Underneath the Purple Stain
Abstract
1. Introduction
2. Materials and Methods
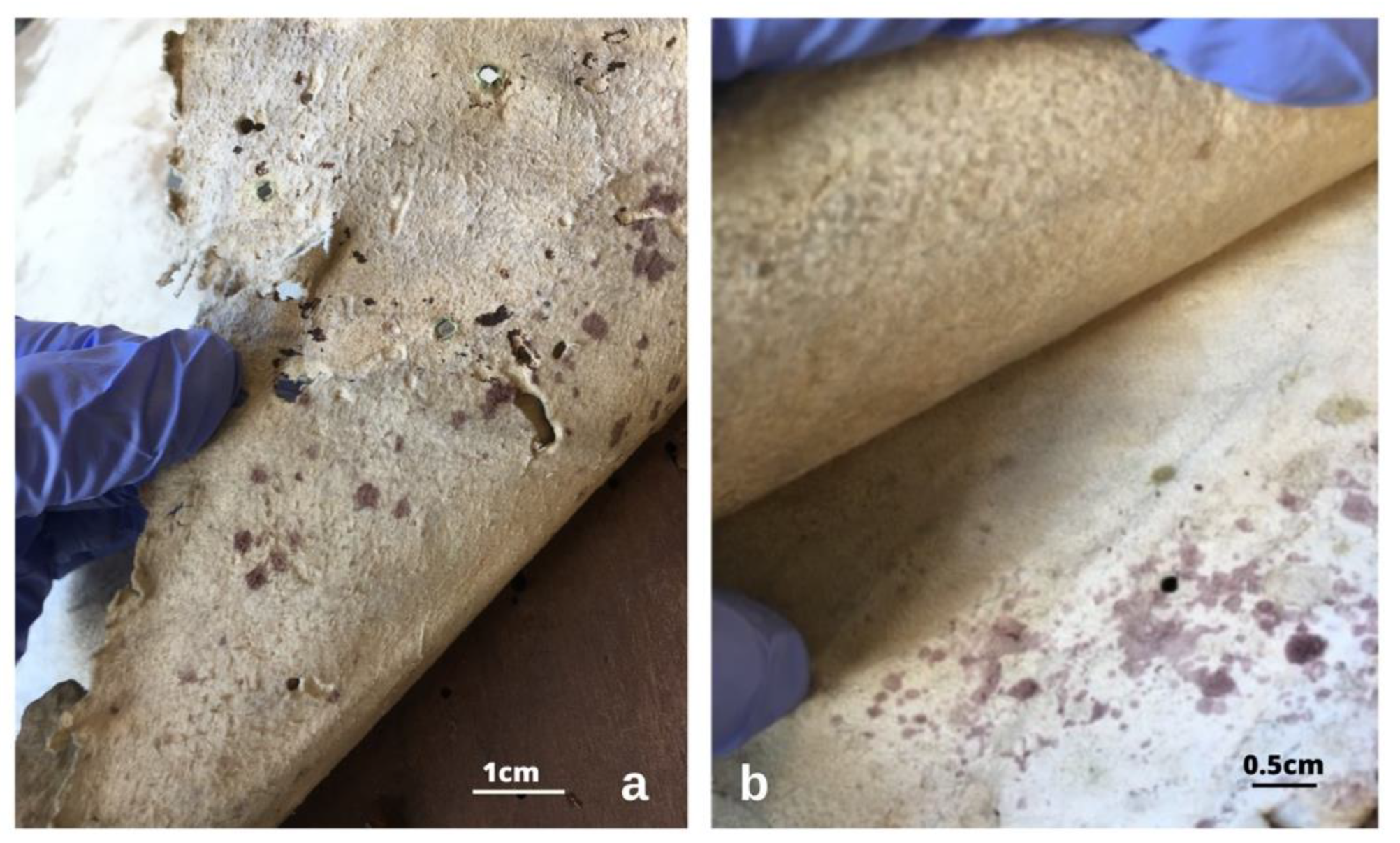
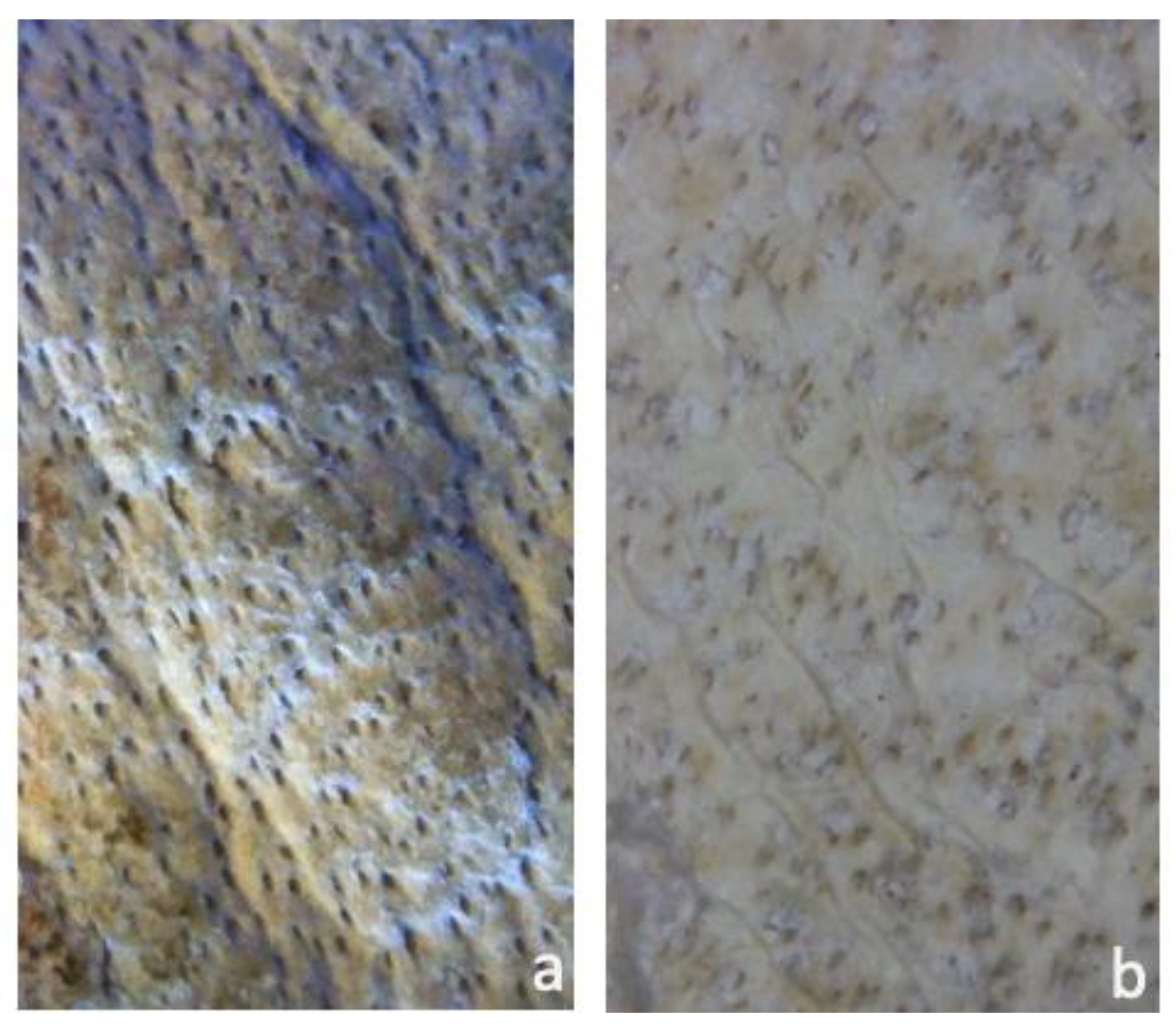
2.1. Visual Inspection
2.2. Fungal Communities
2.3. DNA Extraction and Sequencing
3. Results and Discussion
3.1. General Quality and State of Conservation for Leather and Parchment
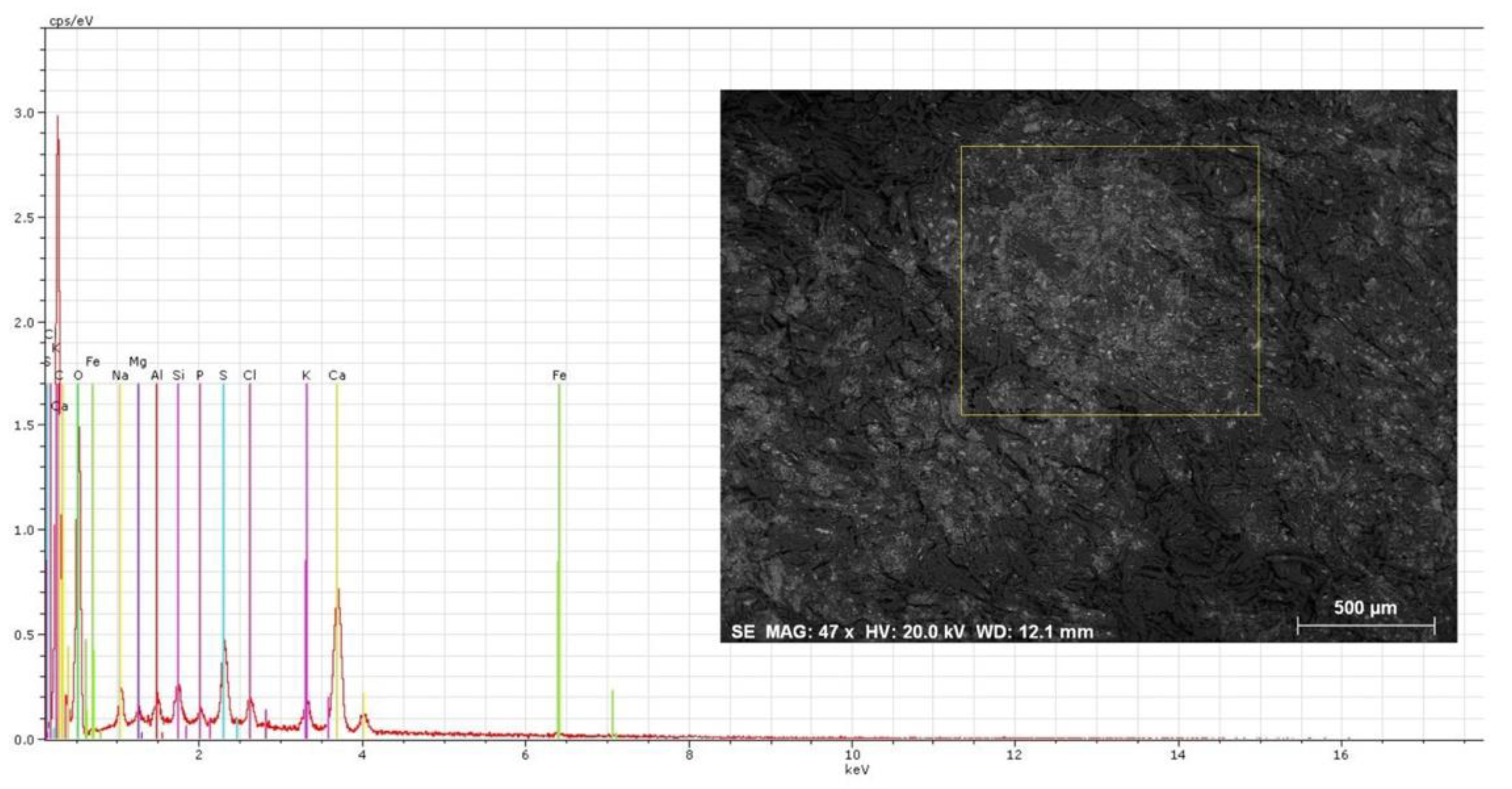
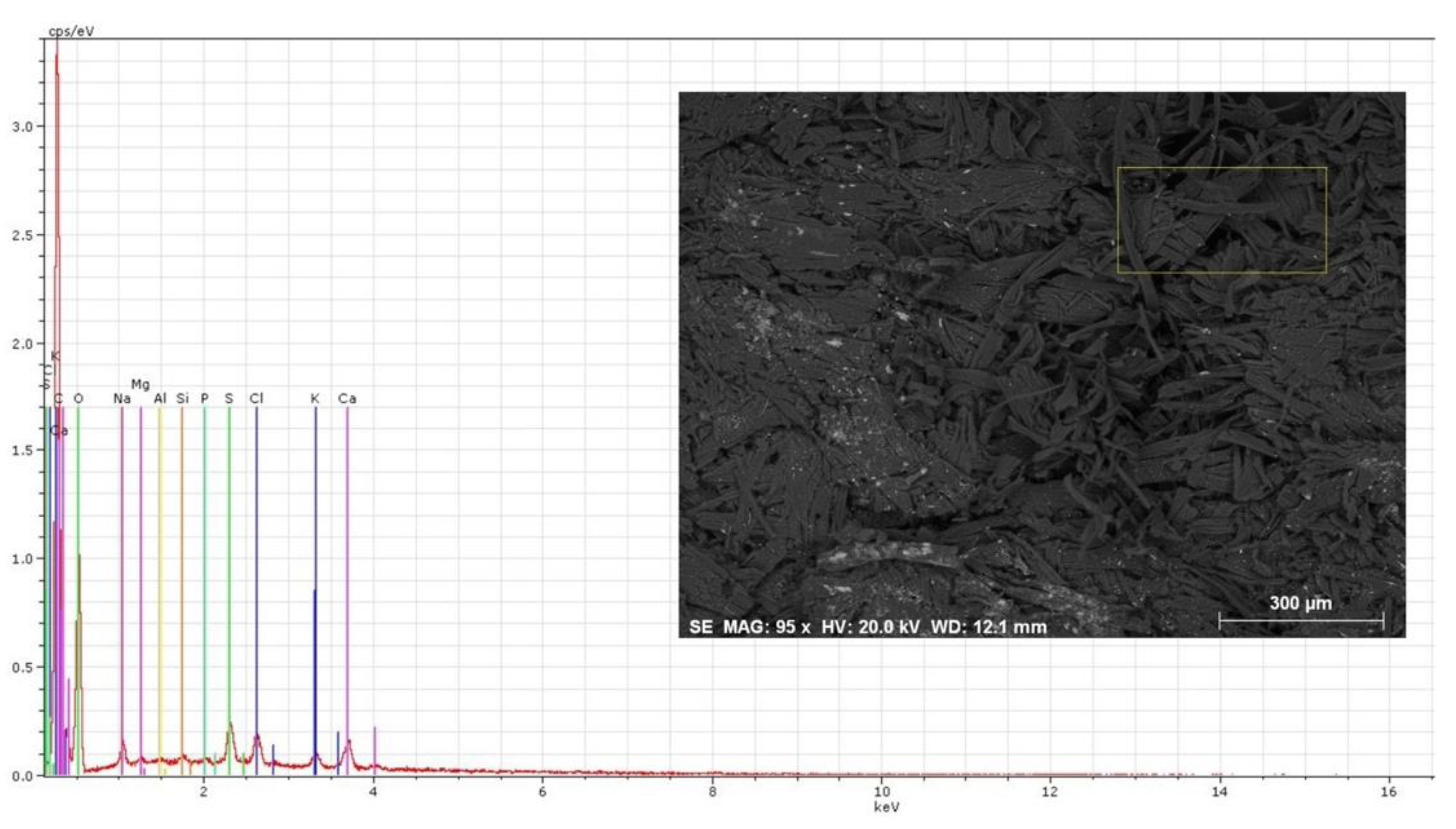
3.2. Chemical Profile by X-ray Energy Dispersive Spectroscopy
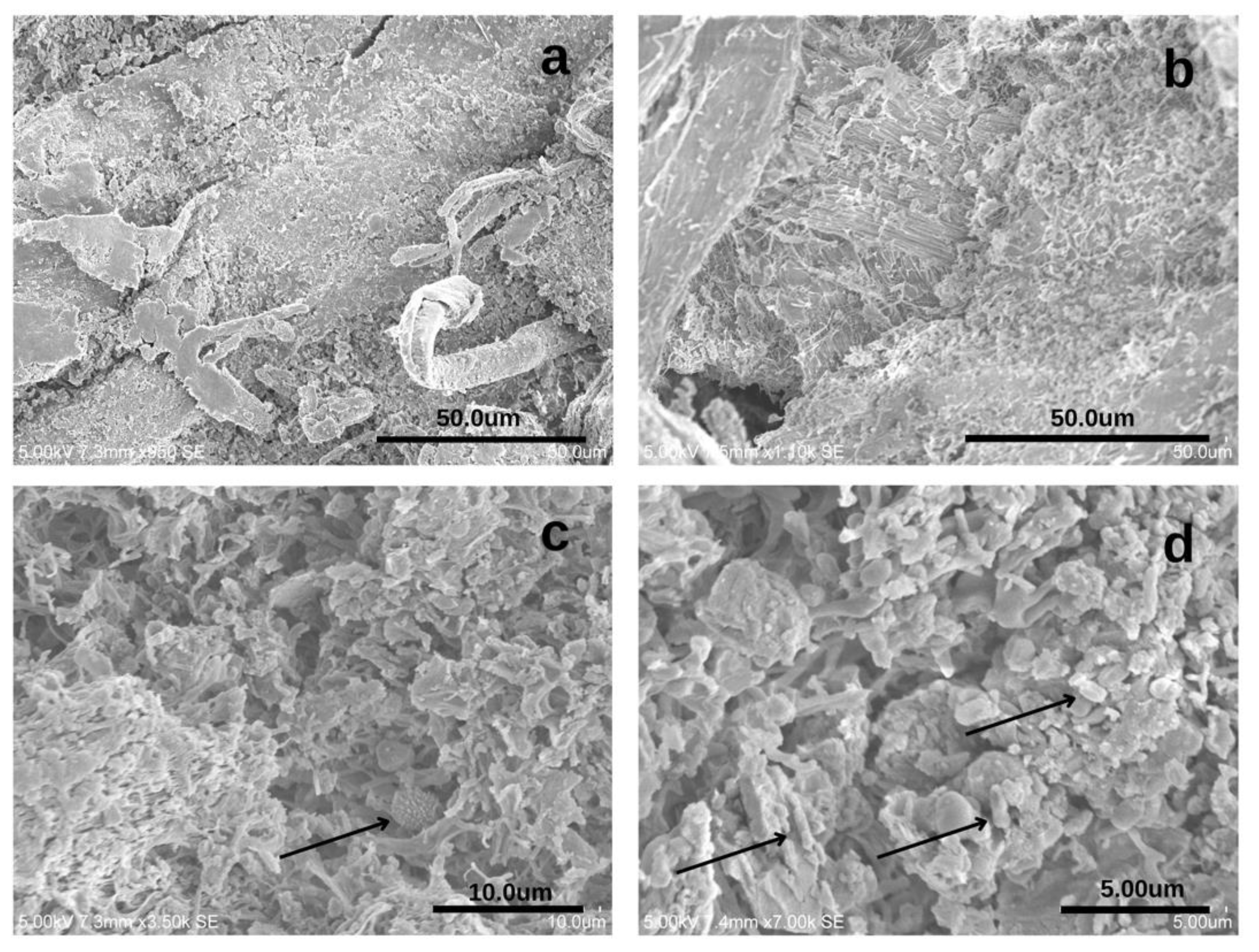
3.3. Biological Communities by Visual Inspection—Scanning Electron Microscopy
3.4. Fungal Cultivated Communities
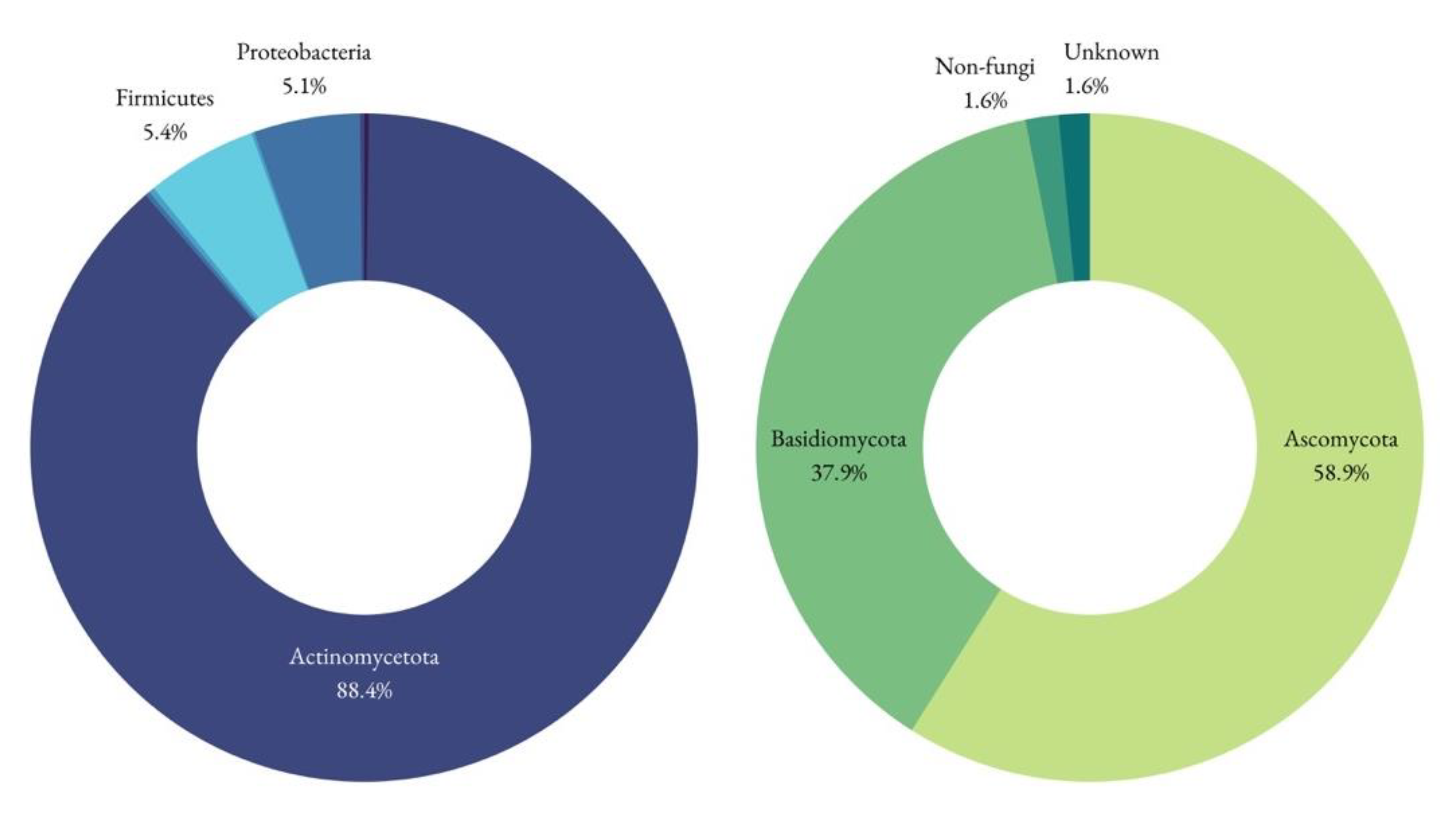
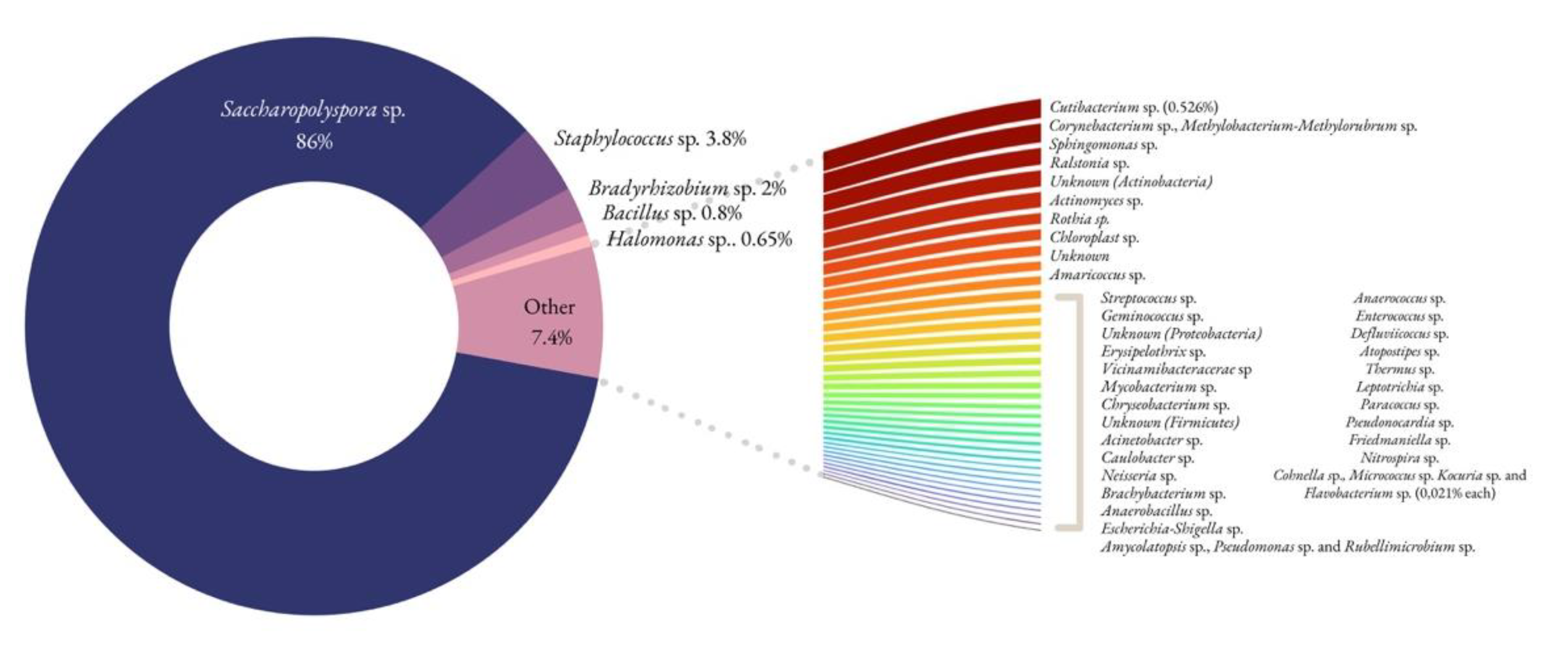
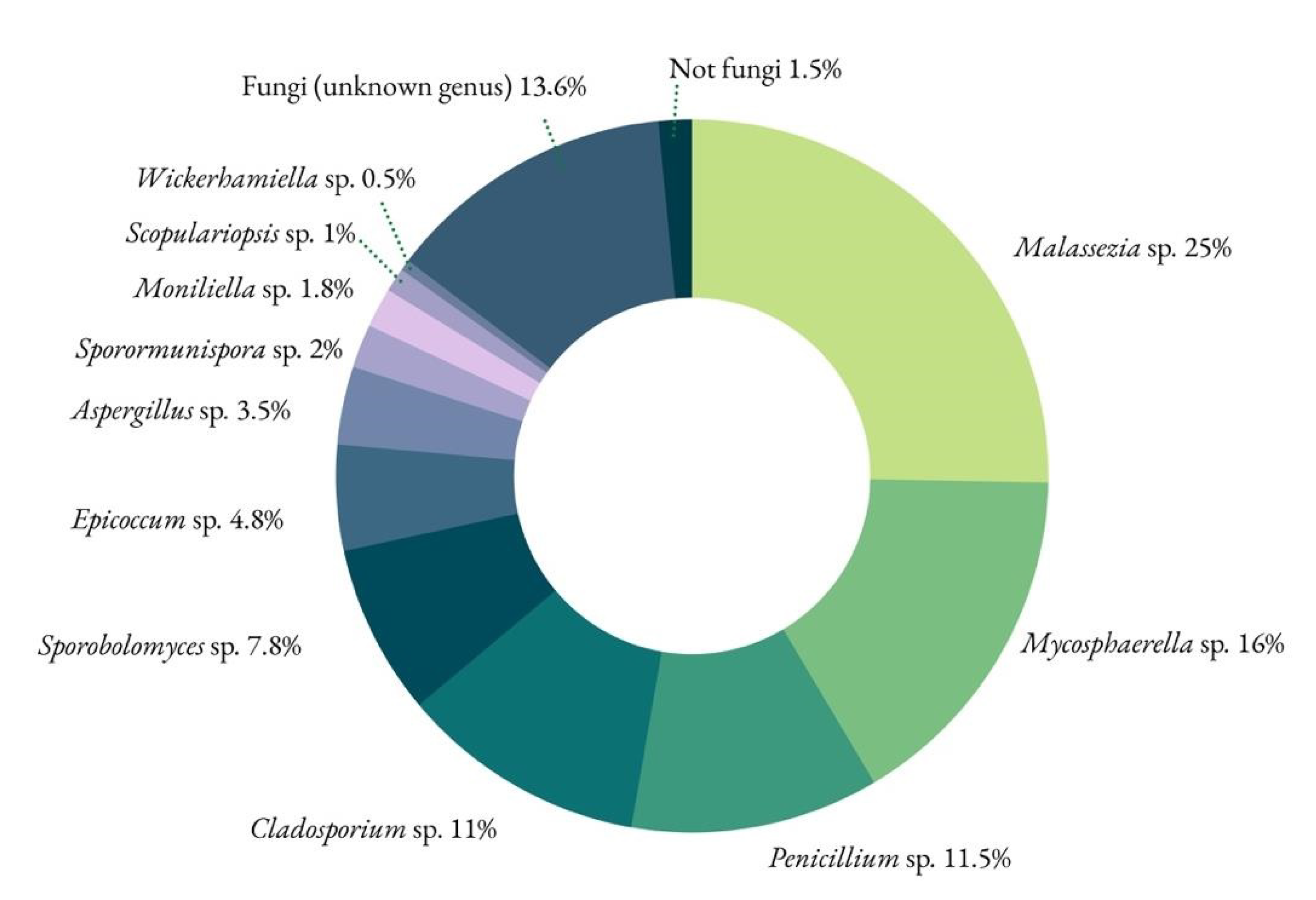
3.5. Biological Communities by NGS
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Miguélez Cavero, A.; Melo, M.J.; Miranda, M.A.; Castro, R.; Casanova, C. Beatus manuscripts under the microscope: The Alcobaça Beatus and the Iberian Cistercian tradition revisited. J. Mediev. Iber Stud. 2016, 8, 217–251. [Google Scholar] [CrossRef]
- Barreira, C. Approaches to the study of a fourteenth-century breviary from the cistercian abbey of Alcobaça. Citeaux 2017, 68, 249–276. [Google Scholar]
- Nascimento, A.A.; Diogo, A.D. Encadernação Portuguesa Medieval—Alcobaça [Medieval Portuguese Bookbinding: Alcobaça]; Imprensa Nacional da Casa da Moeda: Lisbon, Portugal, 1984; 109p. [Google Scholar]
- Tourais, A. Development of a Material Characterization Method for the Medieval Codices of the Collection from the Monastery of Alcobaça; Faculdade de Ciências e Tecnologia da Universidade Nova de Lisboa: Lisboa, Portugal, 2020; Available online: http://hdl.handle.net/10362/114642 (accessed on 17 October 2022).
- Tourais, A.; Casanova, C.; Barreira, C.F. Filling the gap: New approaches to medieval bookbinding studies. J. Mediev. Iber. Stud. 2022, 14, 109–126. [Google Scholar] [CrossRef]
- Larsen, R. Introduction to damage and damage assessment of parchment. In Improved Damage Assessment of Parchment (IDAP), Assessment, Data Collection and Sharing of Knowledge; Larsen, R., Ed.; Office for Official Publications of the European Communities: Luxembourg, 2007; pp. 17–21. [Google Scholar]
- Hallebeek, P.B. Guidelines for the Conservation of Leather and Parchment; Koninklijke Bibliotheek: Amsterdam, The Netherlands, 1997. [Google Scholar]
- Thomas, S. Leathermaking in the Middle Ages. In Leather Manufacture through the Ages; Thomas, S., Clarkson, L., Thomson, R., Eds.; Arkle Print Limited: Northampton, UK, 1983; pp. 1–10. [Google Scholar]
- Clarkson, C. Rediscovering parchment: The nature of the beast. Pap. Conserv. 1992, 16, 5–26. [Google Scholar] [CrossRef]
- Piñar, G.; Sterflinger, K.; Pinzari, F. Unmasking the measles-like parchment discoloration: Molecular and microanalytical approach. Environ. Microbiol. 2015, 17, 427–443. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, S. Role of Cyanobacteria in Biodeterioration of Historical Monuments—A Review. BMR Microbiol. 2014, 1, 1–13. [Google Scholar]
- Warscheid, T.; Braams, J. Biodeterioration of stone: A review. Int. Biodeterior. Biodegrad. 2000, 46, 343–368. [Google Scholar] [CrossRef]
- Sterflinger, K.; Piñar, G. Microbial deterioration of cultural heritage and works of art—Tilting at windmills? Appl. Microbiol. Biotechnol. 2013, 97, 9637–9646. [Google Scholar] [CrossRef]
- Villa, F.; Wu, Y.-L.; Zerboni, A.; Cappitelli, F. In Living Color: Pigment-Based Microbial Ecology at the Mineral–Air Interface. Bioscience 2022, 72, 1156–1175. [Google Scholar] [CrossRef]
- Pinheiro, A.C.; Sequeira, S.O.; Macedo, M.F. Fungi in archives, libraries, and museums: A review on paper conservation and human health. Crit. Rev. Microbiol. 2019, 45, 686–700. [Google Scholar] [CrossRef]
- Karbowska-Berent, J.; Strzelczyk, A. The Role of Streptomycetes in the Biodeterioration of Historic Parchment; Copernicus University Press: Torun, Poland, 2000. [Google Scholar]
- Gallo, F.; Strzelczyk, A. Indagine preliminare sulle alterazioni microbiche della pergamena. Bollett. Ist. Centr. Patologia Libro “Alfonso Gallo.” 1971, 30, 71–87. [Google Scholar]
- Migliore, L.; Thaller, M.C.; Vendittozzi, G.; Mejia, A.Y.; Mercuri, F.; Orlanducci, S.; Rubechini, A. Purple spot damage dynamics investigated by an integrated approach on a 1244 A.D. parchment roll from the Secret Vatican Archive. Sci. Rep. 2017, 7, 9521. [Google Scholar] [CrossRef] [PubMed]
- Perini, N.; Mercuri, F.; Orlanducci, S.; Thaller, M.C.; Migliore, L. The Integration of Metagenomics and Chemical Physical Techniques Biodecoded the Buried Traces of the Biodeteriogens of Parchment Purple Spots. Front. Microbiol. 2020, 11, 598945. [Google Scholar] [CrossRef] [PubMed]
- Perini, N.; Mercuri, F.; Thaller, M.C.; Orlanducci, S.; Castiello, D.; Talarico, V.; Migliore, L. The Stain of the Original Salt: Red Heats on Chrome Tanned Leathers and Purple Spots on Ancient Parchments Are Two Sides of the Same Ecological Coin. Front. Microbiol. 2019, 10, 2459. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Hoekstra, E.S.; Frisvad, J.C.; Filtenborg, O. Introduction to Food- and Airborne Fungi, 7th ed.; Samson, R.A., Hoekstra, E.S., Frisvad, J.C., Filtenborg, O., Eds.; Centraalbureau Voor Schimmelculture: Utrecht, The Netherlands, 2010; 390p. [Google Scholar]
- Pinheiro, C.; Faustino, M.; Tourais, A.; Gonçalves, C.; Casanova, C. The Medieval Leather Bookbinding of Santa Maria de Alcobaça: Technological Approaches. In Medieval Europe in Motion V—Materialities and Devotion (5th–15th Centuries); Barreira, C., Martins, D., Fontes, J.L., Farelo, M., Eds.; 2022; in press. [Google Scholar]
- Pinzari, F.; Cialei, V.; Piñar, G. A case study of ancient parchment biodeterioration using variable pressure and high vacuum scanning electron microscopy. In Historical Technology, Materials and Conservation: SEM and Microanalysis; Archetype Books: London, UK, 2012; pp. 93–99. [Google Scholar]
- Cicero, C.; Pinzari, F.; Mercuri, F. 18th Century knowledge on microbial attacks on parchment: Analytical and historical evidence. Int. Biodeterior. Biodegrad. 2018, 134, 76–82. [Google Scholar] [CrossRef]
- Teasdale, M.D.; Fiddyment, S.; Vnouček, J.; Mattiangeli, V.; Speller, C.; Binois, A.; Carver, M.; Dand, C.; Newfield, T.P.; Webb, C.C.; et al. The York Gospels: A one thousand year biological palimpsest. R. Soc. Open Sci. 2017, 4, 170988. [Google Scholar] [CrossRef]
- Sayed, A.; Abdel-Wahab, N.M.; Hassan, H.; Abdelmohsen, U. Saccharopolyspora: An underexplored source for bioactive natural products. J. Appl. Microbiol. 2020, 128, 314–329. [Google Scholar] [CrossRef]
- Migliore, L.; Perini, N.; Mercuri, F.; Orlanducci, S.; Rubechini, A.; Thaller, M.C. Three ancient documents solve the jigsaw of the parchment purple spot deterioration and validate the microbial succession model. Sci. Rep. 2019, 9, 1623. [Google Scholar] [CrossRef]
- Falkiewicz-Dulík, M. Leather and Leather Products. In Handbook of Material Biodegradation, Biodeterioration, and Biostablization, 2nd ed.; ChemTec Publishing: Toronto, ON, Canada, 2015; pp. 133–256. [Google Scholar] [CrossRef]
- Lech, T. Evaluation of a Parchment Document, the 13th Century Incorporation Charter for the City of Krakow, Poland, for Microbial Hazards. Appl. Environ. Microbiol. 2016, 82, 2620–2631. [Google Scholar] [CrossRef]
- Cappa, F.; Piñar, G.; Brenner, S.; Frühmann, B.; Wetter, W.; Schreiner, M.; Engel, P.; Miklas, H.; Sterflinger, K. The Kiev Folia: An interdisciplinary approach to unravelling the past of an ancient Slavonic manuscript. Int. Biodeterior. Biodegrad. 2022, 167, 105342. [Google Scholar] [CrossRef]
- Green, P.N.; Ardley, J.K. Review of the genus Methylobacterium and closely related organisms: A proposal that some Methylobacterium species be reclassified into a new genus, Methylorubrum gen. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 2727–2748. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, M.Y.; Khan, N.; Tan, L.L.; Yang, S. Potentials, Utilization, and Bioengineering of Plant Growth-Promoting Methylobacterium for Sustainable Agriculture. Sustainability 2021, 13, 3941. [Google Scholar] [CrossRef]
- Balkwill, D.L.; Fredrickson, J.K.; Romine, M.F. Sphingomonas and Related Genera; Pacific Northwest National Lab. (PNNL): Richland, WA, USA, 2003. [Google Scholar]
- Rölleke, S.; Muyzer, G.; Wawer, C.; Wanner, G.; Lubitz, W. Identification of bacteria in a biodegraded wall painting by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 1996, 62, 2059–2065. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, P.M. Refining the DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19451–19452. [Google Scholar] [CrossRef]
- Hossain, H.; Landgraf, V.; Weiss, R.; Mann, M.; Hayatpour, J.; Chakraborty, T.; Mayser, P. Genetic and biochemical characterization of Malassezia pachydermatis with particular attention to pigment-producing subgroups. Med. Mycol. 2007, 45, 41–49. [Google Scholar] [CrossRef]
- Crous, P.W.; Kang, J.-C.; Braun, U. A Phylogenetic Redefinition of Anamorph Genera in Mycosphaerella Based on ITS rDNA Sequence and Morphology. Mycologia 2001, 93, 1081. [Google Scholar] [CrossRef]
- Kot, A.M.; Kieliszek, M.; Piwowarek, K.; Błażejak, S.; Mussagy, C.U. Sporobolomyces and Sporidiobolus—Non-conventional yeasts for use in industries. Fungal Biol. Rev. 2021, 37, 41–58. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Merz, W.G. Infections caused by non-Candida, non-Cryptococcus yeasts. Clin. Mycol. 2009, 10, 251–270. [Google Scholar] [CrossRef]
- Piñar, G.; Tafer, H.; Schreiner, M.; Miklas, H.; Sterflinger, K. Decoding the biological information contained in two ancient Slavonic parchment codices: An added historical value. Environ. Microbiol. 2020, 22, 3218–3233. [Google Scholar] [CrossRef]
- Rölleke, S.; Witte, A.; Wanner, G.; Lubitz, W. Medieval wall paintings—A habitat for archaea: Identification of archaea by denaturing gradient gel electrophoresis (DGGE) of PCR-amplified gene fragments coding for 16S rRNA in a medieval wall painting. Int. Biodeterior. Biodegrad. 1998, 41, 85–92. [Google Scholar] [CrossRef]
- Vest, M. White tawed leather—Aspects of conservation. In Proceedings of the 9th International Congress of IADA, Copenhagen, Denmark, 16–21 August 1999; pp. 67–72. [Google Scholar]
- Casanova, M.C. De Artífice a Cientista. Evolução da Conservação e do Estatuto Profissional do Conservador Restaurador de Documentos Gráficos no AHU (1926–2006); Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa: Lisboa, Portugal, 2012; Available online: http://hdl.handle.net/10362/8777 (accessed on 17 October 2022).
- Caldas, J. História de um Fogo-Morto. Subsídios para uma História Nacional. 1258–1848. Vianna do Castelo; Livraria Chardron: Porto, Portugal, 1903. [Google Scholar]
- Proença, R. A Biblioteca Nacional: Breves Noções Históricas e Descritivas; Publicações da Biblioteca Nacional: Lisbon, Portugal, 1918; Volume 1, pp. 7–57. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinheiro, C.; Miller, A.Z.; Vaz, P.; Caldeira, A.T.; Casanova, C. Underneath the Purple Stain. Heritage 2022, 5, 4100-4113. https://doi.org/10.3390/heritage5040212
Pinheiro C, Miller AZ, Vaz P, Caldeira AT, Casanova C. Underneath the Purple Stain. Heritage. 2022; 5(4):4100-4113. https://doi.org/10.3390/heritage5040212
Chicago/Turabian StylePinheiro, Catarina, Ana Zélia Miller, Patrícia Vaz, Ana Teresa Caldeira, and Conceição Casanova. 2022. "Underneath the Purple Stain" Heritage 5, no. 4: 4100-4113. https://doi.org/10.3390/heritage5040212
APA StylePinheiro, C., Miller, A. Z., Vaz, P., Caldeira, A. T., & Casanova, C. (2022). Underneath the Purple Stain. Heritage, 5(4), 4100-4113. https://doi.org/10.3390/heritage5040212






