pandeya, Y.R.; Lee, J. An instance segmentation model for strawberry diseases based on Mask R-CNN. Sensors 2021, 21, 6565. [Google Scholar] [CrossRef]
- Narla, V.L.; Suresh, G.; Rao, C.S.; Awadh, M.A.; Hasan, N. A multimodal approach with firefly-based CLAHE and multiscale fusion for enhancing underwater images. Sci. Rep. 2024, 14, 27588. [Google Scholar] [CrossRef]
- Islam, T.; Hafiz, M.S.; Jim, J.R.; Kabir, M.M.; Mridha, M.F. A systematic review of deep learning data augmentation in medical imaging: Recent advances and future research directions. Healthc. Anal. 2024, 5, 100340. [Google Scholar] [CrossRef]
- Gao, X.; Xiao, Z.; Deng, Z. High accuracy food image classification via vision transformer with data augmentation and feature augmentation. J. Food Eng. 2024, 365, 111833. [Google Scholar] [CrossRef]
- Sunkari, S.; Sangam, A.; Raman, R.; Rajalakshmi, R. A refined ResNet18 architecture with Swish activation function for diabetic retinopathy classification. Biomed. Signal Process. Control 2024, 88, 105630. [Google Scholar] [CrossRef]
- Javanmardi, S.; Ashtiani, S.H.M. AI-Driven Deep Learning Framework for Shelf Life Prediction of Edible Mushrooms. Postharvest Biol. Technol. 2025, 222, 113396. [Google Scholar] [CrossRef]
- Ramachandran, P.; Zoph, B.; Le, Q.V. Searching for Activation Functions. arXiv 2017, arXiv:1710.05941. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; IEEE: Piscataway, NJ, USA, 2024; pp. 7132–7141. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; IEEE: Piscataway, NJ, USA, 2024; pp. 4510–4520. [Google Scholar]
- Tummala, S.; Kadry, S.; Nadeem, A.; Rauf, H.T.; Gul, N. An explainable classification method based on complex scaling in histopathology images for lung and colon cancer. Diagnostics 2023, 13, 1594. [Google Scholar] [CrossRef]
- Nobel, S.N.; Afroj, M.; Kabir, M.M.; Mridha, M.F. Development of a cutting-edge ensemble pipeline for rapid and accurate diagnosis of plant leaf diseases. Artif. Intell. Agric. 2024, 14, 56–72. [Google Scholar] [CrossRef]
- Mittapalli, P.S.; Tagore, M.R.N.; Reddy, P.A.; Kande, G.B.; Reddy, Y.M. Deep Learning-Based Real-Time Object Detection on Jetson Nano Embedded GPU. In Microelectronics, Circuits and Systems: Select Proceedings of Micro2021; Springer Nature: Singapore, 2023; pp. 511–521. [Google Scholar]
- Mayo, D.; Cummings, J.; Lin, X.; Gutfreund, D.; Katz, B.; Barbu, A. How Hard Are Computer Vision Datasets? Calibrating Dataset Difficulty to Viewing Time. Adv. Neural Inf. Process. Syst. 2023, 36, 11008–11036. [Google Scholar]
- Ochoa-Ornelas, R.; Gudiño-Ochoa, A.; García-Rodríguez, J.A.; Uribe-Toscano, S. Lung and colon cancer detection with InceptionResNetV2: A transfer learning approach. J. Res. Dev. 2024, 10, e11025113. [Google Scholar] [CrossRef]
- Gudiño-Ochoa, A.; García-Rodríguez, J.A.; Ochoa-Ornelas, R.; Ruiz-Velazquez, E.; Uribe-Toscano, S.; Cuevas-Chávez, J.I.; Sánchez-Arias, D.A. Non-invasive multiclass diabetes classification using breath biomarkers and machine learning with explainable AI. Diabetology 2025, 6, 51. [Google Scholar] [CrossRef]
- Tamrakar, N.; Paudel, B.; Karki, S.; Deb, N.C.; Arulmozhi, E.; Kook, J.H.; Kim, H.T. Peduncle Detection of Ripe Strawberry to Localize Picking Point Using DF-Mask R-CNN and Monocular Depth. IEEE Access 2025, 13, 1–12. [Google Scholar] [CrossRef]
- Gudiño-Ochoa, A.; García-Rodríguez, J.A.; Ochoa-Ornelas, R.; Cuevas-Chávez, J.I.; Sánchez-Arias, D.A. Noninvasive Diabetes Detection through Human Breath Using TinyML-Powered E-Nose. Sensors 2024, 24, 1294. [Google Scholar] [CrossRef] [PubMed]
- Samanta, R.; Saha, B.; Ghosh, S.K. TinyML-on-the-Fly: Real-Time Low-Power and Low-Cost MCU-Embedded On-Device Computer Vision for Aerial Image Classification. In Proceedings of the 2024 IEEE Space, Aerospace and Defence Conference (SPACE), Oxford, UK, 8–10 July 2024; IEEE: Piscataway, NJ, USA, 2024; pp. 194–198. [Google Scholar]


| Stage | Operator | Exp. t | Kernel | Stride | SE | |
|---|---|---|---|---|---|---|
| Stem | Conv-BN-ReLU | - | 716 | 2 | - | |
| B1 | IR (Swish) | 1 | 1616 | 1 | ✓ | |
| B2 | IR (Swish) | 6 | 1624 | 2 | ✓ | |
| B3 | IR (Swish) | 6 | 2424 | 1 | ✓ | |
| B4 | IR (Swish) | 6 | 2432 | 2 | ✓ | |
| B5 | IR (Swish) | 6 | 3232 | 1 | ✓ | |
| B6 | IR (Swish) | 6 | 3264 | 2 | ✓ | |
| B7 | IR (Swish) | 6 | 6464 | 1 | ✓ | |
| B8 | IR (Swish) | 6 | 6496 | 1 | ✓ | |
| Head | Conv1 × 1-Swish | - | 96576 | 1 | - | |
| GAP + FC + Drop | - | 576128 | - | - | - | |
| Classifier (Softmax) | - | 1287 | - | - | - |
| Dataset | Class | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Test | Blossom Blight | 1.000 | 1.000 | 1.000 |
| Gray Mold | 0.917 | 0.936 | 0.926 | |
| Powdery Mildew Fruit | 0.957 | 0.898 | 0.927 | |
| Angular Leafspot | 0.990 | 0.952 | 0.971 | |
| Anthracnose Fruit Rot | 0.967 | 1.000 | 0.983 | |
| Powdery Mildew Leaf | 0.989 | 0.978 | 0.983 | |
| Leaf Spot | 0.929 | 0.987 | 0.957 | |
| Train | Blossom Blight | 1.000 | 0.998 | 0.999 |
| Gray Mold | 0.977 | 0.998 | 0.987 | |
| Powdery Mildew Fruit | 0.997 | 0.973 | 0.985 | |
| Angular Leafspot | 0.996 | 0.997 | 0.996 | |
| Anthracnose Fruit Rot | 0.997 | 1.000 | 0.998 | |
| Powdery Mildew Leaf | 1.000 | 0.997 | 0.998 | |
| Leaf Spot | 0.997 | 0.998 | 0.998 | |
| All | Blossom Blight | 1.000 | 0.999 | 0.999 |
| Gray Mold | 0.967 | 0.992 | 0.979 | |
| Powdery Mildew Fruit | 0.992 | 0.963 | 0.977 | |
| Angular Leafspot | 0.994 | 0.992 | 0.993 | |
| Anthracnose Fruit Rot | 0.993 | 1.000 | 0.996 | |
| Powdery Mildew Leaf | 0.998 | 0.995 | 0.996 | |
| Leaf Spot | 0.991 | 0.997 | 0.994 |
| Dataset | Accuracy | Precision | Recall | F1-Score | MCC |
|---|---|---|---|---|---|
| Test | 0.966 | 0.966 | 0.966 | 0.966 | 0.960 |
| Train | 0.995 | 0.995 | 0.995 | 0.994 | 0.994 |
| All | 0.991 | 0.991 | 0.991 | 0.991 | 0.989 |
| Architecture | Params (M) | Size (MB) | MACs (M) | FLOPs (M) | FPS | Latency (ms) | Accuracy | Precision | Recall | F1-Score | MCC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Proposed (Swish, 8 IR blocks) | 0.532 | 2.03 | 143.97 | 287.94 | 131.85 | 7.58 | 0.966 | 0.966 | 0.966 | 0.966 | 0.960 |
| ReLU instead of Swish | 0.533 | 2.03 | 139.89 | 279.78 | 157.43 | 6.35 | 0.901 | 0.913 | 0.901 | 0.903 | 0.886 |
| 7 IR blocks instead of 8 | 0.372 | 1.42 | 127.16 | 254.33 | 124.75 | 8.02 | 0.957 | 0.961 | 0.957 | 0.958 | 0.950 |
| Expansion factor 4 instead of 6 | 0.353 | 1.35 | 91.21 | 182.43 | 159.26 | 6.28 | 0.941 | 0.944 | 0.941 | 0.942 | 0.934 |
| Model | Params (M) | Size (MB) | MACs (M) | FLOPs (M) | FPS | Latency (ms) | Accuracy | Precision | Recall | F1-Score | MCC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EfficientNet-V2L | 118.08 | 450.44 | 12,309.34 | 24,618.68 | ≈6.5 | ≈153.8 | 0.976 | 0.977 | 0.976 | 0.976 | 0.967 |
| EfficientNet-V2M | 53.48 | 204.03 | 5406.72 | 10,813.44 | ≈12.3 | ≈81.3 | 0.975 | 0.976 | 0.975 | 0.975 | 0.966 |
| EfficientNet-V2S | 20.67 | 78.83 | 2877.19 | 5754.38 | ≈18.9 | ≈52.9 | 0.974 | 0.975 | 0.974 | 0.974 | 0.964 |
| MobileNetV3-Small | 1.09 | 4.16 | 59.60 | 119.21 | 205.29 | 4.87 | 0.9742 | 0.9754 | 0.9742 | 0.9744 | 0.970 |
| Xception | 21.39 | 81.62 | 4569.02 | 9138.04 | ≈13.3 | ≈75.0 | 0.973 | 0.974 | 0.972 | 0.972 | 0.962 |
| EfficientNet-B3 | 11.18 | 42.66 | 992.99 | 1985.98 | 219.21 | 30.02 | 0.969 | 0.969 | 0.969 | 0.969 | 0.964 |
| EfficientNet-B0 | 4.38 | 16.72 | 401.46 | 802.93 | 215.98 | 25.76 | 0.968 | 0.968 | 0.968 | 0.965 | 0.963 |
| EfficientNet-B7 | 64.76 | 247.06 | 5264.81 | 10,529.62 | ≈11.5 | ≈86.9 | 0.970 | 0.977 | 0.969 | 0.969 | 0.962 |
| Light- MobileBerryNet | 0.53 | 2.03 | 143.97 | 287.94 | 131.85 | 7.58 | 0.966 | 0.966 | 0.967 | 0.969 | 0.960 |
| DenseNet121 | 7.30 | 27.87 | 2851.11 | 5702.23 | ≈20.1 | ≈49.7 | 0.957 | 0.959 | 0.956 | 0.957 | 0.943 |
| NASNetMobile | 4.55 | 17.34 | 573.88 | 1147.77 | ≈85.5 | ≈11.7 | 0.952 | 0.955 | 0.952 | 0.953 | 0.939 |
| DenseNet169 | 13.08 | 49.88 | 3380.19 | 6760.38 | ≈17.0 | ≈58.8 | 0.949 | 0.951 | 0.949 | 0.949 | 0.935 |
| ResNet50 | 24.12 | 92.02 | 3877.57 | 7755.15 | ≈14.8 | ≈67.6 | 0.941 | 0.945 | 0.941 | 0.941 | 0.921 |
| MobileNetV2 | 2.59 | 9.89 | 307.59 | 615.19 | 210.31 | 13.02 | 0.937 | 0.940 | 0.937 | 0.937 | 0.926 |
| ResNet101 | 43.19 | 164.76 | 7599.18 | 15,198.36 | ≈9.8 | ≈101.9 | 0.927 | 0.930 | 0.926 | 0.926 | 0.905 |
| MobileNetV3-Large | 3.25 | 12.39 | 224.25 | 448.50 | 124.56 | 8.03 | 0.925 | 0.930 | 0.925 | 0.925 | 0.913 |
| Level | Infection Severity | IoU (Mean) | Dice (Mean) | Pointing Game (%) | Energy In (%) | IoU ≥ 0.30 (%) | IoU ≥ 0.50 (%) | Images |
|---|---|---|---|---|---|---|---|---|
| Level 1 | Low-Mid | 0.366 | 0.502 | 58.25 | 47.34 | 62.14 | 30.58 | 206 |
| Level 2 | High | 0.320 | 0.432 | 56.42 | 44.63 | 54.75 | 31.66 | 537 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ochoa-Ornelas, R.; Gudiño-Ochoa, A.; Rodríguez González, A.Y.; Trujillo, L.; Fajardo-Delgado, D.; Puga-Nathal, K.L. Lightweight and Accurate Deep Learning for Strawberry Leaf Disease Recognition: An Interpretable Approach. AgriEngineering 2025, 7, 355. https://doi.org/10.3390/agriengineering7100355
Ochoa-Ornelas R, Gudiño-Ochoa A, Rodríguez González AY, Trujillo L, Fajardo-Delgado D, Puga-Nathal KL. Lightweight and Accurate Deep Learning for Strawberry Leaf Disease Recognition: An Interpretable Approach. AgriEngineering. 2025; 7(10):355. https://doi.org/10.3390/agriengineering7100355
Chicago/Turabian StyleOchoa-Ornelas, Raquel, Alberto Gudiño-Ochoa, Ansel Y. Rodríguez González, Leonardo Trujillo, Daniel Fajardo-Delgado, and Karla Liliana Puga-Nathal. 2025. "Lightweight and Accurate Deep Learning for Strawberry Leaf Disease Recognition: An Interpretable Approach" AgriEngineering 7, no. 10: 355. https://doi.org/10.3390/agriengineering7100355
APA StyleOchoa-Ornelas, R., Gudiño-Ochoa, A., Rodríguez González, A. Y., Trujillo, L., Fajardo-Delgado, D., & Puga-Nathal, K. L. (2025). Lightweight and Accurate Deep Learning for Strawberry Leaf Disease Recognition: An Interpretable Approach. AgriEngineering, 7(10), 355. https://doi.org/10.3390/agriengineering7100355











