1. Introduction
Environmental characteristics and soil site conditions have a significant impact on forest growth and stand development [
1], and as a result, understanding them is critical for forecasting planting limits on specific forest species and their productivity [
2]. Soils, in particular, play an important role in regulating, supporting, and provisioning ecosystem services, and data on soil properties represent critical information for forest planning and management schemes [
3]. Key soil properties such as the texture and organic matter content are essential to estimate site productivity, which is defined as a quantitative estimate of a site’s potential to produce a volume of trees in a given time [
4].
Soil texture has shown to be strongly related to the movement, retention, capacity, and availability of nutrients and water content, as well as the ease with which plant roots may penetrate the ground and absorb water [
5]. Previous research on forest growth has observed that soil texture is one of the most important properties for evaluating soil compaction recovery to guide future planting procedures [
6]. For example, Ref. [
7] found a strong correlation (R
2 = 0.84) between the soil texture content and soil density. Soil organic matter (SOM) is an important chemical property for evaluating forest site fertility because it has a large influence on the soil cation exchange capacity [
8], which is a vital factor for nutrient supply to plants [
9]. Previous research found that increasing the SOM by 1% raised the site index (the height of dominant trees per stand in a target year) by approximately 20% in
Pinus banksiana plantations and by 7% in
Picea glauca plantations [
10]. Additionally, soil depth (SoD), described as the depth of the soil profile from the top surface to the bedrock or root barriers [
11], is also correlated to nutrient capacity and the plant’s available water content, and it controls biological activity [
12]. This soil property has been linked to site index prediction in
Pinus plantations [
13] as well as overall productivity in forest plantations [
14]. Changes in the SoD have also been linked to variations in the basal area within hardwood plantations. Specifically, the loss of soil thickness due to erosion is projected to decrease the basal area from an initial estimate of 18 to 26 m
2/ha to a range of 8 to 21 m
2/ha in future generations of forest plantations [
15].
Understanding the interplay between physical and chemical soil properties that contribute to forest biomass production and carbon storage is vital, particularly in managed forest plantations where the goal is to maximize productivity over short rotation cycles. Enhancing the spatial characterization of soil properties not only facilitates an increased understanding of site productivity [
4] but also optimizes fertilizer applications. Forests planted on sites with high nutrient contents tend to be less responsive to fertilizer applications, potentially eliminating the need for such interventions [
16]. Ref. [
17], for instance, observed a 400% increase in volume in
Pinus ponderosa plantations due to fertilizer applications at sites with low concentrations of organic matter and nitrogen content. Consequently, identifying site fertility is critical for reducing fertilization management costs without significantly compromising potential growth. Furthermore, soil property data helps in optimizing heavy machinery allocation during timber harvesting and site preparation in plantations to minimize soil compaction.
Conventionally, the representation of soil properties and their relationships with environmental characteristics are traditionally obtained through field surveys, soil pit samples, and laboratory analyses [
18]. Several approaches for interpolating the spatial information of soil properties, commonly employing geostatistical models, have emerged in recent decades based on field and laboratory measurements and spatially explicit environmental data [
19]. The introduction of the SCORPAN theoretical model has proven to provide the best approximation for the development of digital soil mapping (DSM), improving the spatial representation of numerous soil properties. This theoretical model posits that soil properties are the result of the interactions between seven factors or variables that describe soil formation: S (previously measured soil information), C (climate), O (land cover), R (topography), P (parent material), A (soil age), and N (spatial position) [
18]. This approach has been frequently used to predict and map soil properties such as depth gradients of soil organic carbon [
9] and soil texture [
20]. These soil maps are designed to provide decision-makers with accurate and precise information, which is necessary in forest plantation management to improve and enable site-specific management operations [
21], mainly in relation to precise fertilization treatments based on granular assessments of soil nutrient deficiencies [
22]. Nonetheless, it is also well recognized that the maps produced using DSM techniques are not error-free [
23].
Uncertainty in digital soil mapping can originate from modeling errors and measurement inaccuracies associated with the input data [
23]. Quantifying uncertainty is a crucial step because it assesses the reliability of the prediction data, typically expressed within a range of confidence intervals [
24]. In the context of precision silviculture, possessing a comprehensive understanding of the confidence intervals for predicted variables, such as soil fertility, holds significant importance when making site-specific fertilization management decisions [
25]. This knowledge provides valuable advantages by allowing resources to be directed towards areas based on the inherent variability in soil properties. However, the majority of digital soil mapping predictions tend to focus solely on the mean of the prediction distribution and often avoid estimating local uncertainty in soil predictions. This is due to inherent challenges, particularly with machine learning models, which are the preferred and most common approach for DSM [
26]. For instance, the Random Forest (RF) method has gained widespread use in digital soil mapping, attributable to its ensemble of regression trees that yield more robust estimations with less biased internal error estimates [
27]. Nevertheless, this approach only retains the mean of the prediction observations while disregarding other valuable information [
28]. Other statistical models, such as the Bayesian Maximum Entropy (BME) model for spatial prediction, address the integration of data with uncertainty into the modeling process, aiming to enhance their predictive capabilities compared with traditional estimation methods [
29]. However, this is normally challenged by computational complexity, sample size limitations [
30], uncertainty in interpretability, and the complexity of parameter estimation [
31].
The Quantile Regression Forest (QRF) [
32] approach is a tree-based ensemble method which has the advantage of allowing the measurement of prediction uncertainties for each soil property as well as the depiction of probability distributions of dependent environmental variables [
28]. A QRF, akin to an RF, offers valuable information regarding both the median and the distribution of the target variable. The principal difference lies in the treatment of observations within each node and tree: while an RF retains only the mean of the observations, thereby discarding additional data, a QRF preserves the values of all observations [
32]. In recent years, QRFs have been gaining popularity in soil investigations, providing accurate estimates of the SOM [
23,
33,
34], clay content, and other soil parameters [
28,
35], including in-depth 3D representations of the SOM, pH, and clay content, among others [
36]. While most studies have centered on comparing the QRF method with other machine learning algorithms concerning their predictive prowess—for example, for soil organic carbon [
37] and soil organic matter [
28,
34]—with favorable results, there remains a gap in terms of its use for management decisions in the soil domain. The application of uncertainty for management is undoubtedly beneficial for decision making. Still, recent research lacks practical examples on harnessing this uncertainty for the benefit of forest managers.
Recognizing the significant gap in current research, this study introduces an innovative approach to soil science by developing probabilistic maps to aid forest management decisions. The ability to visualize a range of possible soil conditions across a given area presents a substantial advancement over conventional methods, which typically rely on less descriptive, average-based data. This probabilistic approach becomes essential for making informed, data-driven decisions crucial for sustainable forest management. By focusing on these maps, this study aims to enhance operational strategies and decision-making processes in forestry. The purpose of this study therefore is to evaluate the spatial uncertainty distribution for soil depth, texture, and organic matter, with the intention of applying these findings to forest management operations at a 10 m spatial resolution. For this, we utilized the Quantile Regression Forest (QRF) model approach. To do so, we investigated (1) the uncertainty of soil property mapping products, (2) elaborating a 3D map for the texture and SOM based on the SoD, and (3) practical management application examples that leverage fine resolution and its associated prediction distribution.
6. Discussion
This study presents a method for obtaining spatial predictions and their associated uncertainties for digital soil mapping. Additionally, it demonstrates how to integrate the distributions of these predictions of soil properties into forest management operations.
Our results show that the soil properties were well represented spatially when using the QRF. The chosen 10 m spatial resolution, selected to observe soil property changes within the forest stand, provided accurate results when compared to other studies on SoD prediction using a regression model (R
2 = 0.74, MAPE = 10.53%, and RMSE = 19.4 cm). Although SoD is a difficult soil property to predict, we obtained a significant improvement by considering all of the SCORPAN input variables when compared to other studies using only topographic variables [
68], indicating an overall good performance in line with the results presented in [
69].
In the validation analysis, the SOM map showed reliable results, with R
2 = 0.61, MAPE = 41.18%, and RMSE = 2.03%. When compared to previous studies, we can observe that the general level of validation accuracy for SOM predictions ranges between R
2 = 0.45 at 10 m using topographic attributes only [
70], R
2 = 0.51 at 30 m [
71], and R
2 = 0.53 using a QRF at 30 m resolution [
34]. In our study, the observed standard deviation of the prediction distribution ranged from 1.7% to 7.7%. In most of the valley and the Andes foothills, the variability hovered at around 17% of the mean. However, in the Andes, it rose to approximately 50%. This trend can potentially be attributed to the fact that as topographic heterogeneity increases, soil properties vary significantly [
72].
Soil texture also showed accurate results given the successful validation of the clay, silt, and sand contents. Clay had the higher accuracy, followed by silt and lastly by sand (
Table 5), with mean standard deviations of 29%, 33%, and 35%, respectively. Similar results were reported by other studies [
20,
73,
74], which account for their good prediction accuracy based on the strength of the used input variables of the SCORPAN approach, emphasizing the importance of integrating key soil formation variables for reliable predictions of soil texture.
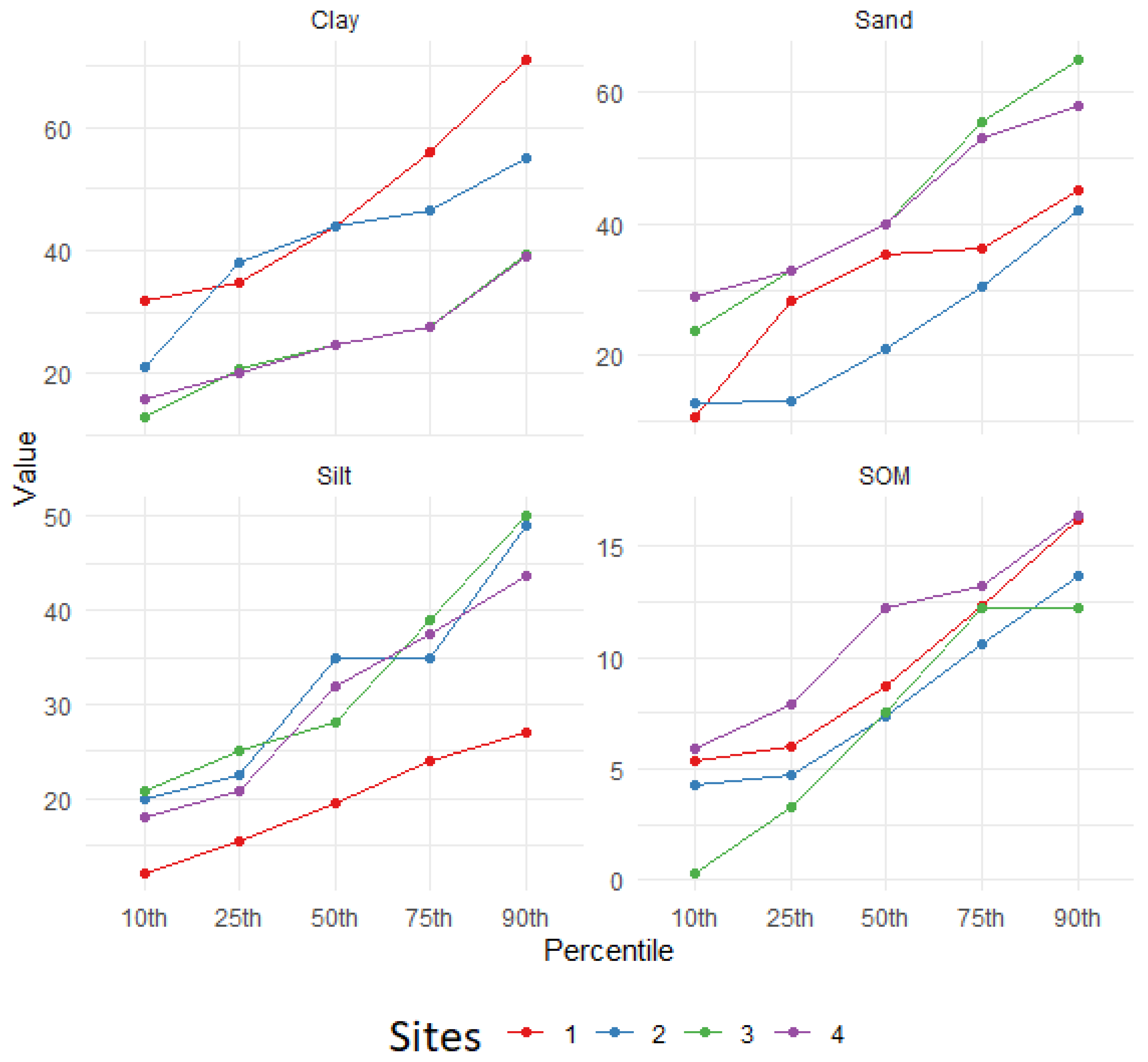
Observing the spatial distribution of standard deviation values and the spread of prediction distributions for the four selected sites depicted in
Figure 1 and explained in
Figure 5, it becomes apparent that there is significant variability in the prediction distributions across all predicted soil properties. This variability is particularly pronounced, as anticipated, on the foothills of the Andes coastal range, where the topography is highly heterogeneous and maximum variation occurs [
75]. For example, the SOM ranges from 3.2% (25th percentile) to 12.2% (75th percentile). Surprisingly, even in the valley, where the topographic relief is relatively consistent, a notably variable prediction distribution of soil properties persists. Following the SOM example, it varies from 7.9% to 13.2% in the 25th and 75th percentiles, respectively. Capturing the spatial predictive distribution of soil properties is crucial to reducing uncertainty and enhancing the quality of digital soil mapping (DSM) products. As Ref. [
25] noted, one direct application of soil prediction uncertainty is in optimizing soil sampling designs, with a focus on areas of high uncertainty. Furthermore, the use of prediction variability and the entire distribution of soil properties is also beneficial to managers in reducing uncertainty in forest growth responses to fertilizer application, as illustrated in the practical management application example. This phenomenon, where responses to fertilizer application are heightened in areas of low soil fertility, has been substantiated by previous research [
16,
17]. Relying solely on the mean for decision-supporting purposes, as exemplified in
Figure 6, and using the same 5% SOM as a threshold to identify areas for fertilization, would imply that in 50% of cases, values will exceed the decided threshold. This translates to 50% of areas exhibiting a low response or no response to fertilizer application. As demonstrated in the example, utilizing the entire distribution range can ensure positive responses of forest growth to management applications, depending on the level of probability chosen for decisions (80% in our example), thereby minimizing resource allocation and maximizing the economic benefits.
As discussed in previous research [
12], detecting the presence of shallow soils is crucial for soil protection management in forested areas. It is vital to understand the forest roots’ capacity to reach available nutrients and to enhance our knowledge of the plants’ available water content and the variation in nutrient availability at different soil depths for tree root uptake. As shown in
Figure 7, the 3D map demonstrates a significant variation in SOM content at different depths. The decrease in SOM with increasing soil depths has been mentioned several times in the literature [
76,
77]. This occurs because SOM is produced near the surface due to superficial leaf decomposition in forested areas and fine-root decomposition in grasslands. This shift in distribution is also linked to varying microbial community structures at different soil depths, which influence the rate of organic matter decomposition [
78]—a reduction rate that can be visualized with the methodology presented in this study. Although there is some variation in soil texture at different depths, it is not significant. Similar results found in [
79] were explained by the soil’s age since most of the horizons were still forming, as may be the case in our study area. Furthermore, particularly around the coast, the soil is classified as clay through several depth levels, likely because they are on alfisols—classified by the USDA soil taxonomy as clay-enriched with a relatively high natural fertility [
61].
Regarding the SCORPAN variables selected for this research, DEM has emerged as one of the top variables related to soil texture and depth. This correlation can primarily be attributed to its association with soil erosion and redistribution, as well as its impact on the SOM accumulation cycle [
75]. Previous research has found that elevation, the topographic wetness index (TWI), plan curvature, the total catchment area, and the channel network base level are the key topography characteristics most closely connected to soil organic carbon concentration in flat-slope locations [
80]. Furthermore, for hydrologic and geomorphic purposes, the Multiresolution Index of Ridge-Top Flatness (MRRTF), Multiresolution Index of Valley-Bottom Flatness (MRVBF), slope, and valley depth have shown strong links with sediment deposits and influence deposit depth [
81]. As observed in [
82], the distance to water channels provides essential information on sediment accumulation, altering the SOM content, SoD, and soil texture, which are also reflected in this study. Besides elevation, this variable has also been recognized as one of the most critical topographic parameters for the digital soil mapping of SOM [
80]. According to the literature, one of the most important environmental variables influencing the rate of weathering and organic decomposition, which cause new layers of soil to accumulate at depth, is the mean annual temperature [
83,
84]. Furthermore, precipitation notably impacts hydrological processes such as surface runoff and groundwater flow, which are vital for organic matter decomposition rates and infiltration from the litter layer to mineral soil, as extensively reported [
45,
46]. This notion is corroborated in this study, wherein these two climatic variables are selected as some of the most significant predictors of among all of the predicted soil properties.
The most significant advantage of the proposed methodology is that it enables the prediction of accurate high-spatial-resolution maps of soil properties and their predictive distributions, based on soil formation factors (SCORPAN). The application of QRFs in DSM, as used in this study, demonstrates the utility of this methodology for the spatial interpolation of key soil properties and its uncertainty within the forest stand, which has been previously applied in SoilGrids250m [
23]. As mentioned in previous research [
28,
35], the QRF model is a promising alternative for observing the probabilistic distribution of soil predictions, especially for precision silviculture tasks. Nonetheless, the primary advantage of using a QRF for high-resolution soil properties lies in its ability to capture variable distributions. This aspect is frequently overlooked in conventional soil prediction models and management applications.
Similar to previous soil property mapping efforts [
85,
86], spatial soil information has proven to be a valuable tool for enhancing our understanding of site productivity variations within forested areas, especially when utilizing the spatial resolution chosen for this study (10 m). This approach to digital soil property analysis, along with its associated uncertainties, enhances fertilizer prescription capabilities by identifying areas with fertility issues, thereby facilitating precision in fertilizer application during forest operations, as demonstrated in the example provided. Moreover, granulometric information at a fine spatial resolution using the QRF method, alongside SoD data, can be employed to improve assessments of water-holding capacity and soil fertility ratings. These enhancements are crucial for process-based models used in predicting forest growth, as well as for precision planning in timber harvesting and site preparation to mitigate soil compaction risks. Integrating predictive soil property distribution into decision management strategies can further contribute to reductions in fertilization costs and increases in productivity. This methodology holds promise for transferability, as it is applicable to various soil properties, soil-related management practices, and diverse locations or countries. Future research should use high-resolution soil property information and its predictive distribution combined with ALS individual tree metrics to develop tree growth models in order to better understand the effects of site productivity on growth.