Two-Stage Ensemble Deep Learning Model for Precise Leaf Abnormality Detection in Centella asiatica
Abstract
1. Introduction
2. Related Literature
2.1. Contemporary Methods for Detecting Leaf Diseases in CAU: Limitations, Challenges, and Our Approach
2.2. Deep Learning-Based Ensemble Techniques in Leaf Disease Detection: Focused Approaches and Advancements
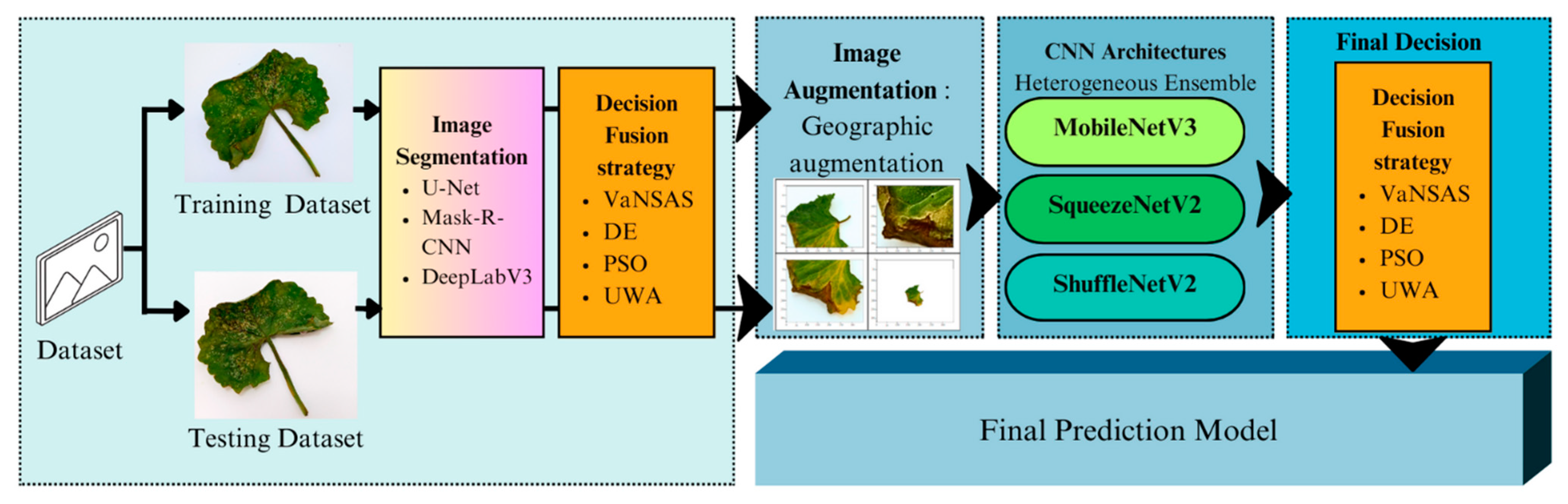
3. Research Methodology
3.1. Dataset Preparation
3.2. Model Building Classification
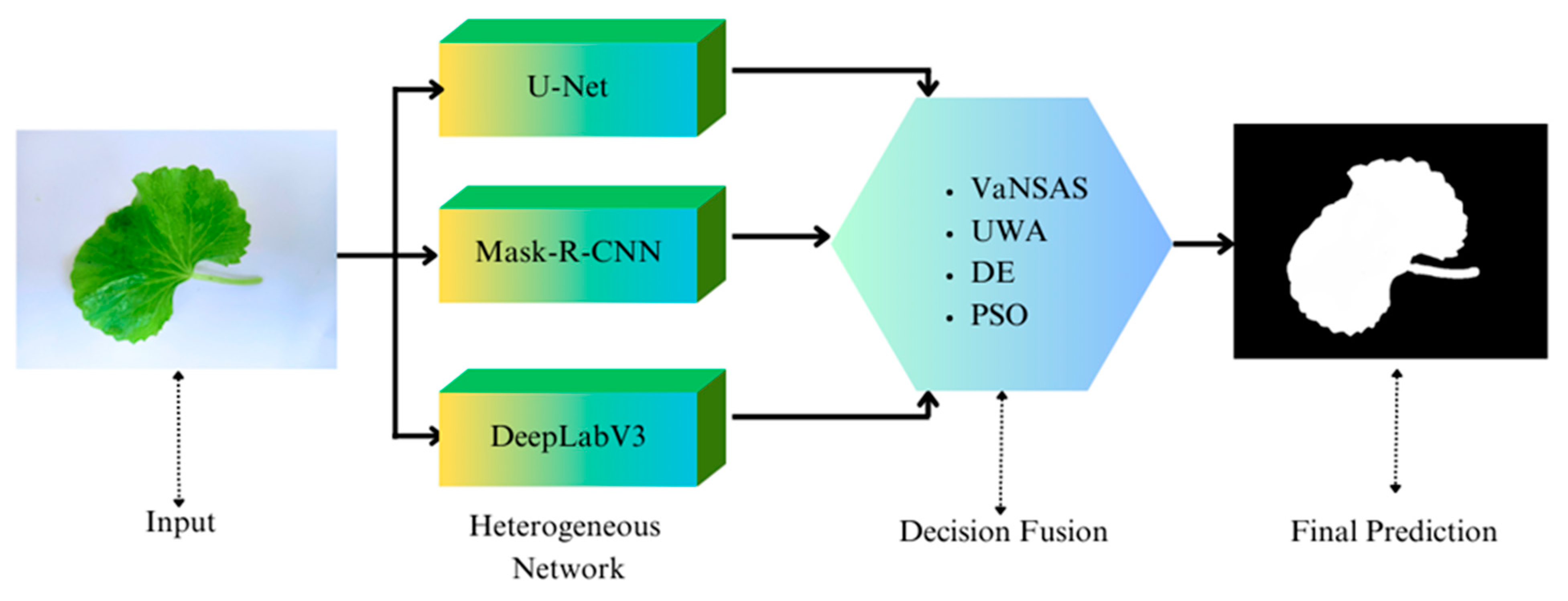
3.2.1. Image Segmentation
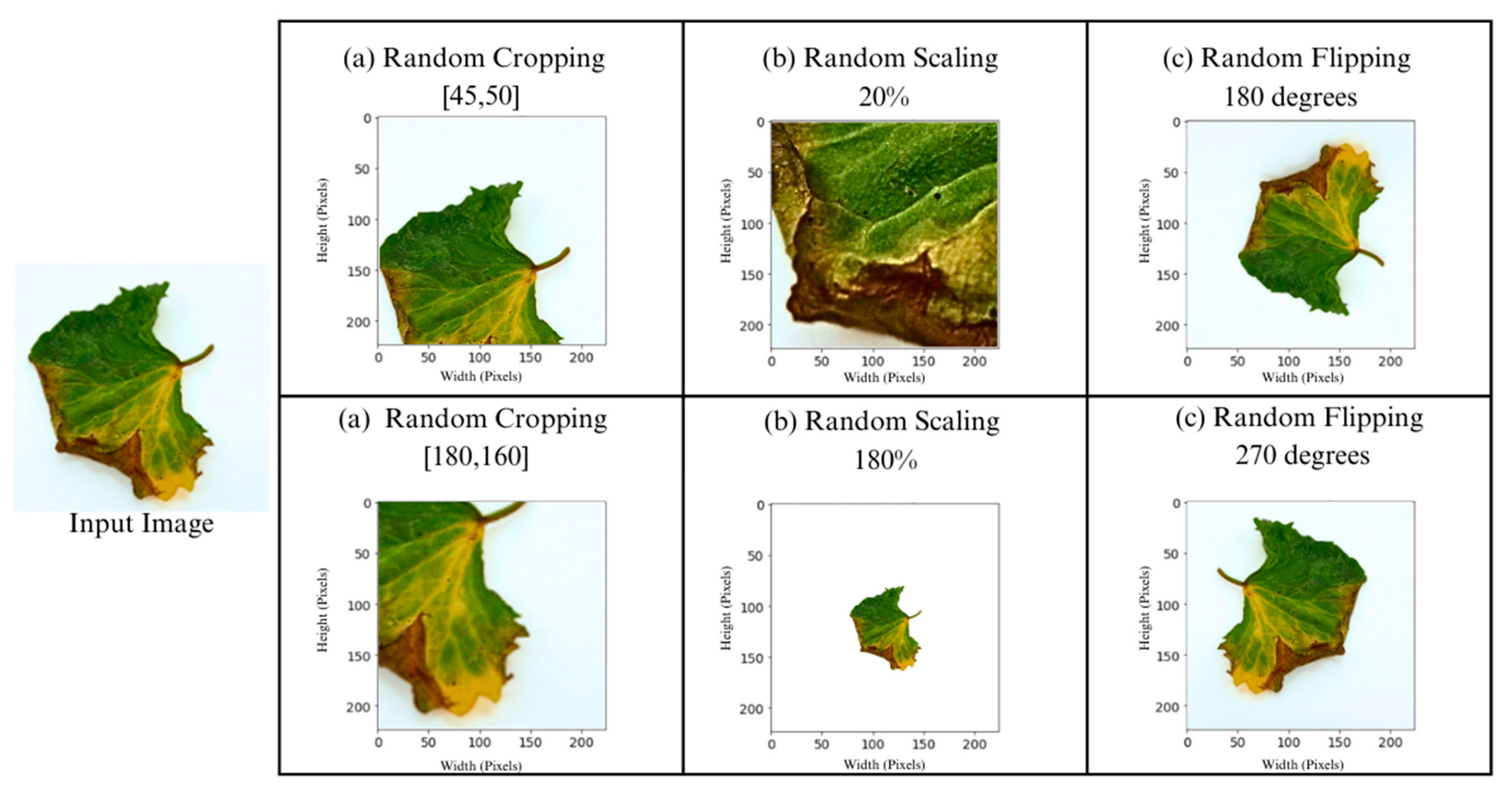
3.2.2. Image Augmentation
3.2.3. CNN Architectures
3.2.4. Optimizing of the Decision Fusion Strategy
3.3. The Initial Tracks Generation
3.4. Track Touring Process
3.5. Probability Update for the IB
3.6. The Comparison Methods
3.7. Evaluation of Performance Metrics
4. Computational Framework and Result
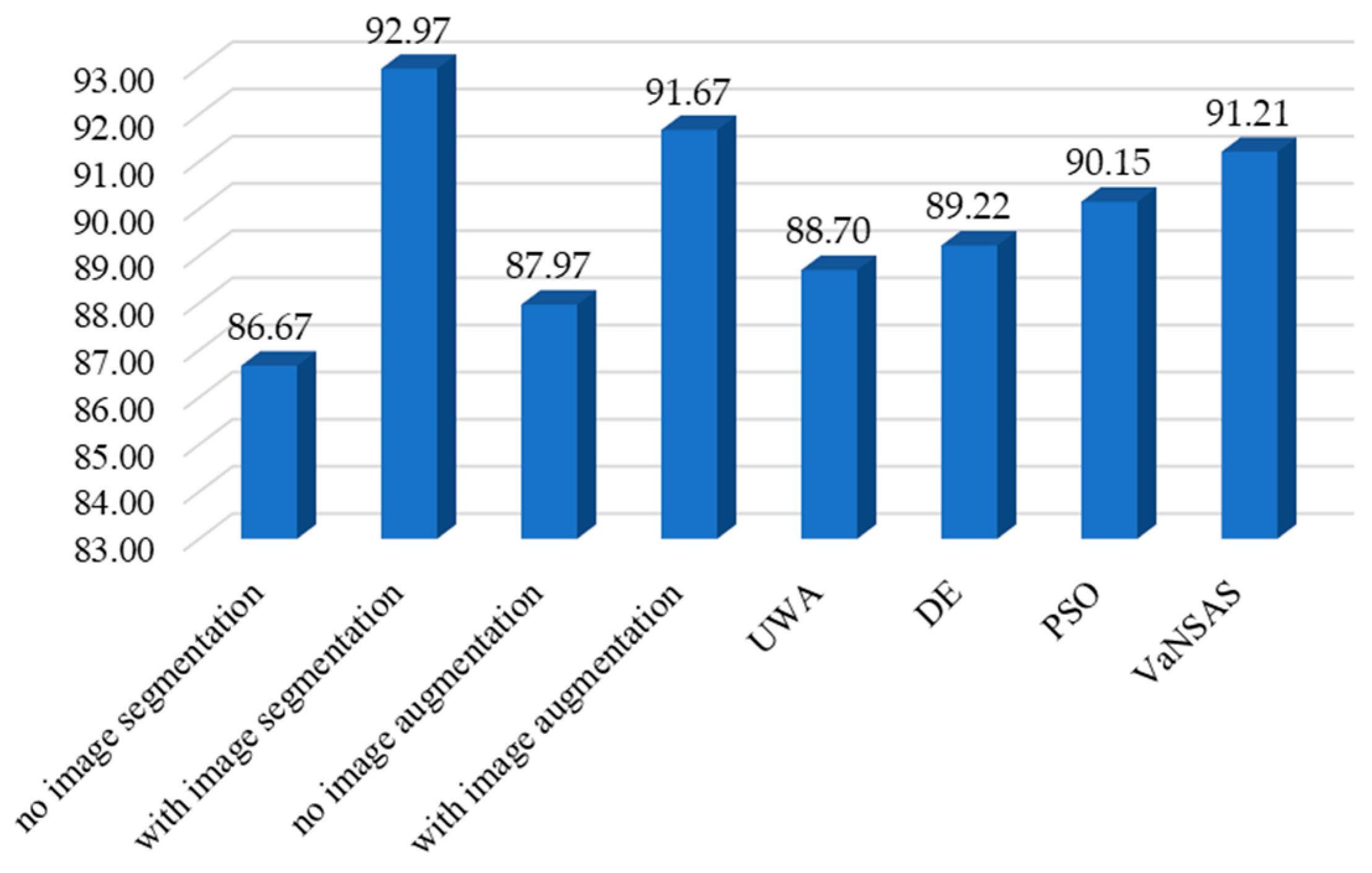
4.1. Optimizing the Model Entities
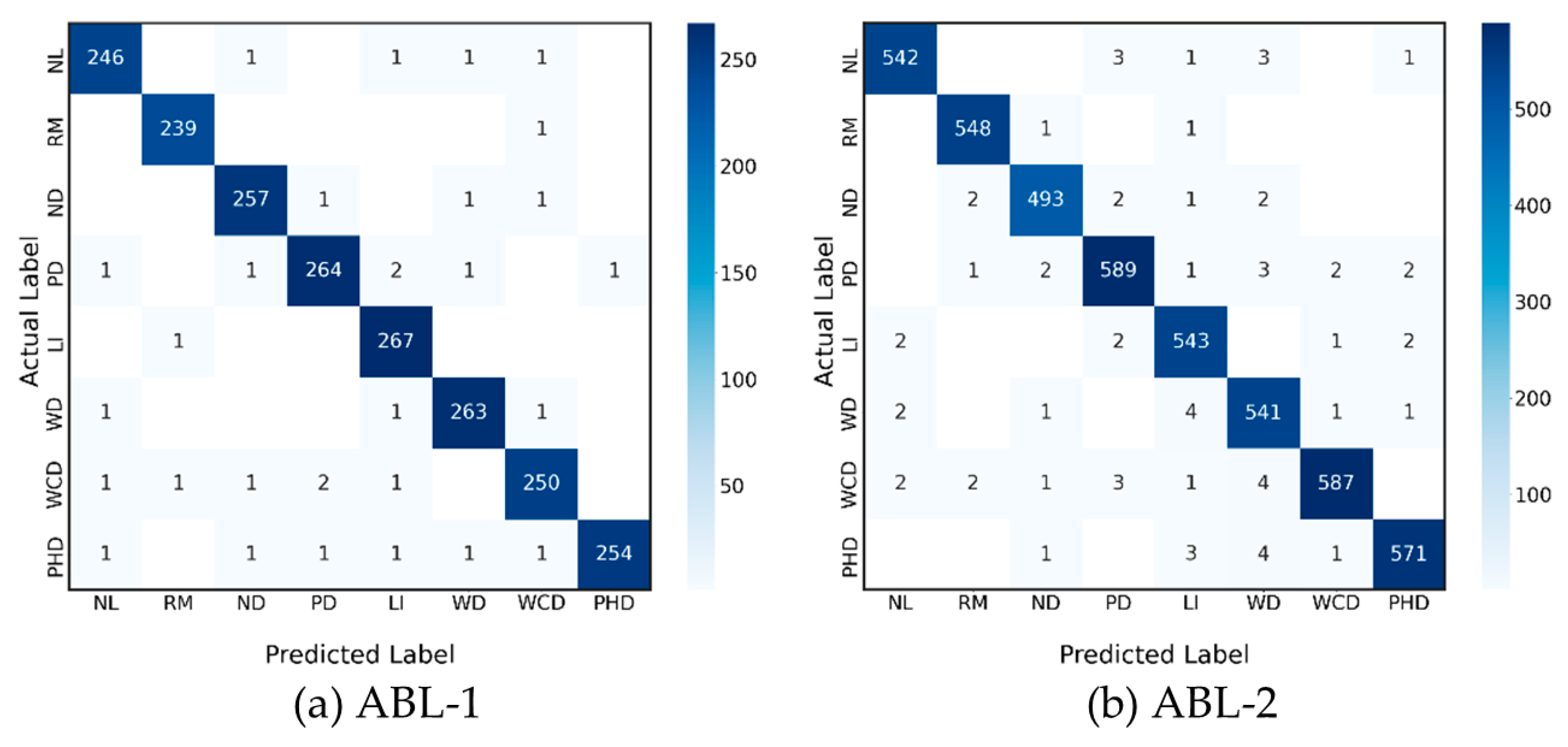
4.2. Comparison of Optimal Proposed Model with the State-of-Art Methods (ABL-1)
4.3. Comparison with the Unseen Dataset (ABL-2)
5. Discussion and Implications
5.1. Enhancing Leaf Abnormality Detection in C. asiatica: An Ensemble Deep Learning Approach
5.2. Advancing Leaf Disease Classification in CAU: A Meta-Learner Guided Ensemble Deep Learning Model
5.3. In-Depth Analysis of Model Performance and Comparative Evaluation
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gleiser, G.; Leme Da Cunha, N.; Sáez, A.; Aizen, M.A. Ecological Correlates of Crop Yield Growth and Interannual Yield Variation at a Global Scale. Web Ecol. 2021, 21, 15–43. [Google Scholar] [CrossRef]
- Hemalatha, R.; Alagar, M.; Rameshbabu, P.; Azhagu Parvathi, A.; Hepzi Pramila Devamani, R. Development of Medicinal Plant (Centella Asiatica–Gotu Kola) Based Proton Conducting Polymer Electrolytes for Electrochemical Device Applications. Mater. Today Proc. 2023, 81, 330–335. [Google Scholar] [CrossRef]
- Ranjith, G.P.; Jisha, S.; Hemanthakumar, A.S.; Saji, C.V.; Shenoi, R.A.; Sabu, K.K. Impact of Potential Stimulants on Asiaticoside and Madecassoside Levels and Expression of Triterpenoid-Related Genes in Axenic Shoot Cultures of Centella asiatica (L.) Urb. Phytochemistry 2021, 186, 112735. [Google Scholar] [CrossRef]
- Gupta, S.; Bhatt, P.; Chaturvedi, P. Determination and Quantification of Asiaticoside in Endophytic Fungus from Centella asiatica (L.) Urban. World J. Microbiol. Biotechnol. 2018, 34, 111. [Google Scholar] [CrossRef]
- Trivedi, N.K.; Gautam, V.; Anand, A.; Aljahdali, H.M.; Villar, S.G.; Anand, D.; Goyal, N.; Kadry, S. Early Detection and Classification of Tomato Leaf Disease Using High-Performance Deep Neural Network. Sensors 2021, 21, 7987. [Google Scholar] [CrossRef] [PubMed]
- Spadaro, D.; Agustí, N.; Ortega, S.F.; Hurtado Ruiz, M.A. Diagnostics and Identification of Diseases, Insects and Mites. In Integrated Pest and Disease Management in Greenhouse Crops; Gullino, M., Albajes, R., Nicot, P., Eds.; Springer: Cham, Switzerland, 2020; pp. 231–258. [Google Scholar] [CrossRef]
- Raina, S.; Gupta, A. A Study on Various Techniques for Plant Leaf Disease Detection Using Leaf Image. In Proceedings of the 2021 International Conference on Artificial Intelligence and Smart Systems (ICAIS), Coimbatore, India, 25 March 2021; pp. 900–905. [Google Scholar]
- Bondre, S.; Sharma, A.K. Review on Leaf Diseases Detection Using Deep Learning. In Proceedings of the 2021 Second International Conference on Electronics and Sustainable Communication Systems (ICESC), Coimbatore, India, 4 August 2021; pp. 1455–1461. [Google Scholar]
- Maheswaran, S.; Sathesh, S.; Rithika, P.; Shafiq, I.M.; Nandita, S.; Gomathi, R.D. Detection and Classification of Paddy Leaf Diseases Using Deep Learning (CNN). In Proceedings of the Computer, Communication, and Signal Processing; Neuhold, E.J., Fernando, X., Lu, J., Piramuthu, S., Chandrabose, A., Eds.; Springer International Publishing: Cham, Switzerland, 2022; Volume 651, pp. 60–74. [Google Scholar]
- Sethanan, K.; Pitakaso, R.; Srichok, T.; Khonjun, S.; Thannipat, P.; Wanram, S.; Boonmee, C.; Gonwirat, S.; Enkvetchakul, P.; Kaewta, C.; et al. Double AMIS-Ensemble Deep Learning for Skin Cancer Classification. Expert Syst. Appl. 2023, 234, 121047. [Google Scholar] [CrossRef]
- Triki, A.; Bouaziz, B.; Gaikwad, J.; Mahdi, W. Deep Leaf: Mask R-CNN Based Leaf Detection and Segmentation from Digitized Herbarium Specimen Images. Pattern Recognit. Lett. 2021, 150, 76–83. [Google Scholar] [CrossRef]
- Wang, C.; Du, P.; Wu, H.; Li, J.; Zhao, C.; Zhu, H. A Cucumber Leaf Disease Severity Classification Method Based on the Fusion of DeepLabV3+ and U-Net. Comput. Electron. Agric. 2021, 189, 106373. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, L. A New Lightweight Network Based on MobileNetV3. KSII Trans. Internet Inf. Syst. 2022, 16, 1–15. [Google Scholar] [CrossRef]
- Setiawan, W.; Ghofur, A.; Hastarita Rachman, F.; Rulaningtyas, R. Deep Convolutional Neural Network AlexNet and Squeezenet for Maize Leaf Diseases Image Classification. Kinet. Game Technol. Inf. Syst. Comput. Netw. Comput. Electron. Control. 2021. [Google Scholar] [CrossRef]
- Zhang, Y.; Xie, W.; Yu, X. Design and Implementation of Liveness Detection System Based on Improved Shufflenet V2. SIViP 2023, 17, 3035–3043. [Google Scholar] [CrossRef]
- Pitakaso, R.; Sethanan, K.; Theeraviriya, C. Variable Neighborhood Strategy Adaptive Search for Solving Green 2-Echelon Location Routing Problem. Comput. Electron. Agric. 2020, 173, 105406. [Google Scholar] [CrossRef]
- Alguliyev, R.; Imamverdiyev, Y.; Sukhostat, L.; Bayramov, R. Plant Disease Detection Based on a Deep Model. Soft Comput. 2021, 25, 13229–13242. [Google Scholar] [CrossRef]
- Sharma, A.; Rajesh, B.; Javed, M. Detection of Plant Leaf Disease Directly in the JPEG Compressed Domain Using Transfer Learning Technique. Adv. Mach. Intell. Signal Process. 2022, 858, 407–418. [Google Scholar] [CrossRef]
- Sophia, S.; Devi, D.; Lakshmi Prabha, K.; Keerthana, V.; Kavin, V. A Novel Method to Detect Disease in Leaf Using Deep Learning Approach. In Proceedings of the 2021 7th International Conference on Advanced Computing and Communication Systems (ICACCS), Coimbatore, India, 19 March 2021; pp. 1558–1562. [Google Scholar]
- Kawatra, M.; Agarwal, S.; Kapur, R. Leaf Disease Detection Using Neural Network Hybrid Models. In Proceedings of the 2020 IEEE 5th International Conference on Computing Communication and Automation (ICCCA), Greater Noida, India, 30 October 2020; pp. 225–230. [Google Scholar]
- Pushpa, B.R.; Ashok, A.; Shree Hari, A.V. Plant Disease Detection and Classification Using Deep Learning Model. In Proceedings of the 2021 Third International Conference on Inventive Research in Computing Applications (ICIRCA), Coimbatore, India, 2 September 2021; pp. 1285–1291. [Google Scholar]
- Kartikeyan, P.; Shrivastava, G. Impact Evaluation of Deep Learning Models in the Context of Plant Disease Detection. In Proceedings of International Conference on Data Science and Applications; Saraswat, M., Roy, S., Chowdhury, C., Gandomi, A.H., Eds.; Springer: Singapore, 2022; Volume 288, pp. 527–540. [Google Scholar]
- Kumar, N.A.; Sathish Kumar, S. Deep Learning-Based Image Preprocessing Techniques for Crop Disease Identification. In Futuristic Communication and Network Technologies; Sivasubramanian, A., Shastry, P.N., Hong, P.C., Eds.; Springer: Singapore, 2022; Volume 792, pp. 1–10. [Google Scholar] [CrossRef]
- Sowmiya, M.; Krishnaveni, S. Deep Learning Techniques to Detect Crop Disease and Nutrient Deficiency—A Survey. In Proceedings of the 2021 International Conference on System, Computation, Automation and Networking (ICSCAN), Puducherry, India, 30 July 2021; pp. 1–5. [Google Scholar]
- Rohit, V.; Boominathan, P. Crop Diseases and Pest Detection Using Deep Learning and Image Processing Techniques. Int. J. Res. Appl. Sci. Eng. Technol. 2021, 9, 372–380. [Google Scholar] [CrossRef]
- Anh, P.T.; Duc, H.T.M. A Benchmark of Deep Learning Models for Multi-Leaf Diseases for Edge Devices. In Proceedings of the 2021 International Conference on Advanced Technologies for Communications (ATC), Ho Chi Minh City, Vietnam, 14 October 2021; pp. 318–323. [Google Scholar]
- Smetanin, A.; Uzhinskiy, A.; Ososkov, G.; Goncharov, P.; Nechaevskiy, A. Deep Learning Methods for The Plant Disease Detection Platform. AIP Conf. Proc. 2021, 2377, 060006. [Google Scholar]
- Ray, A.; Chakraborty, T.; Ghosh, D. Optimized Ensemble Deep Learning Framework for Scalable Forecasting of Dynamics Containing Extreme Events. Chaos Interdiscip. J. Nonlinear Sci. 2021, 31, 111105. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.; Kora, R. An Effective Ensemble Deep Learning Framework for Text Classification. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 8825–8837. [Google Scholar] [CrossRef]
- Sharma, S.; Yadav, N.S. Ensemble-Based Machine Learning Techniques for Attack Detection. In Proceedings of the 2021 9th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions) (ICRITO), Noida, India, 3 September 2021; pp. 1–6. [Google Scholar]
- Ahuja, S.; Panigrahi, B.K.; Gandhi, T.K. Fully Automatic Brain Tumor Segmentation Using DeepLabv3+ with Variable Loss Functions. In Proceedings of the 2021 8th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 26 August 2021; pp. 522–526. [Google Scholar]
- Dogan, R.O.; Dogan, H.; Bayrak, C.; Kayikcioglu, T. A Two-Phase Approach Using Mask R-CNN and 3D U-Net for High-Accuracy Automatic Segmentation of Pancreas in CT Imaging. Comput. Methods Programs Biomed. 2021, 207, 106141. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Wang, H.; Song, W.; Xia, J.; Liu, T. Deep Sea Nodule Mineral Image Segmentation Algorithm Based on Mask R-CNN. In Proceedings of the ACM Turing Award Celebration Conference—China (ACM TURC 2021), Hefei, China, 30 July 2021; pp. 278–284. [Google Scholar]
- Durkee, M.S.; Abraham, R.; Ai, J.; Fuhrman, J.D.; Clark, M.R.; Giger, M.L. Comparing Mask R-CNN and U-Net Architectures for Robust Automatic Segmentation of Immune Cells in Immunofluorescence Images of Lupus Nephritis Biopsies. In Proceedings of the Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues XIX; Leary, J.F., Tarnok, A., Georgakoudi, I., Eds.; SPIE: Bellingham, WA, USA, 2021; p. 23. [Google Scholar]
- Liu, H.; Zhang, Q.; Liu, Y. Image Segmentation of Bladder Cancer Based on DeepLabv3+. In Proceedings of 2021 Chinese Intelligent Systems Conference; Springer: Singapore, 2022; Volume 805, pp. 614–621. [Google Scholar] [CrossRef]
- Minatel, P.G.; Oliveira, B.C.; Albertazzi, A. Comparison of Unet and Mask R-CNN for Impact Damage Segmentation in Lock-in Thermography Phase Images. In Proceedings of the Automated Visual Inspection and Machine Vision IV; Beyerer, J., Heizmann, M., Eds.; SPIE: Bellingham, WA, USA, 2021; p. 26. [Google Scholar]
- Quan, B.; Liu, B.; Fu, D.; Chen, H.; Liu, X. Improved Deeplabv3 for Better Road Segmentation in Remote Sensing Images. In Proceedings of the 2021 International Conference on Computer Engineering and Artificial Intelligence (ICCEAI), Shanghai, China, 27–29 August 2021; pp. 331–334. [Google Scholar]
- Quoc, T.T.P.; Linh, T.T.; Minh, T.N.T. Comparing U-Net Convolutional Network with Mask R-CNN in Agricultural Area Segmentation on Satellite Images. In Proceedings of the 2020 7th NAFOSTED Conference on Information and Computer Science (NICS), Ho Chi Minh City, Vietnam, 26 November 2020; pp. 124–129. [Google Scholar]
- Wang, Z.; Wang, J.; Yang, K.; Wang, L.; Su, F.; Chen, X. Semantic Segmentation of High-Resolution Remote Sensing Images Based on a Class Feature Attention Mechanism Fused with Deeplabv3+. Comput. Geosci. 2022, 158, 104969. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, R.; Gao, F.; Wang, W.; Fu, L.; Li, R. Segmentation of Abnormal Leaves of Hydroponic Lettuce Based on DeepLabV3+ for Robotic Sorting. Comput. Electron. Agric. 2021, 190, 106443. [Google Scholar] [CrossRef]
- Kathawala, A.A.; Chauhan, A.; Chalmal, A.; Ransubhe, O.; Bhosale, A.A. Plant Leaf Disease Detection Using Data Augmentation and CNN. J. Emerg. Technol. Innov. Res. 2021, 8, b896–b901. [Google Scholar]
- Bharati Devi, M.; Amarendra, K. A Convolutional Neural Network Architecture for Tomato Leaf Disease Detection Using Data Augmentation. Smart Comput. Tech. Appl. 2021, 225, 507–516. [Google Scholar] [CrossRef]
- Sethanan, K.; Pitakaso, R.; Srichok, T.; Khonjun, S.; Weerayuth, N.; Prasitpuriprecha, C.; Preeprem, T.; Jantama, S.S.; Gonwirat, S.; Enkvetchakul, P.; et al. Computer-Aided Diagnosis Using Embedded Ensemble Deep Learning for Multiclass Drug-Resistant Tuberculosis Classification. Front. Med. 2023, 10, 1122222. [Google Scholar] [CrossRef]
- Abraham, S.E.; Kovoor, B.C. Residual Decoder Based U-Net for Semantic Segmentation. Int. Conf. Innov. Comput. Commun. 2022, 1388, 699–708. [Google Scholar] [CrossRef]
- Boyina, L.; Sandhya, G.; Vasavi, S.; Koneru, L.; Koushik, V. Weed Detection in Broad Leaves Using Invariant U-Net Model. In Proceedings of the 2021 International Conference on Communication, Control and Information Sciences (ICCISc), Idukki, India, 16 June 2021; pp. 1–4. [Google Scholar]
- Arun Pandian, J.; Geetharamani, G.; Annette, B. Data Augmentation on Plant Leaf Disease Image Dataset Using Image Manipulation and Deep Learning Techniques. In Proceedings of the 2019 IEEE 9th International Conference on Advanced Computing (IACC), Tiruchirappalli, India, 13–14 December 2019; pp. 199–204. [Google Scholar]
- Min, B.; Kim, T.; Shin, D.; Shin, D. Data Augmentation Method for Plant Leaf Disease Recognition. Appl. Sci. 2023, 13, 1465. [Google Scholar] [CrossRef]
- Kant Duggal, J.; El-Sharkawy, M. Shallow SqueezeNext: Real Time Deployment on Bluebox2.0 with 272KB Model Size. JEEE 2020, 8, 127. [Google Scholar] [CrossRef]
- Lv, S.; Liu, J.; Gong, C.; Yang, B.; Gan, X. The High Precision Real-Time Facial Landmark Detection Technique Based on ShufflenetV2. Theor. Comput. Sci. 2021, 1494, 59–71. [Google Scholar] [CrossRef]
- Wan, X.; Zuo, X.; Zhao, X. A Differential Evolution Algorithm Combined with Linear Programming for Solving a Closed Loop Facility Layout Problem. Appl. Soft Comput. 2022, 121, 108725. [Google Scholar] [CrossRef]
- He, Z.; Liu, T.; Liu, H. Improved Particle Swarm Optimization Algorithms for Aerodynamic Shape Optimization of High-Speed Train. Adv. Eng. Softw. 2022, 173, 103242. [Google Scholar] [CrossRef]
- Qin, H.; Zhang, W.; Zhai, H. Cooperative Control of Multiple Intersections Combining Agent and Chaotic Particle Swarm Optimization. Comput. Electr. Eng. 2023, 110, 108875. [Google Scholar] [CrossRef]
- Song, L.; Dong, Y.; Guo, Q.; Meng, Y.; Zhao, G. An Adaptive Differential Evolution Algorithm with DBSCAN for the Integrated Slab Allocation Problem in Steel Industry. Appl. Soft Comput. 2023, 146, 110665. [Google Scholar] [CrossRef]
- Yang, L.; Yu, H.; Cheng, Y.; Mei, S.; Duan, Y.; Li, D.; Chen, Y. A Dual Attention Network Based on efficientNet-B2 for Short-Term Fish School Feeding Behavior Analysis in Aquaculture. Comput. Electron. Agric. 2021, 187, 106316. [Google Scholar] [CrossRef]
- Shafiq, M.; Gu, Z. Deep Residual Learning for Image Recognition: A Survey. Appl. Sci. 2022, 12, 8972. [Google Scholar] [CrossRef]
- Li, G.; Zhang, M.; Li, J.; Lv, F.; Tong, G. Efficient Densely Connected Convolutional Neural Networks. Pattern Recognit. 2021, 109, 107610. [Google Scholar] [CrossRef]
- Tian, Y.; Li, E.; Liang, Z.; Tan, M.; He, X. Diagnosis of Typical Apple Diseases: A Deep Learning Method Based on Multi-Scale Dense Classification Network. Front. Plant Sci. 2021, 12, 698474. [Google Scholar] [CrossRef]
- Sanath Rao, U.; Swathi, R.; Sanjana, V.; Arpitha, L.; Chandrasekhar, K.; Chinmayi; Naik, P.K. Deep Learning Precision Farming: Grapes and Mango Leaf Disease Detection by Transfer Learning. Glob. Transit. Proc. 2021, 2, 535–544. [Google Scholar] [CrossRef]
- Venkatesh, T.; Prathyush, K.; Deepak, S.; Preetham, U.V.S.A.M. Agriculture Crop Leaf Disease Detection Using Image Processing. Int. J. Innov. Technol. Explor. Eng. 2021, 10, 110–114. [Google Scholar] [CrossRef]
- Chainarong, S.; Pitakaso, R.; Sirirak, W.; Srichok, T.; Khonjun, S.; Sethanan, K.; Sangthean, T. Multi-Objective Variable Neighborhood Strategy Adaptive Search for Tuning Optimal Parameters of SSM-ADC12 Aluminum Friction Stir Welding. J. Manuf. Mater. Process. 2021, 5, 123. [Google Scholar] [CrossRef]
- Chainarong, S.; Srichok, T.; Pitakaso, R.; Sirirak, W.; Khonjun, S.; Akararungruangku, R. Variable Neighborhood Strategy Adaptive Search for Optimal Parameters of SSM-ADC 12 Aluminum Friction Stir Welding. Processes 2021, 9, 1805. [Google Scholar] [CrossRef]
- Nagamani, H.S.; Devi, H.S. Leaf Region Segmentation for Plant Leaf Disease Detection Using Color Conversion and Flood Filling. In Proceedings of the 2021 2nd Global Conference for Advancement in Technology (GCAT), Bangalore, India, 1 October 2021; pp. 1–7. [Google Scholar]
- Abusham, E.A. Image Processing Technique for the Detection of Alberseem Leaves Diseases Based on Soft Computing. Artif. Intell. Robot. Dev. J. 2021, 1, 103–115. [Google Scholar] [CrossRef]
- Entuni, C.J.; Afendi Zulcaffle, T.M.; Kipli, K. Severity Estimation of Plant Leaf Diseases Using Segmentation Method. Appl. Sci. Eng. Prog. 2021, 14, 108–119. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Paul, M.; Pal, R.; De, D. Tea Leaf Disease Detection Using Multi-Objective Image Segmentation. Multimed. Tools Appl. 2021, 80, 753–771. [Google Scholar] [CrossRef]
- Trecene, J.K.D. Brassicaceae Leaf Disease Detection Using Image Segmentation Technique. In Proceedings of the IEEE EUROCON 2021—19th International Conference on Smart Technologies, Lviv, Ukraine, 6 July 2021; pp. 30–34. [Google Scholar]
- Narayanapur, A.; Naik, P.; Kori, P.B.; Kalaburgi, N.; IM, R.; MC, M. Leaf Disease Detection of Agricultural Plant Using Image Processing. Int. J. Sci. Res. Comput. Sci. Eng. Inf. Technol. 2020, 6, 432–435. [Google Scholar] [CrossRef]
- Bjånes, A.; De La Fuente, R.; Mena, P. A Deep Learning Ensemble Model for Wildfire Susceptibility Mapping. Ecol. Inform. 2021, 65, 101397. [Google Scholar] [CrossRef]
- Hirasen, D.; Pillay, V.; Viriri, S.; Gwetu, M. Skeletal Age Estimation from Hand Radiographs Using Ensemble Deep Learning. Pattern Recognit. 2021, 12725, 173–183. [Google Scholar] [CrossRef]
- Lee, K.; Laskin, M.; Srinivas, A.; Abbeel, P. Sunrise: A Simple Unified Framework for Ensemble Learning in Deep Reinforcement Learning. Proc. Mach. Learn. Res. 2021, 139, 6131–6141. [Google Scholar]
- Wei, W.; Liu, L. Robust Deep Learning Ensemble Against Deception. IEEE Trans. Dependable Secur. Comput. 2020, 18, 1. [Google Scholar] [CrossRef]

| Name of Leaf Abnormality | Description | Leaf Image Example |
|---|---|---|
| Normal Leaf (NL) | Plant leaves are spreading, green, no lesions. |  |
| Red Mite Disease (RM) | Red mites suck the sap from the leaves, causing the loss of chlorophyll. Red mites cause the upper surfaces of the leaves to become pale yellow spots, burn shoots, wither leaves, and fall off. |  |
| Worm Creep Disease (WCD) | The caterpillars will eat young and old leaves until only the stems and branches remain. |  |
| Low Light Intensity (LI) | Plant leaves reduce chlorophyll production. The leaves turn yellow. |  |
| Nitrogen Deficiency (ND) | The leaves are pale yellow. The tip of the leaf and the edge of the leaf will gradually dry and this will spread continuously until the leaves fall from the tree prematurely. |  |
| Phosphorus Deficiency (PHD) | Phosphorus deficiency manifests as stunted growth, dark green or purple pigmentation in leaves, and underdeveloped root systems in plants. |  |
| Potassium Deficiency (PD) | Older leaves are pale yellow, starting from the leaf margin and leaf tip. |  |
| Water Deficiency (WD) | The leaves wither, and dry leaves cannot spread. |  |
| ABL-1 | ABL-2 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NL | RM | ND | PD | LI | WD | WCD | PHD | NL | RM | ND | PD | LI | WD | WCD | PHD | |
| Training set | 1000 | 960 | 1040 | 1080 | 1072 | 1064 | 1024 | 1040 | - | - | - | - | - | - | - | - |
| Testing set | 250 | 240 | 260 | 270 | 268 | 266 | 256 | 260 | 550 | 550 | 500 | 600 | 550 | 550 | 600 | 580 |
| Total | 1250 | 1200 | 1300 | 1350 | 1340 | 1330 | 1280 | 1300 | 550 | 500 | 600 | 550 | 550 | 600 | 580 | 580 |
| No. | Segmentation | Augmentation | Decision Fusion Strategy | Accuracy | AUC | F1-Score | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No Segment | With Segment | No Augment | With Augment | UWA | DE | PSO | VaNSAS | ||||
| 1 | - | - | - | - | - | 85.65 | 86.06 | 86.74 | |||
| 2 | - | - | - | - | - | 83.81 | 84.17 | 85.09 | |||
| 3 | - | - | - | - | - | 84.02 | 84.23 | 85.11 | |||
| 4 | - | - | - | - | - | 84.59 | 85.94 | 86.32 | |||
| 5 | - | - | - | - | - | 86.71 | 86.82 | 87.03 | |||
| 6 | - | - | - | - | - | 88.28 | 88.40 | 89.55 | |||
| 7 | - | - | - | - | - | 89.85 | 90.03 | 91.28 | |||
| 8 | - | - | - | - | - | 90.48 | 90.85 | 92.47 | |||
| 9 | - | - | - | - | - | 89.63 | 90.58 | 91.38 | |||
| 10 | - | - | - | - | - | 90.41 | 90.96 | 91.57 | |||
| 11 | - | - | - | - | - | 92.19 | 93.17 | 93.26 | |||
| 12 | - | - | - | - | - | 93.46 | 93.63 | 94.02 | |||
| 13 | - | - | - | - | - | 92.82 | 94.01 | 94.23 | |||
| 14 | - | - | - | - | - | 94.39 | 94.48 | 94.75 | |||
| 15 | - | - | - | - | - | 94.54 | 95.01 | 95.34 | |||
| 16 | - | - | - | - | - | 96.31 | 96.47 | 96.69 | |||
| Method | Description | Number of CNN | Total Size (MB) | Training Time (min) | Testing Time (min/image) |
|---|---|---|---|---|---|
| Ho-Mo [13] | Homogenous Ensemble Mobile NetV3 | 7 | 35.0 | 65.38 | 0.62 |
| Ho-SQ [14] | Homogenous Ensemble Squeeze NetV2 | 8 | 38.4 | 67.09 | 0.64 |
| Ho-Sh [15] | Homogenous Ensemble Shuffle NetV2 | 5 | 37 | 66.27 | 0.64 |
| EfficientNet-B2 [54] | Single Model | 1 | 41 | 70.50 | 0.68 |
| EfficientNet-B3 [54] | Single Model | 1 | 44 | 73.94 | 0.70 |
| ResNet-50 [55] | Single Model | 1 | 44 | 74.11 | 0.72 |
| DenseNet121 [56] | Single Model | 1 | 33 | 58.43 | 0.56 |
| Inception-ResNet-v2 [57] | Single Model | 1 | 56 | 78.08 | 0.84 |
| He-Meta (proposed model) | Heterogeneous Ensemble ShuffleNetV2, Squeeze NetV2 and Mobile NetV3 | 6 | 34.4 | 60.41 | 0.61 |
| Method | AUC | F1-Score | Accuracy |
|---|---|---|---|
| Ho-Mo [13] | 96.09 | 95.78 | 94.70 |
| Ho-SQ [14] | 95.42 | 95.01 | 94.81 |
| Ho-Sh [15] | 94.27 | 94.13 | 93.38 |
| EfficientNet-B2 [54] | 95.07 | 94.19 | 93.86 |
| EfficientNet-B3 [54] | 94.51 | 94.40 | 93.92 |
| ResNet-50 [55] | 93.46 | 93.07 | 92.75 |
| DenseNet121 [56] | 93.26 | 92.11 | 91.08 |
| Inception-ResNet-v2 [57] | 97.69 | 97.74 | 97.38 |
| He-Meta (proposed model) | 98.95 | 98.82 | 98.51 |
| Method | 3-cv | 5-cv | ||||
|---|---|---|---|---|---|---|
| AUC | F1-Score | Accuracy | AUC | F1-Score | Accuracy | |
| Ho-Mo [13] | 96.11 ± 0.07 | 95.81 ± 1.81 | 94.71 ± 0.64 | 96.14 ± 0.35 | 95.83 ± 0.27 | 94.70 ± 0.37 |
| Ho-SQ [14] | 95.46 ± 0.76 | 95.05 ± 1.87 | 94.83 ± 0.47 | 95.43 ± 0.65 | 95.03 ± 0.29 | 94.83 ± 0.53 |
| Ho-Sh [15] | 94.32 ± 0.58 | 94.18 ± 0.98 | 93.41 ± 0.72 | 94.31 ± 0.86 | 94.13 ± 0.36 | 93.38 ± 0.41 |
| EfficientNet-B2 [54] | 95.10 ± 0.76 | 94.23 ± 0.85 | 93.88 ± 0.86 | 95.10 ± 0.29 | 94.21 ± 0.85 | 93.89 ± 0.25 |
| EfficientNet-B3 [54] | 94.54 ± 0.48 | 94.41 ± 0.78 | 93.93 ± 1.82 | 94.51 ± 0.67 | 94.43 ± 0.52 | 93.97 ± 0.31 |
| ResNet-50 [55] | 93.51 ± 0.94 | 93.08 ± 0.67 | 92.76 ± 0.98 | 93.46 ± 0.73 | 93.11 ± 0.58 | 92.77 ± 0.26 |
| DenseNet121 [56] | 93.28 ± 0.32 | 92.13 ± 0.56 | 91.12 ± 0.45 | 93.29 ± 0.27 | 92.16 ± 0.97 | 91.10 ± 0.12 |
| Inception-ResNet-v2 [57] | 97.72 ± 0.43 | 97.76 ± 0.42 | 97.39 ± 0.37 | 97.70 ± 0.47 | 97.74 ± 0.12 | 97.41 ± 0.04 |
| He-Meta (proposed model) | 98.97 ± 0.19 | 98.86 ± 0.32 | 98.55 ± 0.29 | 98.99 ± 0.27 | 98.83 ± 0.07 | 98.55 ± 0.08 |
| Method | AUC | F1-Score | Accuracy |
|---|---|---|---|
| Ho-Mo [13] | 96.05 | 95.73 | 94.67 |
| Ho-SQ [14] | 95.39 | 94.97 | 94.74 |
| Ho-Sh [15] | 94.25 | 94.05 | 93.35 |
| EfficientNet-B2 [54] | 95.06 | 94.12 | 93.78 |
| EfficientNet-B3 [54] | 94.48 | 94.32 | 93.87 |
| ResNet-50 [55] | 93.44 | 93.03 | 92.71 |
| DenseNet121 [56] | 93.18 | 92.08 | 91.04 |
| Inception-ResNet-v2 [57] | 97.68 | 97.71 | 97.35 |
| He-Meta (proposed model) | 98.93 | 98.75 | 98.45 |
| Method | 3-cv | 5-cv | ||||
|---|---|---|---|---|---|---|
| AUC | F1-Score | Accuracy | AUC | F1-Score | Accuracy | |
| Ho-Mo [13] | 96.13 ± 2.48 | 95.72 ± 2.03 | 94.64 ± 1.48 | 96.14 ± 1.57 | 95.73 ± 2.02 | 94.65 ± 2.13 |
| Ho-SQ [14] | 95.43 ± 0.81 | 95.05 ± 0.54 | 94.77 ± 1.51 | 95.45 ± 1.86 | 95.05 ± 2.81 | 94.78 ± 2.56 |
| Ho-Sh [15] | 94.22 ± 0.86 | 94.16 ± 0.68 | 93.34 ± 0.74 | 94.23 ± 1.47 | 94.18 ± 1.04 | 93.34 ± 0.85 |
| EfficientNet-B2 [54] | 95.07 ± 1.48 | 94.15 ± 1.67 | 93.88 ± 1.74 | 95.08 ± 2.15 | 94.15 ± 1.51 | 93.89 ± 1.07 |
| EfficientNet-B3 [54] | 94.50 ± 1.85 | 94.36 ± 1.74 | 93.89 ± 0.96 | 94.52 ± 1.16 | 94.38 ± 1.05 | 93.91 ± 0.86 |
| ResNet-50 [55] | 93.44 ± 0.89 | 93.08 ± 0.85 | 92.75 ± 1.47 | 93.44 ± 1.26 | 93.08 ± 1.74 | 92.76 ± 0.94 |
| DenseNet121 [56] | 93.26 ± 0.58 | 92.07 ± 0.53 | 91.10 ± 0.47 | 93.28 ± 0.38 | 92.08 ± 0.31 | 91.12 ± 0.68 |
| Inception-ResNet-v2 [57] | 97.68 ± 0.19 | 97.72 ± 0.24 | 97.41 ± 0.27 | 97.69 ± 0.34 | 97.74 ± 0.19 | 97.41 ± 0.48 |
| He-Meta (proposed model) | 98.98 ± 0.09 | 98.84 ± 0.11 | 98.46 ± 0.18 | 98.98 ± 0.15 | 98.85 ± 0.13 | 98.47 ± 0.07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buakum, B.; Kosacka-Olejnik, M.; Pitakaso, R.; Srichok, T.; Khonjun, S.; Luesak, P.; Nanthasamroeng, N.; Gonwirat, S. Two-Stage Ensemble Deep Learning Model for Precise Leaf Abnormality Detection in Centella asiatica. AgriEngineering 2024, 6, 620-644. https://doi.org/10.3390/agriengineering6010037
Buakum B, Kosacka-Olejnik M, Pitakaso R, Srichok T, Khonjun S, Luesak P, Nanthasamroeng N, Gonwirat S. Two-Stage Ensemble Deep Learning Model for Precise Leaf Abnormality Detection in Centella asiatica. AgriEngineering. 2024; 6(1):620-644. https://doi.org/10.3390/agriengineering6010037
Chicago/Turabian StyleBuakum, Budsaba, Monika Kosacka-Olejnik, Rapeepan Pitakaso, Thanatkij Srichok, Surajet Khonjun, Peerawat Luesak, Natthapong Nanthasamroeng, and Sarayut Gonwirat. 2024. "Two-Stage Ensemble Deep Learning Model for Precise Leaf Abnormality Detection in Centella asiatica" AgriEngineering 6, no. 1: 620-644. https://doi.org/10.3390/agriengineering6010037
APA StyleBuakum, B., Kosacka-Olejnik, M., Pitakaso, R., Srichok, T., Khonjun, S., Luesak, P., Nanthasamroeng, N., & Gonwirat, S. (2024). Two-Stage Ensemble Deep Learning Model for Precise Leaf Abnormality Detection in Centella asiatica. AgriEngineering, 6(1), 620-644. https://doi.org/10.3390/agriengineering6010037










