Regulation of Antimicrobial Pathways by Endogenous Heat Shock Proteins in Gastrointestinal Disorders
Abstract
:1. Introduction
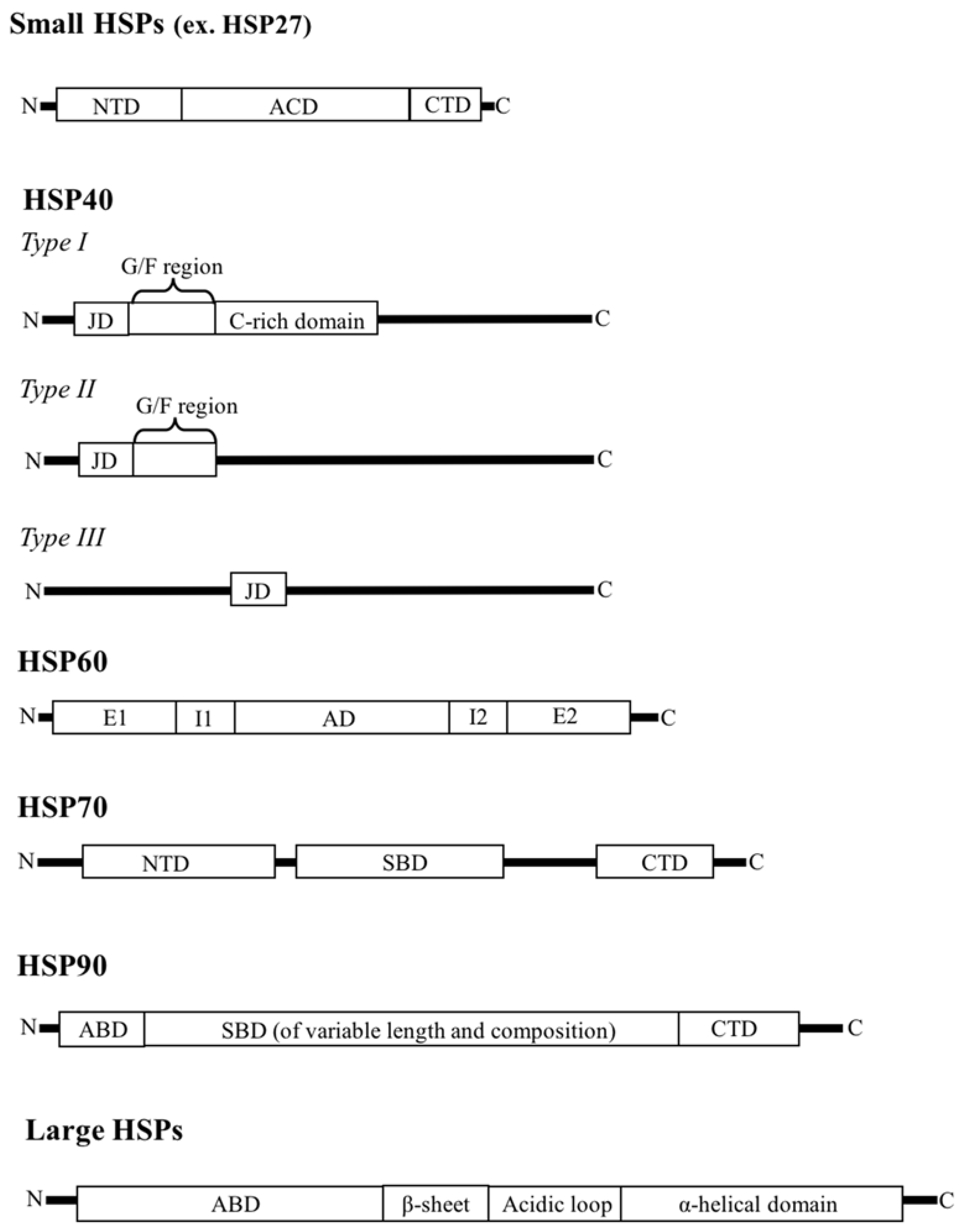
2. HSP Families
2.1. Small HSPs
2.2. HSP40
2.3. HSP60
2.4. HSP70
2.5. HSP90
2.6. Large HSPs
3. HSPs in Gastric and Intestinal Homeostasis
4. HSP Interactions with NOD-Like Receptor Innate Immunity Receptors
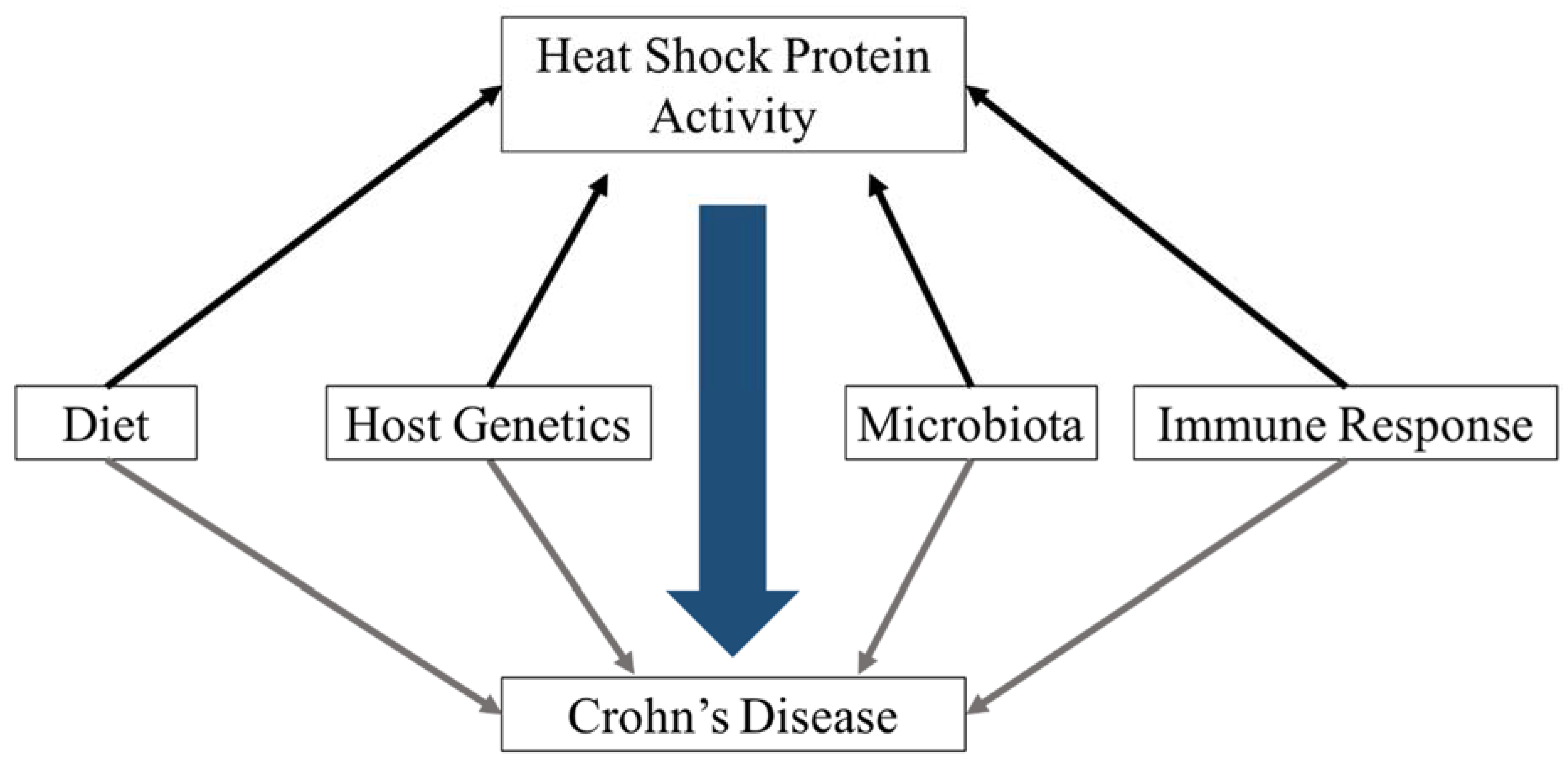
5. Environmental Factors That Influence HSP Abundance
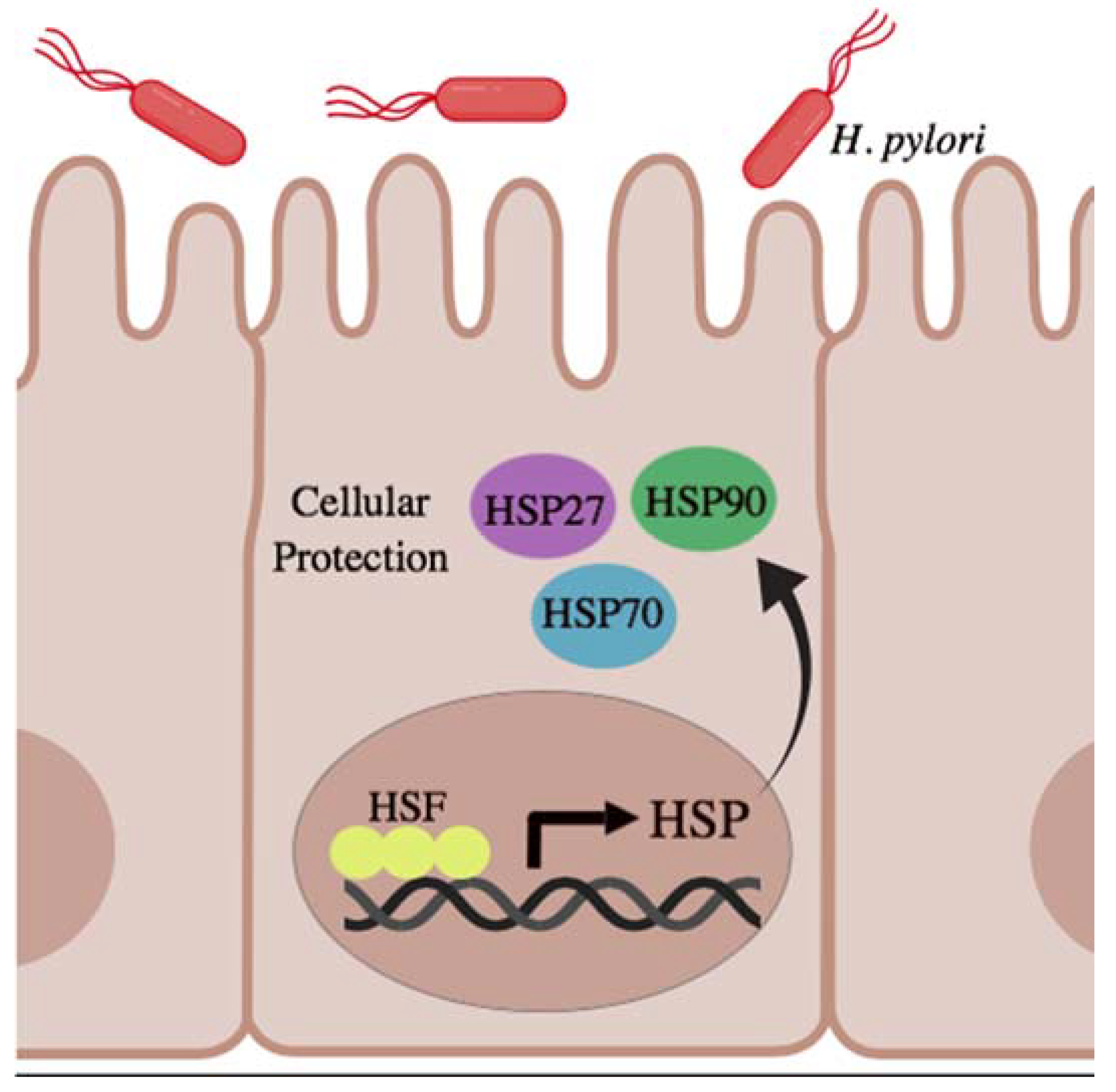
5.1. Microbial Stimuli
5.2. Dietary Stimuli
6. Role of Bacterial HSP in Host Immune Responses
6.1. HtpG
6.2. GroEL
6.3. DnaK
7. Conclusions
Acknowledgments
Conflicts of Interest
References
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet (Lond. Engl.) 2018, 390, 2769–2778. [Google Scholar] [CrossRef]
- Kamm, M.A. Rapid changes in epidemiology of inflammatory bowel disease. Lancet 2017, 390, 2741–2742. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Schumm, L.P.; Sharma, Y.; Anderson, C.A.; et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlig, H.H. Monogenic diseases associated with intestinal inflammation: Implications for the understanding of inflammatory bowel disease. Gut 2013, 62, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.M.; Girardelli, M.; Tommasini, A. Genetics of inflammatory bowel disease from multifactorial to monogenic forms. World J. Gastroenterol. 2015, 21, 12296–12310. [Google Scholar] [CrossRef] [PubMed]
- Momozawa, Y.; Dmitrieva, J.; Théâtre, E.; Deffontaine, V.; Rahmouni, S.; Charloteaux, B.; Crins, F.; Docampo, E.; Elansary, M.; Gori, A.S.; et al. IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Amininejad, L.; Charloteaux, B.; Theatre, E.; Liefferinckx, C.; Dmitrieva, J.; Hayard, P.; Muls, V.; Maisin, J.M.; Schapira, M.; Ghislain, J.M.; et al. Analysis of Genes Associated with Monogenic Primary Immunodeficiency Identifies Rare Variants in XIAP in Patients with Crohn’s Disease. Gastroenterology 2018, 154, 2165–2177. [Google Scholar] [CrossRef] [PubMed]
- Denson, L.A.; Jurickova, I.; Karns, R.; Shaw, K.A.; Cutler, D.J.; Okou, D.T.; Dodd, A.; Quinn, K.; Mondal, K.; Aronow, B.J.; et al. Clinical and Genomic Correlates of Neutrophil Reactive Oxygen Species Production in Pediatric Patients with Crohn’s Disease. Gastroenterology 2018, 154, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Marigorta, U.M.; Denson, L.A.; Hyams, J.S.; Mondal, K.; Prince, J.; Walters, T.D.; Griffiths, A.; Noe, J.D.; Crandall, W.V.; Rosh, J.R.; et al. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn’s disease. Nat. Genet. 2017, 49, 1517–1521. [Google Scholar] [CrossRef] [PubMed]
- Barbatis, C.; Tsopanomichalou, M. Heat shock proteins in inflammatory bowel disease. Ann. Gastroenterol. 2009, 22, 244–247. [Google Scholar]
- Millar, N.L.; Murrell, G.A.C. Heat shock proteins in tendinopathy: Novel molecular regulators. Mediat. Inflamm. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Binder, R.J. Functions of Heat Shock Proteins in Pathways of the Innate and Adaptive Immune System. J. Immunol. 2014, 193, 5765–5771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Menoret, A.; Srivastava, P. Roles of heat-shock proteins in antigen presentation and cross-presentation. Curr. Opin. Immunol. 2002, 14, 45–51. [Google Scholar] [CrossRef]
- Broere, F.; Van Der Zee, R.; Van Eden, W. Heat shock proteins are no DAMPs, rather “DAMPERs.”. Nat. Rev. Immunol. 2011, 11, 565. [Google Scholar] [CrossRef] [PubMed]
- Pockley, A.G.; Henderson, B. Extracellular cell stress (Heat shock) proteins—Immune responses and disease: An overview. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [PubMed]
- Samborski, P.; Grzymisławski, M. The role of HSP70 Heat shock proteins in the pathogenesis and treatment of inflammatory bowel diseases. Adv. Clin. Exp. Med. 2015, 24, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, D.; Stahl, M.; Ibrahim, E.T.; Wenzel, B.E.; Drabicki, D.; Wecke, A.; Fellermann, K.; Stange, E.F. Enhanced intestinal expression of heat shock protein 70 in patients with inflammatory bowel diseases. Dig. Dis. Sci. 1999, 44, 1440–1447. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.B.; Aherne, C.M.; Yeckes, A.; Pound, K.; Eltzschig, H.K.; Jedlicka, P.; De Zoeten, E.F. Inhibition of N-terminal ATPase on HSP90 attenuates colitis through enhanced Treg function. Mucosal Immunol. 2013, 6, 960–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betancourt, A.B.; Lyu, Q.; Broere, F.; Sijts, A.; Rutten, V.P.M.G.; Van Eden, W. T cell-mediated chronic inflammatory diseases are candidates for therapeutic tolerance induction with heat shock proteins. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Edkins, A.L.; Price, J.T.; Pockley, A.G.; Blatch, G.L. Heat shock proteins as modulators and therapeutic targets of chronic disease: An integrated perspective. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20160521. [Google Scholar] [CrossRef] [PubMed]
- Van Eden, W.; Jansen, M.A.A.; Ludwig, I.; van Kooten, P.; van der Zee, R.; Broere, F. The Enigma of Heat Shock Proteins in Immune Tolerance. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudeja, V.; Vickers, S.M.; Saluja, A.K. The role of heat shock proteins in gastrointestinal diseases. Gut 2009, 58, 1000–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaspard, E.; Hunault, G. sHSPdb: A database for the analysis of small Heat Shock Proteins. BMC Plant Biol. 2016, 16, 135. [Google Scholar] [CrossRef] [PubMed]
- Kappé, G.; Leunissen, J.; de Jong, W. Evolution and diversity of prokaryotic small heat shock proteins. In Small Stress Proteins; Springer: Berlin/Heidelberg, Germany, 2001; pp. 1–17. [Google Scholar]
- Huang, L.-H.; Wang, H.-S.; Kang, L. Different evolutionary lineages of large and small heat shock proteins in eukaryotes. Cell Res. 2008, 18, 1074–1076. [Google Scholar] [CrossRef] [PubMed]
- Narberhaus, F. Crystallin-Type Heat Shock Proteins: Socializing Minichaperones in the Context of a Multichaperone Network. Microbiol. Mol. Biol. Rev. 2002, 66, 64–93. [Google Scholar] [CrossRef] [PubMed]
- Shashidharamurthy, R.; Koteiche, H.A.; Dong, J.; Mchaourab, H.S. Mechanism of Chaperone Function in Small Heat Shock Proteins. J. Biol. Chem. 2005, 280, 5281–5289. [Google Scholar] [CrossRef] [PubMed]
- Haslbeck, M.; Franzmann, T.; Weinfurtner, D.; Buchner, J. Some like it hot: The structure and function of small heat-shock proteins. Nat. Struct Mol. Biol. 2005, 12, 842–846. [Google Scholar] [CrossRef] [PubMed]
- Delbecq, S.P.; Klevit, R.E. One size does not fit all: The oligomeric states of alphaB crystallin. FEBS Lett. 2013, 587, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Benesch, J.L.P.; Ayoub, M.; Robinson, C.V.; Aquilina, J.A. Small heat shock protein activity is regulated by variable oligomeric substructure. J. Biol. Chem. 2008, 283, 28513–28517. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Burns, T.F. Targeting heat shock proteins in cancer: A promising therapeutic approach. Int. J. Mol. Sci. 2017, 18, 1978. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.B.; Shao, Y.M.; Miao, S.; Wang, L. The diversity of the DNAJ/HSP40 family, the crucial partners for HSP70 chaperones. Cell. Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Walsh, P.; Bursać, D.; Law, Y.C.; Cyr, D.; Lithgow, T. The J-protein family: Modulating protein assembly, disassembly and translocation. EMBO Rep. 2004, 5, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, M.E.; Caplan, A.J. Structure, function and evolution of DNAJ: Conservation and adaptation of chaperone function. Cell Stress Chaperones 1998, 3, 28–36. [Google Scholar] [CrossRef]
- Cajo, G.C.; Horne, B.E.; Kelley, W.L.; Schwager, F.; Georgopoulos, C.; Genevaux, P. The role of the DIF motif of the DNAJ (HSP40) co-chaperone in the regulation of the DNAK (HSP70) chaperone cycle. J. Biol. Chem. 2006, 281, 12436–12444. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Hageman, J.; Vos, M.J.; Kubota, H.; Tanguay, R.M.; Bruford, E.A.; Cheetham, M.E.; Chen, B.; Hightower, L.E. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 2009, 14, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.Y.; Lee, S.; Cyr, D.M. Mechanisms for regulation of HSP70 function by HSP40. Cell Stress Chaperones 2003, 8, 309–316. [Google Scholar] [CrossRef]
- Cheng, M.Y.; Hartl, F.U.; Martin, J.; Pollock, R.A.; Kalousek, F.; Neuper, W.; Hallberg, E.M.; Hallberg, R.L.; Horwich, A.L. Mitochondrial heat-shock protein HSP60 is essential for assembly of proteins imported into yeast mitochondria. Nature 1989, 337, 620–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parnas, A.; Nisemblat, S.; Weiss, C.; Levy-Rimler, G.; Pri-Or, A.; Zor, T.; Lund, P.A.; Bross, P.; Azem, A. Identification of Elements That Dictate the Specificity of Mitochondrial HSP60 for Its Co-Chaperonin. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Brocchieri, L.; Karlin, S. Conservation among HSP60 sequences in relation to structure, function, and evolution. Protein Sci. 2008, 9, 476–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koll, H.; Guiard, B.; Rassow, J.; Ostermann, J.; Horwich, A.L.; Neupert, W.; Hartl, F.U. Antifolding activity of HSP60 couples protein import into the mitochondrial matrix with export to the intermembrane space. Cell 1992, 68, 1163–1175. [Google Scholar] [CrossRef]
- Shi, J.; Fu, M.; Zhao, C.; Zhou, F.; Yang, Q.; Qiu, L. Characterization and function analysis of HSP60 and HSP10 under different acute stresses in black tiger shrimp, Penaeus monodon. Cell Stress Chaperones 2016, 21, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, T.; Ishida, R.; Yamamoto, H.; Tanabe-Ishida, M.; Haga, A.; Takahashi, H.; Takahashi, K.; Goto, D.; Grave, E.; Itoh, H. Functional structure and physiological functions of mammalian wild-type HSP60. Arch. Biochem. Biophys. 2015, 586, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Motojima, F. How do chaperonins fold protein? Biophysics 2015, 11, 93–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinker, A.; Pfeifer, G.; Kerner, M.J.; Naylor, D.J.; Hartl, F.U.; Hayer-Hartl, M. Dual function of protein confinement in chaperonin-assisted protein folding. Cell 2001, 107, 223–233. [Google Scholar] [CrossRef]
- Georgescauld, F.; Popova, K.; Gupta, A.J.; Bracher, A.; Engen, J.R.; Hayer-Hartl, M.; Hartl, F.U. GroEL/ES chaperonin modulates the mechanism and accelerates the rate of TIM-barrel domain folding. Cell 2014, 157, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P. HSP70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 2013, 38, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.E. The HSP70 family and cancer. Carcinogenesis 2013, 34, 1181–1188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Ulrich Hartl, F. Molecular Chaperone Functions in Protein Folding and Proteostasis. Ann. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.C.; Tang, Y.C.; Hayer-Hartl, M.; Hartl, F.U. SnapShot: Molecular Chaperones, Part I. Cell 2007, 128, 212-e1–212-e2. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.C.; Chang, H.C.; Hayer-Hartl, M.; Hartl, F.U. SnapShot: Molecular Chaperones, Part II. Cell 2007, 128, 412-e1–412-e2. [Google Scholar] [CrossRef] [PubMed]
- Bertelsen, E.B.; Chang, L.; Gestwicki, J.E.; Zuiderweg, E.R.P. Solution conformation of wild-type E. coli HSP70 (DNAK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. USA 2009, 106, 8471–8476. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.M.; DeLuca-Flaherty, C.; McKay, D.B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 1990, 346, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Pearl, L.H.; Prodromou, C. Structure and Mechanism of the HSP90 Molecular Chaperone Machinery. Ann. Rev. Biochem. 2006, 75, 271–294. [Google Scholar] [CrossRef] [PubMed]
- Street, T.O.; Lavery, L.A.; Agard, D.A. Substrate Binding Drives Large-Scale Conformational Changes in the HSP90 Molecular Chaperone. Mol. Cell 2011, 42, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Karagöz, G.E.; Rüdiger, S.G.D. HSP90 interaction with clients. Trends Biochem. Sci. 2015, 40, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Easton, D.P.; Kaneko, Y.; Subjeck, J.R. The HSP110 and GRP170 stress proteins: Newly recognized relatives of the HSP70s. Cell Stress Chaperones 2000, 5, 276–290. [Google Scholar] [CrossRef]
- Yasuda, K.; Nakai, A.; Hatayama, T.; Nagata, K. Cloning and expression of murine high molecular mass heat shock proteins, HSP105. J. Biol. Chem. 1995, 270, 29718–29723. [Google Scholar] [PubMed]
- Saito, Y.; Yamagishi, N.; Hatayama, T. Nuclear localization mechanism of HSP105beta and its possible function in mammalian cells. J. Biochem. 2009, 145, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Sciandra, J.J.; Subjeck, J.R.; Hughes, C.S. Induction of glucose-regulated proteins during anaerobic exposure and of heat-shock proteins after reoxygenation. Proc. Natl. Acad. Sci. USA 1984, 81, 4843–4847. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Masso-Welch, P.; Di, Y.P.; Cai, J.W.; Shen, J.W.; Subjeck, J.R. The 170-kDa glucose-regulated stress protein is an endoplasmic reticulum protein that binds immunoglobulin. Mol. Biol. Cell 1993, 4, 1109–1119. [Google Scholar] [CrossRef] [PubMed]
- Gething, M.J.; Sambrook, J. Protein folding in the cell. Nature 1992, 355, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Craig, E.A.; Gambill, B.D.; Nelson, R.J. Heat shock proteins: Molecular chaperones of protein biogenesis. Microbiol. Rev. 1993, 57, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Zuo, D.; Subjeck, J.; Wang, X.-Y. Unfolding the Role of Large Heat Shock Proteins: New Insights and Therapeutic Implications. Front. Immunol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Evgrafov, O.V.; Mersiyanova, I.; Irobi, J.; Van Den Bosch, L.V.; Dierick, I.; Leung, C.L.; Schagina, O.; Verpoorten, N.; Van Impe, K.; Fedotov, V.; et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat. Genet. 2004, 36, 602–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamin, I.J.; McMillan, D.R. Stress (Heat Shock) Proteins: Molecular Chaperones in Cardiovascular Biology and Disease. Circ. Res. 1998, 83, 117–132. [Google Scholar] [CrossRef] [PubMed]
- Wyttenbach, A.; Carmichael, J.; Swartz, J.; Furlong, R.A.; Narain, Y.; Rankin, J.; Rubinsztein, D.C. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington’s disease. Proc. Natl. Acad. Sci. USA 2000, 97, 2898–2903. [Google Scholar] [CrossRef] [PubMed]
- Pockley, A.G.; Wu, R.; Lemne, C.; Kiessling, R.; de Faire, U.; Frostegård, J. Circulating heat shock protein 60 is associated with early cardiovascular disease. Hypertension 2000, 36, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.Y.; Kim, N.; Kim, J.S.; Lim, S.H.; Jung, H.C.; Song, I.S. Heat shock protein gene 70-2 polymorphism is differentially associated with the clinical phenotypes of ulcerative colitis and Crohn’s disease. J. Gastroenterol. Hepatol. 2007, 22, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Christians, E.S.; Benjamin, I.J. Heat shock response: Lessons from mouse knockouts. Handb. Exp. Pharmacol. 2006, 172, 139–152. [Google Scholar] [CrossRef]
- Liu, H.; Dicksved, J.; Lundh, T.; Lindberg, J. Heat Shock Proteins: Intestinal Gatekeepers that Are Influenced by Dietary Components and the Gut Microbiota. Pathogens 2014, 3, 187–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Chauhan, N.R.; Chowdhury, D.; Singh, A.; Meena, R.C.; Chakrabarti, A.; Singh, S.B. Heat stress modulated gastrointestinal barrier dysfunction: Role of tight junctions and heat shock proteins. Scand. J. Gastroenterol. 2017, 52. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Andreoletti, G.; Chen, R.; Munehira, Y.; Batra, A.; Afzal, N.A.; Beattie, R.M.; Bernstein, J.A.; Ennis, S.; Snyder, M. De novo and rare mutations in the HSPA1L heat shock gene associated with inflammatory bowel disease. Genome Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Tahara, T.; Shibata, T.; Okubo, M.; Ishizuka, T.; Kawamura, T.; Yamashita, H.; Nakamura, M.; Nakagawa, Y.; Nagasaka, M.; Arisawa, T.; et al. Heat-shock protein 70-2 BB genotype is associated with reduced risks of the steroid-dependent and refractory phenotypes of ulcerative colitis. Biomed. Rep. 2014, 2, 555–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esaki, M.; Furuse, M.; Matsumoto, T.; Furuse, M.; Matsumoto, T.; Aoyagi, K.Y.J.; Yamagata, H.; Nakano, H.; Fujishima, M. Polymorphism of Heat-Shock Protein Gene HSP70-2 in Crohn Disease: Possible Genetic Marker for Two Forms of Crohn Disease. Scand. J. Gastroenterol. 1999, 34, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Debler, J.; Schiemann, U.; Seybold, U.; Mussack, T.; Landauer, N.; Ladurner, R.; Gross, M. Heat-shock protein HSP70-2 genotypes in patients with Crohn’s disease: A more severe clinical course with intestinal complications in presence of the PstI-polymorphism. Eur. J. Med. Res. 2003, 8, 120–124. [Google Scholar] [PubMed]
- Klausz, G.; Molnár, T.; Nagy, F.; Gyulai, Z.; Boda, K.; Lonovics, J.; Mándi, Y. Polymorphism of the heat-shock protein gene HSP70-2, but not polymorphisms of the IL-10 and CD14 genes, is associated with the outcome of Crohn’s disease. Scand. J. Gastroenterol. 2005, 40, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ren, J.; Gu, G.; Wang, G.; Wu, X.; Yan, D.; Liu, S.; Li, J. Crohn’s disease and polymorphism of heat shock protein gene HSP70-2 in the Chinese population. J. Gastroenterol. Hepatol. 2013, 28, 814–818. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, T.; Rios, Z.; Mei, Q.; Lin, X.; Cao, S. Heat Shock Proteins and Cancer. Trends Pharmacol. Sci. 2017, 38, 226–256. [Google Scholar] [CrossRef] [PubMed]
- Lasry, A.; Zinger, A.; Ben-Neriah, Y. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 2016, 17, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Desantis, C.; Jemal, A. Colorectal Cancer Statistics, 2014. CA Cancer J. Clin. 2014, 64, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Canadian Cancer Society’s Advisory Committee on Cancer Statistics Canadian Cancer Statistics 2017. Available online: http://www.cancer.ca/~/media/cancer.ca/CW/cancer%20information/cancer%20101/Canadian%20cancer%20statistics/Canadian-Cancer-Statistics-2017-EN.pdf (accessed on 13 August 2018).
- Yu, Z.; Zhi, J.; Peng, X.; Zhong, X.; Xu, A. Clinical significance of HSP27 expression in colorectal cancer. Mol. Med. Rep. 2010, 3, 953–958. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Zou, H.; Huang, Z. Effects of Helicobacter pylori and Heat Shock Protein 70 on the Proliferation of Human Gastric Epithelial Cells. Gastroenterol. Res. Pract. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Asea, A.A.A. HSP 70 in Human Diseases & Disorders; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2018; ISBN 978-3-319-89550-5. [Google Scholar]
- Tao, Y.; Hart, J.; Lichtenstein, L.; Joseph, L.J.; Ciancio, M.J.; Hu, S.; Chang, E.B.; Bissonnette, M. Inducible heat shock protein 70 prevents multifocal flat dysplastic lesions and invasive tumors in an inflammatory model of colon cancer. Carcinogenesis 2009, 30, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Zhu, X.; Triggs, J.R.; Tao, Y.; Wang, Y.; Lichtenstein, L.; Bissonnette, M.; Musch, M.W.; Chang, E.B. Inflammation-induced, 3’UTR-dependent translational inhibition of HSP70 mRNA impairs intestinal homeostasis. AJP Gastrointest. Liver Physiol. 2009, 296, G1003–G1011. [Google Scholar] [CrossRef] [PubMed]
- Jagadish, N.; Parashar, D.; Gupta, N.; Agarwal, S.; Suri, V.; Kumar, R.; Suri, V.; Sadasukhi, T.C.; Gupta, A.; Ansari, A.S.; et al. Heat shock protein 70-2 (HSP70-2) is a novel therapeutic target for colorectal cancer and is associated with tumor growth. BMC Cancer 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Shevtsov, M.; Multhoff, G. Heat shock protein-Peptide and HSP-based immunotherapies for the treatment of cancer. Front. Immunol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Jhaveri, K.; Ochiana, S.O.; Dunphy, M.P.; Gerecitano, J.F.; Corben, A.D.; Peter, R.I.; Janjigian, Y.Y.; Gomes-DaGama, E.M.; Koren, J., III; Modi, S.; et al. Heat shock protein 90 inhibitors in the treatment of cancer: Current status and future directions. Expert. Opin. Investig. Drugs 2014, 23, 611–628. [Google Scholar] [CrossRef] [PubMed]
- Chionh, Y.T.; Arulmuruganar, A.; Venditti, E.; Ng, G.Z.; Han, J.-X.; Entwisle, C.; Ang, C.-S.; Colaco, C.A.; McNulty, S.; Sutton, P. Heat shock protein complex vaccination induces protection against Helicobacter pylori without exogenous adjuvant. Vaccine 2014, 32, 2350–2358. [Google Scholar] [CrossRef] [PubMed]
- Caruso, R.; Warner, N.; Inohara, N.; Núñez, G. NOD1 and NOD2: Signaling, host defense, and inflammatory disease. Immunity 2014, 41, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Biasci, D.; Roberts, R.; Gearry, R.B.; Mansfield, J.C.; Ahmad, T.; Prescott, N.J.; Satsangi, J.; Wilson, D.C.; Jostins, L.; et al. Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat. Genet. 2017, 49, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Al Nabhani, Z.; Dietrich, G.; Hugot, J.P.; Barreau, F. Nod2: The intestinal gate keeper. PLoS Pathog. 2017, 13, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Hsu, L.; Liu, H.; Bankston, L.A.; Iimura, M.; Kagnoff, M.F. Nod2 Mutation in Crohn’s Disease Potentiates NF-κB Activity and IL-1 b Processing. Science 2005, 307, 734–738. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J. Beyond peptidoglycan for Nod2. Nat. Immunol. 2009, 10, 1053–1054. [Google Scholar] [CrossRef] [PubMed]
- Hedl, M.; Li, J.; Cho, J.H.; Abraham, C. Chronic stimulation of NOD2 mediates tolerance to bacterial products. Proc. Natl. Acad. Sci. USA 2007, 104, 19440–19445. [Google Scholar] [CrossRef] [PubMed]
- Balzola, F.; Bernstein, C.; Van Assche, G. Nod2 is required for the regulation of commensal microbiota in the intestine: Commentary. Inflamm. Bowel Dis. Monit. 2010, 10, 100–101. [Google Scholar]
- Ramanan, D.; Tang, M.S.; Bowcutt, R.; Loke, P.; Cadwell, K. Bacterial sensor NOD2 prevents inflammation of the small intestine by restricting the expansion of the commensal bacteroides vulgatus. Immunity 2014, 41, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Balzola, F.; Bernstein, C.; Ho, G.T.; Lees, C. Induction and rescue of NOD2-dependent TH1-driven granulomatous inflammation of the ileum: Commentary. Inflamm. Bowel Dis. Monit. 2011, 11, 123–124. [Google Scholar]
- Kobayashi, K.S.K. Nod2-Dependent Regulation of Innate and Adaptive Immunity in the Intestinal Tract. Science 2005, 307, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Mayor, A.; Martinon, F.; De Smedt, T.; Pétrilli, V.; Tschopp, J. A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat. Immunol. 2007, 8, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Villani, A.C.; Lemire, M.; Fortin, G.; Louis, E.; Silverberg, M.S.; Collette, C.; Baba, N.; Libioulle, C.; Belaiche, J.; Bitton, A.; et al. Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat. Genet. 2009, 41, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Kitani, A.; Similuk, M.; Oler, A.J.; Albenberg, L.; Kelsen, J.; Aktay, A.; Quezado, M.; Yao, M.; Montgomery-Recht, K.; et al. Loss-of-function CARD8 mutation causes NLRP3 inflammasome activation and Crohn’s disease. J. Clin. Investig. 2018, 128, 1793–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Dong, Y.; Ye, M.; Jin, S.; Yang, J.; Joosse, M.E.; Sun, Y.; Zhang, J.; Lazarev, M.; Brant, S.R.; et al. The Pathogenic Role of NLRP3 Inflammasome Activation in Inflammatory Bowel Diseases of Both Mice and Humans. J. Crohns Colitis 2016, jjw219. [Google Scholar] [CrossRef] [PubMed]
- Zaki, M.H.; Boyd, K.L.; Vogel, P.; Kastan, M.B.; Lamkanfi, M.; Kanneganti, T.D. The NLRP3 Inflammasome Protects against Loss of Epithelial Integrity and Mortality during Experimental Colitis. Immunity 2010, 32, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-H.; Biswas, A.; Liu, Y.-J.; Kobayashi, K.S. Proteasomal Degradation of NOD2 Protein Mediates Tolerance to Bacterial Cell Wall Components. J. Biol. Chem. 2012, 287, 39800–39811. [Google Scholar] [CrossRef] [PubMed]
- Boyle, J.P.; Parkhouse, R.; Monie, T.P. Insights into the molecular basis of the NOD2 signalling pathway. Open Biol. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Sutterwala, F.S.; Haasken, S.; Cassel, S.L. Mechanism of NLRP3 inflammasome activation. Ann. N. Y. Acad. Sci. 2014, 1319, 82–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohanan, V.; Grimes, C.L. The molecular chaperone HSP70 binds to and stabilizes NOD2, an important protein involved in crohn disease. J. Biol. Chem. 2014, 289, 18987–18998. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.K.; Wastyk, H.C.; Mohanan, V.; Hou, C.W.; Lauro, M.L.; Melnyk, J.E.; Burch, J.M.; Grimes, C.L. Crohn’s Disease Variants of Nod2 Are Stabilized by the Critical Contact Region of HSP70. Biochemistry 2017, 56, 4445–4448. [Google Scholar] [CrossRef] [PubMed]
- Inohara, N.; Ogura, Y.; Fontalba, A.; Gutierrez, O.; Pons, F.; Crespo, J.; Fukase, K.; Inamura, S.; Kusumoto, S.; Hashimoto, M.; et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2: Implications for Crohn’s disease. J. Biol. Chem. 2003, 278, 5509–5512. [Google Scholar] [CrossRef] [PubMed]
- Åkerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haslbeck, M.; Walke, S.; Stromer, T.; Ehrnsperger, M.; White, H.E.; Chen, S.; Saibil, H.R.; Buchner, J.; Allen, S.; Polazzi, J.; et al. HSP26: A temperature-regulated chaperone. EMBO J. 1999, 18, 6744–6751. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus-Steinmetz, U.; Rensing, L. Heat shock protein induction by certain chemical stressors is correlated with their cytotoxicity, lipophilicity and protein-denaturing capacity. Toxicology 1997, 123, 185–195. [Google Scholar] [CrossRef]
- Beere, H.M. “The stress of dying”: The role of heat shock proteins in the regulation of apoptosis. J. Cell Sci. 2004, 117, 2641–2651. [Google Scholar] [CrossRef] [PubMed]
- Arvans, D.; Vavricka, S.; Ren, H.; Musch, M.; Kang, L.; Rocha, F.; Lucioni, A.; Turner, J.; Alverdy, J.; Chang, E. Luminal bacterial flora determines physiological expression of intestinal epithelial cytoprotective heat shock proteins 25 and 72. AJP Gastrointest. Liver Physiol. 2005, 288, G696–G704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, K.; Musch, M.W.; Ren, H.; Boone, D.L.; Hendrickson, B.A.; Ma, A.; Chang, E.B. Enteric flora and lymphocyte-derived cytokines determine expression of heat shock proteins in mouse colonic epithelial cells. Gastroenterology 2003, 124, 1395–1407. [Google Scholar] [CrossRef]
- Hu, S.; Wang, Y.; Lichtenstein, L.; Tao, Y.; Musch, M.W.; Jabri, B.; Antonopoulos, D.; Claud, E.C.; Chang, E.B. Regional differences in colonic mucosa-associated microbiota determine the physiological expression of host heat shock proteins. Am. J. Physiol.-Gastrointest. Liver Physiol. 2010, 299, G1266–G1275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, K.; Musch, M.W.; Ropeleski, M.J.; Boone, D.L.; Ma, A.; Chang, E.B. Escherichia coli LPS induces heat shock protein 25 in intestinal epithelial cells through MAP kinase activation. Am. J. Physiol.-Gastrointest. Liver Physiol. 2004, 286, G645–G652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, H.; Musch, M.W.; Kojima, K.; Boone, D.; Ma, A.; Chang, E.B. Short-chain fatty acids induce intestinal epithelial heat shock protein 25 expression in rats and IEC 18 cells. Gastroenterology 2001, 121, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Koninkx, J.F.J.G.; Tooten, P.C.J.; Malago, J.J. Probiotic bacteria induced improvement of the mucosal integrity of enterocyte-like Caco-2 cells after exposure to Salmonella enteritidis 857. J. Funct. Foods 2010, 2, 225–234. [Google Scholar] [CrossRef]
- Tao, Y.; Drabik, K.A.; Waypa, T.S.; Musch, M.W.; Alverdy, J.C.; Schneewind, O.; Chang, E.B.; Petrof, E.O. Soluble factors from Lactobacillus GG activate MAPKs and induce cytoprotective heat shock proteins in intestinal epithelial cells. Am. J. Physiol.-Cell Physiol. 2006, 290, C1018–C1030. [Google Scholar] [CrossRef] [PubMed]
- Fujiya, M.; Musch, M.W.; Nakagawa, Y.; Hu, S.; Alverdy, J.; Kohgo, Y.; Schneewind, O.; Jabri, B.; Chang, E.B. The Bacillus subtilis Quorum-Sensing Molecule CSF Contributes to Intestinal Homeostasis via OCTN2, a Host Cell Membrane Transporter. Cell Host Microbe 2007, 1, 299–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueno, N.; Fujiya, M.; Segawa, S.; Nata, T.; Moriichi, K.; Tanabe, H.; Mizukami, Y.; Kobayashi, N.; Ito, K.; Kohgo, Y. Heat-killed body of lactobacillus brevis SBC8803 ameliorates intestinal injury in a murine model of colitis by enhancing the intestinal barrier function. Inflamm. Bowel Dis. 2011, 17, 2235–2250. [Google Scholar] [CrossRef] [PubMed]
- Paszti-Gere, E.; Szeker, K.; Csibrik-Nemeth, E.; Csizinszky, R.; Marosi, A.; Palocz, O.; Farkas, O.; Galfi, P. Metabolites of lactobacillus plantarum 2142 prevent oxidative stress-induced overexpression of proinflammatory cytokines in IPEC-J2 cell line. Inflammation 2012, 35, 1487–1499. [Google Scholar] [CrossRef] [PubMed]
- Malago, J.J.; Koninkx, J.F.J.G.; Ovelgönne, H.H.; van Asten, F.J.A.M.; Swennenhuis, J.F.; van Dijk, J.E. Expression levels of heat shock proteins in enterocyte-like Caco-2 cells after exposure to Salmonella enteritidis. Cell Stress Chaperones 2003, 8, 194–203. [Google Scholar] [CrossRef]
- Okamoto, K.; Fujiya, M.; Nata, T.; Ueno, N.; Inaba, Y.; Ishikawa, C.; Ito, T.; Moriichi, K.; Tanabe, H.; Mizukami, Y.; et al. Competence and sporulation factor derived from Bacillus subtilis improves epithelial cell injury in intestinal inflammation via immunomodulation and cytoprotection. Int. J. Colorectal Dis. 2012, 27, 1039–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, K.J.; Ullman, T.A.; Ford, A.C.; Abreu, M.T.; Abadir, A.; Marshall, J.K.; Talley, N.J.; Moayyedi, P. Antibiotic Therapy in Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 2011, 106, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Francino, M.P. Antibiotics and the human gut microbiome: Dysbioses and accumulation of resistances. Front. Microbiol. 2016, 6, 1543. [Google Scholar] [CrossRef] [PubMed]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Zare, A.; Hajhashemi, M.; Hassan, Z.M.; Zarrin, S.; Pourpak, Z.; Moin, M.; Salarilak, S.; Masudi, S.; Shahabi, S. Effect of Ramadan fasting on serum heat shock protein 70 and serum lipid profile. Sing. Med. J. 2011, 52, 491–495. [Google Scholar]
- Lallès, J.P.; David, J.C. Fasting and refeeding modulate the expression of stress proteins along the gastrointestinal tract of weaned pigs. J. Anim. Physiol. Anim. Nutr. 2011, 95, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Grongnet, J.F.; David, J.C. Reciprocal variations of nNOS and HSP90 are associated with fasting in gastrointestinal tract of the piglet. Digest. Dis. Sci. 2003, 48, 365–372. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singleton, K.D.; Wischmeyer, P.E. Glutamine’s protection against sepsis and lung injury is dependent on heat shock protein 70 expression. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 292, R1839–R1845. [Google Scholar] [CrossRef] [PubMed]
- Moura, C.S.; Lollo, P.C.B.; Morato, P.N.; Amaya-Farfan, J. Dietary nutrients and bioactive substances modulate heat shock protein (HSP) expression: A review. Nutrients 2018, 10, 683. [Google Scholar] [CrossRef] [PubMed]
- Lenaerts, K.; Renes, J.; Bouwman, F.G.; Noben, J.P.; Robben, J.; Smit, E.; Mariman, E.C. Arginine deficiency in preconfluent intestinal Caco-2 cells modulates expression of proteins involved in proliferation, apoptosis, and heat shock response. Proteomics 2007, 7, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Phanvijhitsiri, K.; Musch, M.W.; Ropeleski, M.J.; Chang, E.B. Heat induction of heat shock protein 25 requires cellular glutamine in intestinal epithelial cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 291, C290-9. [Google Scholar] [CrossRef] [PubMed]
- Baird, C.H.; Niederlechner, S.; Beck, R.; Kallweit, A.R.; Wischmeyer, P.E. L-Threonine induces heat shock protein expression and decreases apoptosis in heat-stressed intestinal epithelial cells. Nutrition 2013, 29, 1404–1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Moura, C.S.; Lollo, P.C.B.; Morato, P.N.; Carneiro, E.M.; Amaya-Farfan, J. Whey protein hydrolysate enhances the exercise-induced heat shock protein (HSP70) response in rats. Food Chem. 2013, 136, 1350–1357. [Google Scholar] [CrossRef] [PubMed]
- Ovelgonne, J.; Koninkx, J.; Pusztai, A.; Bardocz, S.; Kok, W.; Ewen, S.; Hendriks, H.; van Dijk, J.E. Decreased levels of heat shock proteins in gut epithelial cells after exposure to plant lectins. Gut 2000, 46, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Ogata, M.; Van Hung, T.; Tari, H.; Arakawa, T.; Suzuki, T. Dietary psyllium fiber increases intestinal heat shock protein 25 expression in mice. Nutr Res. 2017, 39, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Hirayoshi, K.; Nakai, A.; Hosokawa, Y.; Marui, N.; Yoshida, M.; Sakai, T.; Nishino, H.; Aoike, A.; Kawai, K.; et al. Flavonoids inhibit the expression of heat shock proteins. Cell Struct. Funct. 1990, 15, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, L.S.; Catalão, C.H.R.; Felippotti, T.T.; De Oliveira-Pelegrin, G.R.; Petenusci, S.; De Freitas, L.A.P.; Rocha, M.J.A. Curcumin suppresses inflammatory cytokines and heat shock protein 70 release and improves metabolic parameters during experimental sepsis. Pharm. Biol. 2017, 55, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Wieten, L.; Van Der Zee, R.; Spiering, R.; Wagenaar-Hilbers, J.; Van Kooten, P.; Broere, F.; Van Eden, W. A novel heat-shock protein coinducer boosts stress protein HSP70 to activate T cell regulation of inflammation in autoimmune arthritis. Arthritis Rheum. 2010, 62, 1026–1035. [Google Scholar] [CrossRef] [PubMed]
- Arnal, M.E.; Lallés, J.P. Gut epithelial inducible heat-shock proteins and their modulation by diet and the microbiota. Nutr. Rev. 2016, 74, 181–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruemmele, F.M.; Veres, G.; Kolho, K.L.; Griffiths, A.; Levine, A.; Escher, J.C.; Amil Dias, J.; Barabino, A.; Braegger, C.P.; Bronsky, J.; et al. Consensus guidelines of ECCO/ESPGHAN on the medical management of pediatric Crohn’s disease. J. Crohns Colitis 2014, 8, 1179–1207. [Google Scholar] [CrossRef] [PubMed]
- Connors, J.; Basseri, S.; Grant, A.; Giffin, N.; Mahdi, G.; Noble, A.; Rashid, M.; Otley, A.; Van Limbergen, J. Exclusive enteral nutrition therapy in paediatric Crohn’s disease results in long-term avoidance of corticosteroids: Results of a propensity-score matched cohort analysis. J. Crohns Colitis 2017, 11, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- MacLellan, A.; Connors, J.; Grant, S.; Cahill, L.; Langille, M.G.I.; Van Limbergen, J. The impact of exclusive enteral nutrition (EEN) on the gut microbiome in Crohn’s disease: A review. Nutrients 2017, 9, 447. [Google Scholar] [CrossRef] [PubMed]
- Otley, A.R.; Russell, R.K.; Day, A.S. Nutritional therapy for the treatment of pediatric Crohns disease. Expert Rev. Clin. Immunol. 2010, 6, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Wine, E. Effects of enteral nutrition on Crohn’s Disease: Clues to the impact of diet on disease pathogenesis. Inflamm. Bowel Dis. 2013, 19, 1322–1329. [Google Scholar] [CrossRef] [PubMed]
- Day, A.S.; Lopez, R.N. Exclusive enteral nutrition in children with crohn’s disease. World J. Gastroenterol. 2015, 21, 6809–6816. [Google Scholar] [CrossRef] [PubMed]
- Shinnick, T.M. Heat shock proteins as antigens of bacterial and parasitic pathogens. In Heat Shock Proteins and Immune Response; Springer-Verlag: Berlin, Germany, 1990; pp. 145–160. ISBN 3540528571. [Google Scholar]
- Kaul, G.; Thippeswamy, H. Role of Heat Shock Proteins in Diseases and Their Therapeutic Potential. Ind. J. Microbiol. 2011, 51, 124–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, J. Evolution of heat shock protein and immunity. Dev. Comp. Immunol. 2003, 27, 449–464. [Google Scholar] [CrossRef]
- Verbrugghe, E.; Van Parys, A.; Leyman, B.; Boyen, F.; Haesebrouck, F.; Pasmans, F. HtpG contributes to Salmonella Typhimurium intestinal persistence in pigs. Vet. Res. 2015, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, A.M.; Pretre, G.; Bartpho, T.; Sermswan, R.W.; Toma, C.; Suzuki, T.; Eshghi, A.; Picardeau, M.; Adler, B.; Murray, G.L. High-temperature protein G is an essential virulence factor of Leptospira interrogans. Infect. Immun. 2014, 82, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Dang, W.; Hu, Y.H.; Sun, L. HtpG is involved in the pathogenesis of Edwardsiella tarda. Vet. Microbiol. 2011, 152, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Weiss, D.S.; Brotcke, A.; Henry, T.; Margolis, J.J.; Chan, K.; Monack, D.M. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. USA 2007, 104, 6037–6042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelburne, C.E.; Coopamah, M.D.; Sweier, D.G.; An, F.Y.P.; Lopatin, D.E. HtpG, the Porphyromonas gingivalis HSP-90 homologue, induces the chemokine CXCL8 in human monocytic and microvascular vein endothelial cells. Cell. Microbiol. 2007, 9, 1611–1619. [Google Scholar] [CrossRef] [PubMed]
- Shelburne, C.E.; Shelburne, P.S.; Dhople, V.M.; Sweier, D.G.; Giannobile, W.V.; Kinney, J.S.; Coulter, W.A.; Mullally, B.H.; Lopatin, D.E. Serum antibodies to Prophyromonas gingivalis chaperon HtpG predict health in periodontitis susceptible patients. PLoS ONE 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Dunn, K.A.; Moore-Connors, J.; Macintyre, B.; Stadnyk, A.; Thomas, N.A.; Noble, A.; Mahdi, G.; Rashid, M.; Otley, A.R.; Bielawski, J.P.; et al. The Gut Microbiome of Pediatric Crohn’s Disease Patients Differs from Healthy Controls in Genes That Can Influence the Balance between a Healthy and Dysregulated Immune Response. Inflamm. Bowel Dis. 2016, 22, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Tokuriki, N.; Tawfik, D.S. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 2009, 459, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Wyganowski, K.T.; Kaltenbach, M.; Tokuriki, N. GroEL/ES buffering and compensatory mutations promote protein evolution by stabilizing folding intermediates. J. Mol. Biol. 2013, 425, 3403–3414. [Google Scholar] [CrossRef] [PubMed]
- Ohue, R.; Hashimoto, K.; Nakamoto, M.; Furukawa, Y.; Masuda, T.; Kitabatake, N.; Tani, F. Bacterial heat shock protein 60, GroEL, can induce the conversion of naive T cells into a CD4 CD25(+) Foxp3-expressing phenotype. J. Innate Immun. 2011, 3, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Nalbant, A.; Kant, M. Bacterial heat shock protein GroEL (HSP64) exerts immunoregulatory effects on T cells by utilizing apoptosis. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Tabona, P.; Reddi, K.; Khan, S.; Nair, S.P.; Crean, S.J.; Meghji, S.; Wilson, M.; Preuss, M.; Miller, A.D.; Poole, S.; et al. Homogeneous Escherichia coli chaperonin 60 induces IL-1 beta and IL-6 gene expression in human monocytes by a mechanism independent of protein conformation. J. Immunol. 1998, 161, 1414–1421. [Google Scholar] [PubMed]
- Retzlaff, C.; Yamamoto, Y.; Hoffman, P.S.; Friedman, H.; Klein, T.W. Bacterial heat shock proteins directly induce cytokine mRNA and interleukin-1 secretion in macrophage cultures. Infect. Immun. 1994, 62, 5689–5693. [Google Scholar] [PubMed]
- Arora, G.; Sajid, A.; Virmani, R.; Singhal, A.; Kumar, C.M.S.; Dhasmana, N.; Khanna, T.; Maji, A.; Misra, R.; Molle, V.; et al. Ser/Thr protein kinase PrkC-mediated regulation of GroEL is critical for biofilm formation in Bacillus anthracis. NPJ Biofilms Microbiomes 2017, 3. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.-M.; Pinn, M.L.; Karakousis, P.C.; Hung, C.-F. Intranasal Immunization with DNAK Protein Induces Protective Mucosal Immunity against Tuberculosis in CD4-Depleted Mice. Front. Cell. Infect. Microbiol. 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Okuda, J.; Yamane, S.; Nagata, S.; Kunikata, C.; Suezawa, C.; Yasuda, M. The pseudomonas aeruginosa dnaK gene is involved in bacterial translocation across the intestinal epithelial cell barrier. Microbiology (UK) 2017, 163, 1208–1216. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Finlayson-Trick, E.; Connors, J.; Stadnyk, A.; Van Limbergen, J. Regulation of Antimicrobial Pathways by Endogenous Heat Shock Proteins in Gastrointestinal Disorders. Gastrointest. Disord. 2019, 1, 39-56. https://doi.org/10.3390/gidisord1010005
Finlayson-Trick E, Connors J, Stadnyk A, Van Limbergen J. Regulation of Antimicrobial Pathways by Endogenous Heat Shock Proteins in Gastrointestinal Disorders. Gastrointestinal Disorders. 2019; 1(1):39-56. https://doi.org/10.3390/gidisord1010005
Chicago/Turabian StyleFinlayson-Trick, Emma, Jessica Connors, Andrew Stadnyk, and Johan Van Limbergen. 2019. "Regulation of Antimicrobial Pathways by Endogenous Heat Shock Proteins in Gastrointestinal Disorders" Gastrointestinal Disorders 1, no. 1: 39-56. https://doi.org/10.3390/gidisord1010005





