COVID-19-Omics Report: From Individual Omics Approaches to Precision Medicine
Abstract
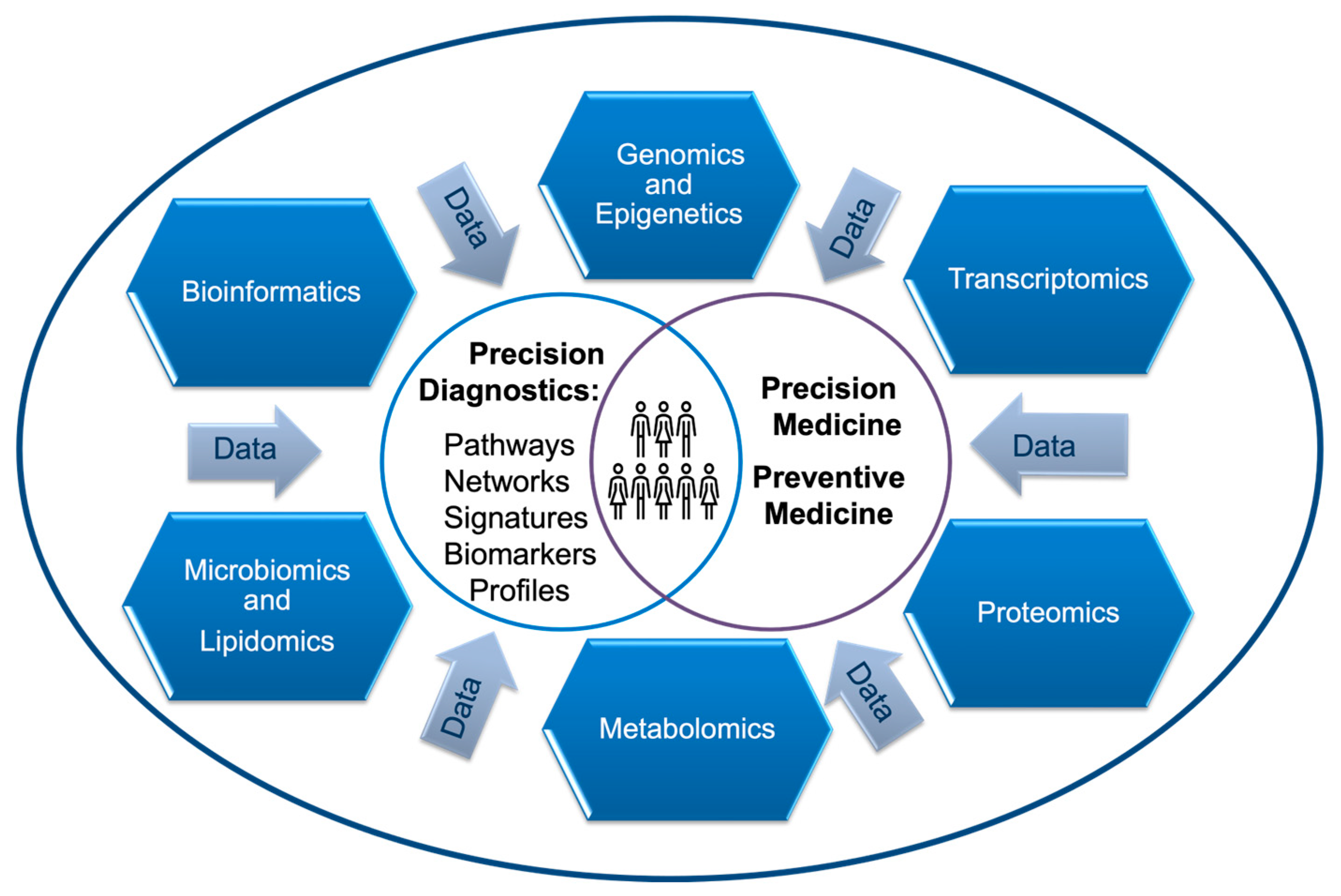
1. Introduction
2. Population-Centric Approach: Demographics of COVID-19 Vulnerable Population
2.1. Age Vulnerability
2.2. Immunocompromised Population
2.3. Pregnant Women and Individuals with Chronic Conditions
3. Genomics and Epigenomics of Host Susceptibility to COVID-19
3.1. Host Genomics
3.2. Host Epigenomics
4. Individual Changes in Metabolome, Lipidome, and Microbiome during SARS-CoV-2 Infection
4.1. Host Metabolomic Response
4.2. Microbiomics
5. Transcriptomics and Proteomics of SARS-CoV-2 Infection
5.1. Host Transcriptomics
5.2. Host Proteomic Response
6. Multiomics and Cross-Ome Comparisons
6.1. One Patient Multi-Omics
6.2. Cross-Ome Bioinformatic, Data Mining and Analytics
7. Precision Medicine: Repurposed and Novel Drugs for COVID-19 Patients
7.1. Precision Antiviral Drugs and Monoclonal Antibodies
7.2. Immunomodulators Targeting Host Responce
| Omics Target | Drug(s) | NCT | Efficacy/Outcomes | Reference |
|---|---|---|---|---|
| IL6R | Tocilizumab | NCT04381936 | Reduction in mortality in patients with high-baseline IL6 | [183] |
| Tocilizumab | IRCT20150303021315N17 | Reduction in the risk of death when treatment begins in early stages of respiratory failure | [184] | |
| Tocilizumab | Retrospective analysis | Shortened time to hospital discharge from intensive care unit regardless of the respiratory support | [185] | |
| Sarilumab | NCT04327388 | Did not improve survival and ventilatory outcomes in critical patients (study was underpowered) | [186] | |
| IL6 | Siltuximab i.v. | NCT04322188 | Downregulated IL8 and pentraxin 3 in the blood; reduced mortality in patients on ventilation support | [187] |
| IFNB1a | Nebulized interferon-β1a alone | NCT04732949 | Reduced mortality but not in-hospital time or time to full recovery | [188] |
| Nebulized interferon-β1a with antivirals | NCT04343768 | Shortened time to significant clinical improvement | [181] | |
| INFAb | Nebulized interferon-α1a | Retrospective analysis | Associated with reduced mortality and accelerated recovery | [189] |
| IFNB1a versus IFNB1b | Interferon-β1a s.c. or Interferon-β1b | NCT04343768 | Significant reduction in time to clinical improvement (TTCI) in IFNβ1a arm | [181] |
| NLRP3 | Tranilast | IRCT20200419047128N1 | Reduced length of hospitalization and mortality | [190] |
| IL1β | Canakinumab | Case-control study | Decreased need for mechanical ventilation; earlier hospital discharge | [191] |
| IL1R | Anakinra i.v. | NCT04318366 | Associated with clinical improvement in 72% of patients | [192] |
| NCT04357366 | Lowered incidence of severe respiratory failure; increased survival rate | [193] | ||
| JAK 1/2 | Baricitinib | NCT04381936 | Reduced mortality in hospitalized COVID-19 patients | [194] |
| Baricitinib with antivirals | NCT04401579 | Improved clinical status among patients on high-oxygen or noninvasive ventilation support | [195,196] | |
| JAK 1/3 | Tofacitinib | NCT04469114 | Lowered risk of death and/or respiratory failure | [197] |
| TNFα | Adalimumab | GRA physician registry | Decreased odds of hospitalization in patients with rheumatic disease treated with adalimumab | [198,199] |
| Infliximab | ISRCTN40580903 | Inferior to namilumab | [200] | |
| GMCSF | Namilumab | ISRCTN40580903 | Significant reduction in inflammation in hospitalized COVID-19 patients | [200] |
| Lenzilumab | NCT04351152 | Improved survival without invasive mechanical ventilation in hospitalized patients with COVID-19 | [201] | |
| TKI | Nintedanib | jRCTs051200036 | Shorter length of mechanical ventilation Reduction in pulmonary fibrosis on imaging tests | [202] |
| CD6 | Itolizumab | CTRI/2020/05/024959 | Improved lung oxygenation parameters and blood cytokine profiles in ARDS patients | [203] |
| IL17 | Netakimab | NCT05302947 | Lowered mortality rate; shortened time to hospital discharge; superior to baricitinib | [204] |
8. Preventive Medicine: Vaccination
8.1. Vaccines Breakthroughs
8.2. Vaccine Side Effects
9. Summary and Future Directions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACE2 | Angiotensin-converting enzyme 2 |
| ARDS | Acute respiratory distress syndrome |
| CDC | Centers for Disease Control and Prevention |
| CEACAM | Carcinoembryonic antigen-related cell adhesion molecule |
| GWAS | Genome-wide association study |
| HLA | Human leucocyte antigen |
| IL | Interleukin |
| ADAMTS13 | A disintegrin and metalloprotease (domain) of metallopeptidase with thrombospondin type 1 motif 13 |
| TLR | Toll-like receptor |
| Gal-9 | Galectin 9 |
| CRP | C-reactive protein |
| PRRs | Pattern recognition receptors |
| SNPs | single nucleotide polymorphism |
| cGAS-STING | cyclic GMP–AMP synthase (cGAS) stimulator of interferon gene (STING) |
| HIST1H2BO | Histone H2B type 1-O |
| HMGB1 | High mobility group box 1 gene |
| NLRP3 | NLR (nucleotide-binding domain and leucine-rich repeat containing) family pyrin domain containing 3 |
| TMPRSS2 | Transmembrane protease serine 2 |
| TCRA/D region | T cell receptor alpha/delta genes |
| TIMP1 | Metalloproteinase inhibitor 1 |
| IFNγ | Interferon gamma |
| IFIH | interferon induced with helicase C domain gene family |
| IRF | interferon regulatory factor gene family |
| PGE2 | Prostaglandin E2 |
| MMP | Matrix metalloproteinase |
| AP-1 | Activator protein 1 |
| J&J | Janssen Biotech Inc. and Johnson & Johnson |
| PBNT | Pfizer-BioNTech |
| VEGFA | Vascular endothelial growth factor A |
| GMCSF | Granulocyte–macrophage colony-stimulating factor |
| JAK 1/2 | Janus kinases |
| SIGLEC | Sialic acid–binding, immunoglobulin (Ig)-like lectin |
| TKI | Tyrosine kinase inhibitor |
| IL6Rα | Interleukin receptor alpha |
| IL1R | Interleukin 1 receptor |
| AP-1 | Activator protein-1 |
| MAPKs | Mitogen-associated protein kinase |
| ERKs | Extracellular signal-regulated kinase |
References
- Louis, I.V.; Gorzalski, A.; Pandori, M. Diagnostic Applications for RNA-Seq Technology and Transcriptome Analyses in Human Diseases Caused by RNA Viruses. In Applications of RNA-Seq in Biology and Medicine; IntechOpen: London, UK, 2021; pp. 122–138. [Google Scholar]
- Gorzalski, A.J.; Kerwin, H.; Verma, S.; Hess, D.C.; Sevinsky, J.; Libuit, K.; Louis, I.V.; Siao, D.; Siao, L.; Buñuel, D.; et al. Rapid Lineage Assignment of Severe Acute Respiratory Syndrome Coronavirus 2 Cases through Automated Library Preparation, Sequencing, and Bioinformatic Analysis. J. Mol. Diagn. 2023, 25, 191–196. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Zhao, R.; Gao, L.J.; Gao, X.F.; Wang, D.P.; Cao, J.M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef] [PubMed]
- Cen, X.; Wang, F.; Huang, X.; Jovic, D.; Dubee, F.; Yang, H.; Li, Y. Towards precision medicine: Omics approach for COVID-19. Biosaf. Health 2023, 5, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Statista. Distribution of Total COVID-19 Deaths in the United States as of 26 April 2023, by Age Group. Available online: https://www.statista.com/statistics/1254488/us-share-of-total-covid-deaths-by-age-group/ (accessed on 26 May 2023).
- Malesevic, S.; Sievi, N.A.; Baumgartner, P.; Roser, K.; Sommer, G.; Schmidt, D.; Vallelian, F.; Jelcic, I.; Clarenbach, C.F.; Kohler, M. Impaired health-related quality of life in long-COVID syndrome after mild to moderate COVID-19. Sci. Rep. 2023, 13, 7717. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef]
- Yüce, M.; Filiztekin, E.; Özkaya, K.G. COVID-19 Diagnosis—A review of current methods. Biosens. Bioelectron. 2021, 172, 112752. [Google Scholar] [CrossRef]
- Louis, I.V. Introductory Chapter: RNA Drugs Development and Commercialization. In RNA Therapeutics—History, Design, Manufacturing, and Applications; IntechOpen: Vienna, Austria, 2023; pp. 2–7. ISBN 978-1-80355-658-1. [Google Scholar]
- Oran, D.P.; Topol, E.J. Prevalence of asymptomatic SARS-CoV-2 infection. A narrative review. Ann. Intern. Med. 2020, 173, 362–368. [Google Scholar] [CrossRef]
- Garg, M.; Maralakunte, M.; Garg, S.; Dhooria, S.; Sehgal, I.; Bhalla, A.S.; Vijayvergiya, R.; Grover, S.; Bhatia, V.; Jagia, P.; et al. The conundrum of ‘long-covid-19ʹ: A narrative review. Int. J. Gen. Med. 2021, 14, 2491–2506. [Google Scholar] [CrossRef]
- Ma, Q.; Liu, J.; Liu, Q.; Kang, L.; Liu, R.; Jing, W.; Wu, Y.; Liu, M. Global Percentage of Asymptomatic SARS-CoV-2 Infections among the Tested Population and Individuals with Confirmed COVID-19 Diagnosis: A Systematic Review and Meta-analysis. JAMA Netw. Open 2021, 4, e2137257. [Google Scholar] [CrossRef]
- Elder, C. Prevalence of Asymptomatic SARS-CoV-2 Infection. Ann. Intern. Med. 2021, 174, 285–286. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.C.; Burgers, W.A. SARS-CoV-2 evolution and vaccines: Cause for concern? Lancet Respir. Med. 2021, 9, 333–335. [Google Scholar] [CrossRef] [PubMed]
- Van Elslande, J.; Vermeersch, P.; Vandervoort, K.; Wawina-Bokalanga, T.; Vanmechelen, B.; Wollants, E.; Laenen, L.; André, E.; Van Ranst, M.; Lagrou, K.; et al. Symptomatic Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Reinfection by a Phylogenetically Distinct Strain. Clin. Infect. Dis. 2020, 73, 354–356. [Google Scholar] [CrossRef] [PubMed]
- Peter, A.E.; Sandeep, B.V.; Rao, B.G.; Kalpana, V.L. Calming the Storm: Natural Immunosuppressants as Adjuvants to Target the Cytokine Storm in COVID-19. Front. Pharmacol. 2021, 11, 583777. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xie, X.; Tu, Z.; Fu, J.; Xu, D.; Zhou, Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Target. Ther. 2021, 6, 255. [Google Scholar] [CrossRef]
- India TRC. BCG Vaccine in Reducing Morbidity and Mortality in Elderly Individuals in COVID-19 Hotspots. NCT04475302. 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04475302 (accessed on 6 June 2023).
- Zeynali Bujani, M.; Behnampour, M.; Rahimi, N.; Safari, T.; Khazaei Feizabad, A.; Hossein Sarbazi, A.; Baniasadi, M.; Rezaei, N.; Ansari Moghaddam, A. The effect of influenza vaccination on covid-19 morbidity, severity and mortality: Systematic review and meta-analysis. Malays. J. Med. Sci. 2021, 28, 20–31. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, V.W.; Stray-Pedersen, A.; Hanson, I.C.; Forbes, L.R.; de la Morena, M.T.; Chinn, I.K.; Gorman, E.; Mendelsohn, N.J.; Pozos, T.; et al. Rapid molecular diagnostics of severe primary immunodeficiency determined by using targeted next-generation sequencing. J. Allergy Clin. Immunol. 2016, 138, 1142–1151.e2. [Google Scholar] [CrossRef][Green Version]
- Strand, J.; Gul, K.A.; Erichsen, H.C.; Lundman, E.; Berge, M.C.; Trømborg, A.K.; Sørgjerd, L.K.; Ytre-Arne, M.; Hogner, S.; Halsne, R.; et al. Second-Tier Next Generation Sequencing Integrated in Nationwide Newborn Screening Provides Rapid Molecular Diagnostics of Severe Combined Immunodeficiency. Front. Immunol. 2020, 11, 1417. [Google Scholar] [CrossRef]
- IDF (Immunodeficiency Foundation). Severe Combined Immunodeficiency and Combined Immunodeficiency. 2018. Available online: https://primaryimmune.org/scid-compass/disease/severe-combined-immunodeficiency-and-combined-immunodeficiency (accessed on 7 May 2021).
- O’Keefe, A.W.; Halbrich, M.; Ben-Shoshan, M.; McCusker, C. Primary immunodeficiency for the primary care provider. Paediatr. Child. Health 2016, 21, e10–e14. [Google Scholar] [CrossRef]
- Orange, J.S. Inborn Errors of Type I IFN Immunity in Patients With Life-Threatening COVID-19. Pediatrics 2021, 148, S72–S73. [Google Scholar] [CrossRef]
- van der Burg, M.; Mahlaoui, N.; Gaspar, H.B.; Pai, S.Y. Universal Newborn Screening for Severe Combined Immunodeficiency (SCID). Front. Pediatr. 2019, 7, 373. [Google Scholar] [CrossRef] [PubMed]
- CDC. Newborn Screening Portal. Available online: https://www.cdc.gov/newbornscreening/index.html (accessed on 6 May 2021).
- ClinicalTrials. Analysis of the Immune Response to COVID-19 Vaccination and Outcomes in Individuals With and Without Immune Deficiencies and Dysregulations. NCT04852276. 2022. Available online: https://clinicaltrials.gov/ct2/show/NCT04852276 (accessed on 2 June 2023).
- Liang, M.; Luo, N.; Chen, M.; Chen, C.; Singh, S.; Singh, S.; Tan, S. Prevalence and Mortality due to COVID-19 in HIV Co-Infected Population: A Systematic Review and Meta-Analysis. Infect. Dis. Ther. 2021, 10, 1267–1285. [Google Scholar] [CrossRef]
- Baek, M.S.; Lee, M.T.; Kim, W.Y.; Choi, J.C.; Jung, S.Y. COVID-19-related outcomes in immunocompromised patients: A nationwide study in Korea. PLoS ONE 2021, 16, e0257641. [Google Scholar] [CrossRef] [PubMed]
- Tamuzi, J.L.; Ayele, B.T.; Shumba, C.S.; Adetokunboh, O.O.; Uwimana-Nicol, J.; Haile, Z.T.; Inugu, J.; Nyasulu, P.S. Implications of COVID-19 in high burden countries for HIV/TB: A systematic review of evidence. BMC Infect. Dis. 2020, 20, 744. [Google Scholar] [CrossRef]
- Hwang, J.K.; Zhang, T.; Wang, A.Z.; Li, Z. COVID-19 vaccines for patients with cancer: Benefits likely outweigh risks. J. Hematol. Oncol. 2021, 14, 38. [Google Scholar] [CrossRef]
- Duly, K.; Farraye, F.A.; Bhat, S. COVID-19 vaccine use in immunocompromised patients: A commentary on evidence and recommendations. Am. J. Health Pharm. 2022, 79, 63–71. [Google Scholar] [CrossRef]
- John Leon Singh, H.; Couch, D.; Yap, K. Mobile Health Apps That Help With COVID-19 Management: Scoping Review. JMIR Nurs. 2020, 3, e20596. [Google Scholar] [CrossRef]
- Flannery, D.D.; Gouma, S. Assessment of Maternal and Neonatal Cord Blood SARS-CoV-2 Antibodies and Placental Transfer Ratios. JAMA Pediatr. 2021, 175, 594–600. [Google Scholar] [CrossRef]
- Edlow, A.G.; Li, J.Z.; Collier, A.R.Y.; Atyeo, C.; James, K.E.; Boatin, A.A.; Gray, K.J.; Bordt, E.A.; Shook, L.L.; Yonker, L.M.; et al. Assessment of Maternal and Neonatal SARS-CoV-2 Viral Load, Transplacental Antibody Transfer, and Placental Pathology in Pregnancies During the COVID-19 Pandemic. JAMA Netw. Open 2020, 3, e2030455. [Google Scholar] [CrossRef]
- Chen, L.; Li, Q.; Zheng, D.; Jiang, H.; Wei, Y.; Zou, L.; Feng, L.; Xiong, G.; Sun, G.; Wang, H.; et al. Clinical Characteristics of Pregnant Women with Covid-19 in Wuhan, China. N. Engl. J. Med. 2020, 382, e100. [Google Scholar] [CrossRef] [PubMed]
- Antoun, L.; Taweel, N.E.; Ahmed, I.; Patni, S.; Honest, H. Maternal COVID-19 infection, clinical characteristics, pregnancy, and neonatal outcome: A prospective cohort study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 252, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.Q.; Bilodeau-Bertrand, M.; Liu, S.; Auger, N. The impact of COVID-19 on pregnancy outcomes: A systematic review and meta-analysis. CMAJ 2021, 193, E540–E548. [Google Scholar] [CrossRef] [PubMed]
- Kharbanda, E.O.; Haapala, J.; Lipkind, H.S.; DeSilva, M.B.; Zhu, J.; Vesco, K.K.; Daley, M.F.; Donahue, J.G.; Getahun, D.; Hambidge, S.J.; et al. COVID-19 Booster Vaccination in Early Pregnancy and Surveillance for Spontaneous Abortion. JAMA Netw. Open 2023, 6, e2314350. [Google Scholar] [CrossRef] [PubMed]
- CDC. Centers for Disease Control and Prevention Interim Clinical Considerations for Use of COVID-19 Vaccines Currently Approved or Authorized in the United States. 2022. Available online: https://www.cdc.gov/vaccines/covid-19/clinical-considerations/covid-19-vaccines-us.html#pregnant (accessed on 25 May 2023).
- Feldstein, L.R.; Rose, E.B.; Horwitz, S.M.; Collins, J.P.; Newhams, M.M.; Son, M.B.F.; Newburger, J.W.; Kleinman, L.C.; Heidemann, S.M.; Martin, A.A.; et al. Multisystem Inflammatory Syndrome in U.S. Children and Adolescents. N. Engl. J. Med. 2020, 383, 334–346. [Google Scholar] [CrossRef]
- Grier, J.T.; Cron, R.Q.; Dos, S.; Faccion, R.S.; Amendola, F.; Carvalho, A.D.; Campelo, D.; Cox, B.; Celia, M.; Zuma, C.; et al. Rare genetic variants involved in multisystem inflammatory syndrome in children: A multicenter Brazilian cohort study. Front. Cell. Infect. Microbiol. 2023, 13, 1182257. [Google Scholar] [CrossRef]
- NCT04644146. Genetic Predisposition to Severe Forms of COVID-19 (SARS-CoV2 Infection) (COVIDGEN). 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04644146 (accessed on 4 June 2023).
- NCT04384250. Genetic and Immunologic Basis of COVID-19 Infection. 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04384250 (accessed on 5 June 2023).
- Niemi, M.E.K.; Daly, M.J.; Ganna, A. The human genetic epidemiology of COVID-19. Nat. Rev. Genet. 2022, 23, 533–546. [Google Scholar] [CrossRef]
- COVID-19 Host Genetics Initiative. A first update on mapping the human genetic architecture of COVID-19. Nature 2022, 608, E1–E10. [Google Scholar] [CrossRef]
- Zeberg, H. The major genetic risk factor for severe COVID-19 is associated with protection against HIV. Proc. Natl. Acad. Sci. USA 2022, 119, 17–19. [Google Scholar] [CrossRef]
- Zietz, M.; Zucker, J.; Tatonetti, N.P. Associations between blood type and COVID-19 infection, intubation, and death. Nat. Commun. 2020, 11, 5761. [Google Scholar] [CrossRef]
- Langton, D.J.; Bourke, S.C.; Lie, B.A.; Reiff, G.; Natu, S.; Darlay, R.; Burn, J.; Echevarria, C. The influence of HLA genotype on the severity of COVID-19 infection. HLA 2021, 98, 14–22. [Google Scholar] [CrossRef]
- Vigó, L.I.; Galá, M.I.; Torres, M.; Martín-Galiano, A.J.; Rodríguez-Mora, S.; Mateos, E.; Corona, M.; Malo, R.; Navarro, C.; Ará nzazu Murciano-Antó, M.; et al. Association between HLA-C alleles and COVID-19 severity in a pilot study with a Spanish Mediterranean Caucasian cohort. PLoS ONE 2022, 17, e0272867. [Google Scholar] [CrossRef]
- Nguyen, A.; David, J.K.; Maden, S.K.; Wood, M.A.; Weeder, B.R.; Nellore, A.; Thompson, R.F. Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2. J. Virol. 2020, 94, e00510-20. [Google Scholar] [CrossRef] [PubMed]
- Dieter, C.; de Almeida Brondani, L.; Bauermann Leitão, C.; Gerchman, F.; lia Emerim Lemos, N.; Crispim, D.I. Genetic polymorphisms associated with susceptibility to COVID-19 disease and severity: A systematic review and meta-analysis. PLoS ONE 2022, 17, e0270627. [Google Scholar] [CrossRef] [PubMed]
- Saengsiwaritt, W.; Jittikoon, J.; Chaikledkaew, U.; Udomsinprasert, W. Genetic polymorphisms of ACE1, ACE2, and TMPRSS2 associated with COVID-19 severity: A systematic review with meta-analysis. Rev. Med. Virol. 2022, 32, e2323. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ke, Y.; Xia, X.; Wang, Y.; Cheng, F.; Liu, X.; Jin, X.; Li, B.; Xie, C.; Liu, S.; et al. Genome-wide association study of COVID-19 severity among the Chinese population. Cell Discov. 2021, 7, 2–16. [Google Scholar] [CrossRef]
- Pasko, B.E.; Abbott, D.; Bocsi, G.T.; Draper, N.L. ABO Blood Groups Are Not Associated With COVID-19 Disease Incidence and Severity When Correcting for Ethnicity Differences in Blood Type. Am. J. Clin. Pathol. 2022, 158, 249–253. [Google Scholar] [CrossRef]
- Kuijpers, Y.; Chu, X.; Jaeger, M.; Moorlag, S.J.C.F.M.; Koeken, V.A.C.M.; Zhang, B.; Nooijer, A.d.; Grondman, I.; Gupta, M.K.; Janssen, N.; et al. The Genetic Risk for COVID-19 Severity Is Associated With Defective Immune Responses. Front. Immunol. 2022, 13, 859387. [Google Scholar] [CrossRef]
- Elek, Z.; Losoncz, E.; Maricza, K.; Fülep, Z.; Bánlaki, Z.; Kovács-Nagy, R.; Keszler, G.; Rónai, Z. Missense Variants of von Willebrand Factor in the Background of COVID-19 Associated Coagulopathy. Genes 2023, 14, 617. [Google Scholar] [CrossRef]
- Giannini, G.; Velez, J.C.Q.; May, R.M.; Sharma, S.G.; Mohamed, M.M.; Cassol, C.A.; Larsen, C.P.; Caza, T.N. Renal Prognosis of COVID-19 Associated Nephropathy. Kidney Int. Rep. 2022, 7, 2722–2725. [Google Scholar] [CrossRef]
- Cappadona, C.; Rimoldi, V.; Paraboschi, E.M.; Asselta, R. Genetic susceptibility to severe COVID-19. Infect. Genet. Evol. 2023, 110, 105426. [Google Scholar] [CrossRef]
- Ishak, A.; Mehendale, M.; AlRawashdeh, M.M.; Sestacovschi, C.; Sharath, M.; Pandav, K.; Marzban, S. The association of COVID-19 severity and susceptibility and genetic risk factors: A systematic review of the literature. Gene 2022, 836, 146674. [Google Scholar] [CrossRef]
- Pecoraro, V.; Cuccorese, M.; Trenti, T. Genetic polymorphisms of ACE1, ACE2, IFTM3, TMPRSS2 and TNFα genes associated with susceptibility and severity of SARS-CoV-2 infection: A systematic review and meta-analysis. Clin. Exp. Med. 2023; online ahead of print. [Google Scholar] [CrossRef]
- Deb, P.; Zannat, K.e.; Talukder, S.; Bhuiyan, A.H.; Jilani, M.S.A.; Saif-Ur-Rahman, K.M. Association of HLA gene polymorphism with susceptibility, severity, and mortality of COVID-19: A systematic review. HLA 2022, 99, 281–312. [Google Scholar] [CrossRef]
- van der Made, C.I.; Netea, M.G.; van der Veerdonk, F.L.; Hoischen, A. Clinical implications of host genetic variation and susceptibility to severe or critical COVID-19. Genome Med. 2022, 14, 96. [Google Scholar] [CrossRef] [PubMed]
- Severe COVID-19 GWAS Group. Genomewide association study of severe COVID-19 with respiratory failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Dorado, G.; Duarte, E.L.; David-Bosne, S.; Trigueiro-Louro, J.; Rebelo-de-Andrade, H. COVID-19: Impact on Public Health and hypothesis-driven investigations on genetic susceptibility and severity. Immunogenetics 2022, 74, 381–407. [Google Scholar] [CrossRef] [PubMed]
- Atlante, S.; Mongelli, A.; Barbi, V.; Martelli, F.; Farsetti, A.; Gaetano, C. The epigenetic implication in coronavirus infection and therapy. Clin. Epigenetics 2020, 12, 156. [Google Scholar] [CrossRef] [PubMed]
- Saksena, N.; Bonam, S.R.; Miranda-Saksena, M. Epigenetic Lens to Visualize the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) Infection in COVID-19 Pandemic. Front. Genet. 2021, 12, 581726. [Google Scholar] [CrossRef]
- Crimi, E.; Benincasa, G.; Figueroa-Marrero, N.; Galdiero, M.; Napoli, C. Epigenetic susceptibility to severe respiratory viral infections and its therapeutic implications: A narrative review. Br. J. Anaesth. 2020, 125, 1002–1017. [Google Scholar] [CrossRef]
- Sawalha, A.H.; Zhao, M.; Coit, P.; Lu, Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin. Immunol. 2020, 215, 2–4. [Google Scholar] [CrossRef]
- Sang, E.R.; Tian, Y.; Miller, L.C.; Sang, Y. Epigenetic evolution of ACE2 and IL-6 genes: Non-canonical interferon-stimulated genes correlate to COVID-19 susceptibility in vertebrates. Genes 2021, 12, 154. [Google Scholar] [CrossRef]
- Sharif-Zak, M.; Abbasi-Jorjandi, M.; Asadikaram, G.; Ghoreshi, Z.-A.-S.; Rezazadeh-Jabalbarzi, M.; Rashidinejad, H. Influence of Disease Severity and Gender on HLA-C Methylation in COVID-19 Patients. Iran. J. Sci. Technol. Trans. A Sci. 2022, 46, 1309–1316. [Google Scholar] [CrossRef]
- Khan, M.A.A.K.; Islam, A.B.M.M.K. SARS-CoV-2 Proteins Exploit Host’s Genetic and Epigenetic Mediators for the Annexation of Key Host Signaling Pathways. Front. Mol. Biosci. 2021, 7, 598583. [Google Scholar] [CrossRef] [PubMed]
- Cassiano, L.M.G.; Cavalcante-Silva, V.; Oliveira, M.S.; Prado, B.V.O.; Cardoso, C.G.; Salim, A.C.M.; Franco, G.R.; D’Almeida, V.; Francisco, S.C.; Coimbra, R.S. Vitamin B12 attenuates leukocyte inflammatory signature in COVID-19 via methyl-dependent changes in epigenetic markings. Front. Immunol. 2023, 14, 1048790. [Google Scholar] [CrossRef] [PubMed]
- Ali Ahmed Eid, M. Differential Expression of Cytokines, Transcriptome and miRNA in Coronavirus Disease 2019 (COVID-19) Egyptian’s Patients. 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT04583566 (accessed on 17 May 2023).
- Salgado-Albarrán, M.; Navarro-Delgado, E.I.; Del Moral-Morales, A.; Alcaraz, N.; Baumbach, J.; González-Barrios, R.; Soto-Reyes, E. Comparative transcriptome analysis reveals key epigenetic targets in SARS-CoV-2 infection. NPJ Syst. Biol. Appl. 2021, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Roberts, I.; Muelas, M.W.; Taylor, J.M.; Davison, A.S.; Xu, Y.; Grixti, J.M.; Gotts, N.; Sorokin, A.; Goodacre, R.; Kell, D.B. Untargeted metabolomics of COVID-19 patient serum reveals potential prognostic markers of both severity and outcome. Metabolomics 2022, 18, 6. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef] [PubMed]
- Danlos, F.X.; Grajeda-Iglesias, C.; Durand, S.; Sauvat, A.; Roumier, M.; Cantin, D.; Colomba, E.; Rohmer, J.; Pommeret, F.; Baciarello, G.; et al. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 2021, 12, 258. [Google Scholar] [CrossRef]
- Meoni, G.; Ghini, V.; Maggi, L.; Vignoli, A.; Mazzoni, A.; Salvati, L.; Capone, M.; Vanni, A.; Tenori, L.; Fontanari, P.; et al. Metabolomic/lipidomic profiling of COVID-19 and individual response to tocilizumab. PLoS Pathog. 2021, 17, e1009243. [Google Scholar] [CrossRef]
- Ghini, V.; Meoni, G.; Pelagatti, L.; Celli, T.; Veneziani, F.; Petrucci, F.; Vannucchi, V.; Bertini, L.; Luchinat, C.; Landini, G.; et al. Profiling metabolites and lipoproteins in COMETA, an Italian cohort of COVID-19 patients. PLoS Pathog. 2022, 18, e1010443. [Google Scholar] [CrossRef]
- Barberis, E.; Timo, S.; Amede, E.; Vanella, V.V.; Puricelli, C.; Cappellano, G.; Raineri, D.; Cittone, M.G.; Rizzi, E.; Pedrinelli, A.R.; et al. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to sars-cov-2. Int. J. Mol. Sci. 2020, 21, 8623. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Zhang, T.; Cheng, Z.J.; Guo, B.; Zeng, Y.; Lin, R.; Zheng, P.; Liu, M.; Hu, F.; Li, F.; et al. Effect of a Functional Phospholipid Metabolome-Protein Association Pathway on the Mechanism of COVID-19 Disease Progression. Int. J. Biol. Sci. 2022, 18, 4618–4628. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Baylink, D.J.; Chen, C.S.; Reeves, M.E.; Xiao, J.; Lacy, C.; Lau, E.; Cao, H. The importance of Vitamin D metabolism as a potential prophylactic, immunoregulatory and neuroprotective treatment for COVID-19. J. Transl. Med. 2020, 18, 322. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, P.M.; Griffin, T.P.; Brennan, M.; Mulkerrin, E.C. COVID-19: The older adult and the importance of vitamin D sufficiency. J. Nutr. Sci. 2020, 9, e40. [Google Scholar] [CrossRef]
- Infusino, F.; Marazzato, M.; Mancone, M.; Fedele, F.; Mastroianni, C.M.; Severino, P.; Ceccarelli, G.; Santinelli, L.; Cavarretta, E.; Marullo, A.G.M.; et al. Diet supplementation, probiotics, and nutraceuticals in SARS-CoV-2 infection: A scoping review. Nutrients 2020, 12, 1718. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Shankar, E.M. SARS-CoV-2-Indigenous Microbiota Nexus: Does Gut Microbiota Contribute to Inflammation and Disease Severity in COVID-19? Front. Cell. Infect. Microbiol. 2021, 11, 590874. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Cai, G.; Chen, F.; Christiani, D.C.; Zhang, Z.; Wang, M. Multiomics Evaluation of Gastrointestinal and Other Clinical Characteristics of COVID-19. Gastroenterology 2020, 158, 2298–2301. [Google Scholar] [CrossRef]
- Merenstein, C.; Liang, G.; Whiteside, S.A.; Cobián-Güemes, A.G.; Merlino, M.S.; Taylor, L.J.; Glascock, A.; Bittinger, K.; Tanes, C.; Graham-Wooten, J.; et al. Signatures of COVID-19 Severity and Immune Response in the Respiratory Tract Microbiome. MBio 2021, 12, e0177721. [Google Scholar] [CrossRef]
- Gohil, K.; Samson, R.; Dastager, S.; Dharne, M. Probiotics in the prophylaxis of COVID-19: Something is better than nothing. 3 Biotech 2021, 11, 1. [Google Scholar] [CrossRef]
- Rajput, S.; Paliwal, D.; Naithani, M.; Kothari, A.; Meena, K.; Rana, S. COVID-19 and Gut Microbiota: A Potential Connection. Indian. J. Clin. Biochem. 2021, 36, 266–277. [Google Scholar] [CrossRef]
- Baghbani, T.; Nikzad, H.; Azadbakht, J.; Izadpanah, F.; Haddad Kashani, H. Dual and mutual interaction between microbiota and viral infections: A possible treat for COVID-19. Microb. Cell Fact. 2020, 19, 217. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, J.; Li, F.; Shi, Y. Main Clinical Features of COVID-19 and Potential Prognostic and Therapeutic Value of the Microbiota in SARS-CoV-2 Infections. Front. Microbiol. 2020, 11, 1302. [Google Scholar] [CrossRef] [PubMed]
- Hirota, M.; Tamai, M.; Yukawa, S.; Taira, N.; Matthews, M.M.; Toma, T.; Seto, Y.; Yoshida, M.; Toguchi, S.; Miyagi, M.; et al. Human immune and gut microbial parameters associated with inter-individual variations in COVID-19 mRNA vaccine-induced immunity. Commun. Biol. 2023, 6, 368. [Google Scholar] [CrossRef] [PubMed]
- Leisman, D.E.; Ronner, L.; Pinotti, R.; Taylor, M.D.; Sinha, P.; Calfee, C.S.; Hirayama, A.V.; Mastroiani, F.; Turtle, C.J.; Harhay, M.O.; et al. Cytokine elevation in severe and critical COVID-19: A rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir. Med. 2020, 8, 30404–30405. [Google Scholar] [CrossRef]
- Jabaudon, M.; Blondonnet, R.; Ware, L.B. Biomarkers in acute respiratory distress syndrome. Curr. Opin. Crit. Care 2021, 27, 46–54. [Google Scholar] [CrossRef]
- Aschenbrenner, A.C.; Mouktaroudi, M.; Krämer, B.; Antonakos, N.; Oestreich, M.; Gkizeli, K.; Nuesch-Germano, M.; Saridaki, M.; Bonaguro, L.; Reusch, N.; et al. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. Genome Med. 2020, 13, 7. [Google Scholar] [CrossRef]
- Kuri-Cervantes, L.; Pampena, M.B.; Meng, W.; Rosenfeld, A.M.; Ittner, C.A.G.; Weisman, A.R.; Agyekum, R.S.; Mathew, D.; Baxter, A.E.; Vella, L.A.; et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020, 5, eabd7114. [Google Scholar] [CrossRef]
- Ren, X.; Wen, W.; Fan, X.; Hou, W.; Su, B.; Cai, P.; Li, J.; Liu, Y.; Tang, F.; Zhang, F.; et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell 2021, 184, 1895–1913. [Google Scholar] [CrossRef]
- Neufeldt, C.J.; Cerikan, B.; Cortese, M.; Frankish, J.; Lee, J.Y.; Plociennikowska, A.; Heigwer, F.; Prasad, V.; Joecks, S.; Burkart, S.S.; et al. SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-κB. Commun. Biol. 2022, 5, 45. [Google Scholar] [CrossRef]
- Schultze, J.L.; Aschenbrenner, A.C. COVID-19 and the human innate immune system. Cell 2021, 184, 1671–1692. [Google Scholar] [CrossRef]
- Cheng, L.C.; Kao, T.J.; Phan, N.N.; Chiao, C.C.; Yen, M.C.; Chen, C.F.; Hung, J.H.; Jiang, J.Z.; Sun, Z.; Wang, C.Y.; et al. Novel signaling pathways regulate SARS-CoV and SARS-CoV-2 infectious disease. Medicine 2021, 100, e24321. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, H.Z.; Madhavarapu, S.; Almuqamam, M. Varying Illness Severity in Patients with MyD88 Deficiency Infected with Coronavirus SARS-CoV-2; American Academy of Pediatrics (AAP): Itasca, IL, USA, 2021; pp. 453–454. [Google Scholar]
- Shah, A. Novel Coronavirus-Induced NLRP3 Inflammasome Activation: A Potential Drug Target in the Treatment of COVID-19. Front. Immunol. 2020, 11, 1021. [Google Scholar] [CrossRef] [PubMed]
- Lage, S.L.; Amaral, E.P.; Hilligan, K.L.; Laidlaw, E.; Rupert, A.; Namasivayan, S.; Rocco, J.; Galindo, F.; Kellogg, A.; Kumar, P.; et al. Persistent Oxidative Stress and Inflammasome Activation in CD14highCD16− Monocytes From COVID-19 Patients. Front. Immunol. 2022, 12, 799558. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.Z.; Islam, S.; Matsumoto, K.; Kawai, T. SARS-CoV-2 infection initiates interleukin-17-enriched transcriptional response in different cells from multiple organs. Sci. Rep. 2021, 11, 16814. [Google Scholar] [CrossRef]
- Hariharan, A.; Hakeem, A.R.; Radhakrishnan, S.; Reddy, M.S.; Rela, M. The Role and Therapeutic Potential of NF-kappa-B Pathway in Severe COVID-19 Patients. Inflammopharmacology 2021, 29, 91–100. [Google Scholar] [CrossRef]
- Sharma, A. Randomized trial drug controlled compendious transcriptome analysis supporting broad and phase specific therapeutic potential of multiple candidates in COVID-19. Cytokine 2021, 148, 155719. [Google Scholar] [CrossRef]
- Cusato, J.; Manca, A.; Palermiti, A.; Mula, J.; Costanzo, M.; Antonucci, M.; Trunfio, M.; Corcione, S.; Chiara, F.; De Vivo, E.D.; et al. COVID-19: A Possible Contribution of the MAPK Pathway. Biomedicines 2023, 11, 1459. [Google Scholar] [CrossRef]
- Zhou, Y.; Fu, B.; Zheng, X.; Wang, D.; Zhao, C.; Qi, Y.; Sun, R.; Tian, Z.; Xu, X.; Wei, H. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020, 7, 998–1002. [Google Scholar] [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. InvestIG. 2020, 130, 2620–2629. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, H.K. Re-analysis of Single Cell Transcriptome Reveals That the NR3C1-CXCL8-Neutrophil Axis Determines the Severity of COVID-19. Front. Immunol. 2020, 11, 2145. [Google Scholar] [CrossRef]
- Xiong, Y.; Liu, Y.; Cao, L.; Wang, D.; Guo, M.; Jiang, A.; Guo, D.; Hu, W.; Yang, J.; Tang, Z.; et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 761–770. [Google Scholar] [CrossRef]
- Pan, T.; Cao, G.; Tang, E.; Zhao, Y.; Penaloza-MacMaster, P.; Fang, Y.; Huang, J. A single-cell atlas reveals shared and distinct immune responses and metabolic profiles in SARS-CoV-2 and HIV-1 infections. Front. Genet. 2023, 14, 1309–1316. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Schughart, K.; Pelaia, T.M.; Chew, T.; Kim, K.; Karvunidis, T.; Knippenberg, B.; Teoh, S.; Phu, A.L.; Short, K.R.; et al. Blood transcriptome responses in patients correlate with severity of COVID-19 disease. Front. Immunol. 2023, 13, 1043219. [Google Scholar] [CrossRef] [PubMed]
- Prokop, J.W.; Hartog, N.L.; Chesla, D.; Faber, W.; Love, C.P.; Karam, R.; Abualkheir, N.; Feldmann, B.; Teng, L.; McBride, T.; et al. High-Density Blood Transcriptomics Reveals Precision Immune Signatures of SARS-CoV-2 Infection in Hospitalized Individuals. Front. Immunol. 2021, 12, 694243. [Google Scholar] [CrossRef]
- Su, Y.; Chen, D.; Yuan, D.; Lausted, C.; Choi, J.; Dai, C.L.; Voillet, V.; Duvvuri, V.R.; Scherler, K.; Troisch, P.; et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 2020, 183, 1479–1495. [Google Scholar] [CrossRef]
- Stephenson, E.; Reynolds, G.; Botting, R.A.; Calero-Nieto, F.J.; Morgan, M.D.; Tuong, Z.K.; Bach, K.; Sungnak, W.; Worlock, K.B.; Yoshida, M.; et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021, 27, 904–916. [Google Scholar] [CrossRef]
- Overmyer, K.A.; Shishkova, E.; Miller, I.J.; Balnis, J.; Bernstein, M.N.; Peters-Clarke, T.M.; Meyer, J.G.; Quan, Q.; Muehlbauer, L.K.; Trujillo, E.A.; et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2021, 12, 23–40. [Google Scholar] [CrossRef]
- Zarkovic, N.; Jakovcevic, A.; Mataic, A.; Jaganjac, M.; Vukovic, T.; Waeg, G.; Zarkovic, K. Post-mortem Findings of Inflammatory Cells and the Association of 4-Hydroxynonenal with Systemic Vascular and Oxidative Stress in Lethal COVID-19. Cells 2022, 11, 444. [Google Scholar] [CrossRef]
- Wang, H.; Liu, C.; Xie, X.; Niu, M.; Wang, Y.; Cheng, X.; Zhang, B.; Zhang, D.; Liu, M.; Sun, R.; et al. Resource Multi-omics blood atlas reveals unique features of immune and platelet responses to SARS-CoV-2 Omicron breakthrough infection. Immunity 2023, 56, 1410–1428.e8. [Google Scholar] [CrossRef]
- Vlasova-St Louis, I.; Musubire, A.K.; Meya, D.B.; Nabeta, H.W.; Mohei, H.; Boulware, D.R.; Bohjanen, P.R. Transcriptomic biomarker pathways associated with death in HIV-infected patients with cryptococcal meningitis. BMC Med. Genom. 2021, 14, 108. [Google Scholar] [CrossRef] [PubMed]
- Kellampalli, U.; Mohei, H.; Louis, I.V. Immune Restoration Disorders in Patients with AIDS and Tuberculosis: Novel Treatment Approaches. ACTA Sci. Microbiol. 2021, 4, 133–139. [Google Scholar]
- Louis, I.V.; Chang, C.C.; Shahid, S.; French, M.A.; Bohjanen, P.R. Transcriptomic predictors of paradoxical cryptococcosis-associated immune reconstitution inflammatory syndrome. Open Forum Infect. Dis. 2018, 5, ofy157. [Google Scholar] [CrossRef] [PubMed]
- Mohei, H.; Kellampalli, U.; Louis, I.V. Immune Reconstitution Disorders: Spotlight on Interferons. Int. J. Biomed. Investig. 2019, 2, 119. [Google Scholar] [CrossRef]
- Guo, L.; Louis, I.V.; Bohjanen, P.R. Post-transcriptional regulation of cytokine expression and signaling. Curr. Trends Immunol. 2018, 19, 33–40. [Google Scholar]
- Dean, M.J.; Ochoa, J.B.; Sanchez-Pino, M.D.; Zabaleta, J.; Garai, J.; Del Valle, L.; Wyczechowska, D.; Baiamonte, L.B.; Philbrook, P.; Majumder, R.; et al. Severe COVID-19 Is Characterized by an Impaired Type I Interferon Response and Elevated Levels of Arginase Producing Granulocytic Myeloid Derived Suppressor Cells. Front. Immunol. 2021, 12, 695972. [Google Scholar] [CrossRef]
- Louis, I.V.; Bohjanen, P.R. Post-transcriptional Regulation of Cytokine Signaling During Inflammatory Responses. In Post-Transcriptional Mechanisms in Endocrine Regulation; Goldstrohm, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 55–70. [Google Scholar]
- Whetton, A.D.; Preston, G.W.; Abubeker, S.; Geifman, N. Proteomics and Informatics for Understanding Phases and Identifying Biomarkers in COVID-19 Disease. J. Proteome Res. 2020, 19, 4219–4232. [Google Scholar] [CrossRef]
- Zoodsma, M.; de Nooijer, A.H.; Grondman, I.; Gupta, M.K.; Bonifacius, A.; Koeken, V.A.C.M.; Kooistra, E.; Kilic, G.; Bulut, O.; Gödecke, N.; et al. Targeted proteomics identifies circulating biomarkers associated with active COVID-19 and post-COVID-19. Front. Immunol. 2022, 13, 1027122. [Google Scholar] [CrossRef]
- Shu, T.; Ning, W.; Wu, D.; Xu, J.; Han, Q.; Huang, M.; Zou, X.; Yang, Q.; Yuan, Y.; Bie, Y.; et al. Plasma Proteomics Identify Biomarkers and Pathogenesis of COVID-19. Immunity 2020, 53, 1108–1122. [Google Scholar] [CrossRef]
- D’Alessandro, A.; Thomas, T.; Dzieciatkowska, M.; Hill, R.C.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hod, E.A.; Spitalnik, S.L.; Hansen, K.C. Serum Proteomics in COVID-19 Patients: Altered Coagulation and Complement Status as a Function of IL-6 Level. J. Proteome Res. 2020, 19, 4417–4427. [Google Scholar] [CrossRef]
- Notz, Q.; Schmalzing, M.; Wedekink, F.; Schlesinger, T.; Gernert, M.; Herrmann, J.; Sorger, L.; Weismann, D.; Schmid, B.; Sitter, M.; et al. Pro- and Anti-Inflammatory Responses in Severe COVID-19-Induced Acute Respiratory Distress Syndrome—An Observational Pilot Study. Front. Immunol. 2020, 11, 581338. [Google Scholar] [CrossRef] [PubMed]
- del Molino del Barrio, I.; Hayday, T.S.; Laing, A.G.; Hayday, A.C.; Di Rosa, F. COVID-19: Using high-throughput flow cytometry to dissect clinical heterogeneity. Cytom. Part A 2023, 103, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Caterino, M.; Fedele, R.; Cevenini, A.; Pontillo, M.; Barra, L.; Ruoppolo, M. COVIDomics: The Proteomic and Metabolomic Signatures of COVID-19. Int. J. Mol. Sci. 2022, 23, 2414. [Google Scholar] [CrossRef]
- Medina, L.M.P.; Dzidic, M.; Garcia, M.; Maleki, K.T.; Unge, C.; Lourda, M.; Kvedaraite, E.; Chen, P.; Muvva, J.R.; Cornillet, M.; et al. Targeted plasma proteomics reveals signatures discriminating COVID-19 from sepsis with pneumonia. Respir. Res. 2023, 24, 62. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.A.; Knauer, M.J.; Nicholson, M.; Daley, M.; Van Nynatten, L.R.; Cepinskas, G.; Fraser, D.D. Organ and cell-specific biomarkers of Long-COVID identified with targeted proteomics and machine learning. Mol. Med. 2023, 29, 26. [Google Scholar] [CrossRef]
- Bozorgmehr, N.; Mashhouri, S.; Rosero, E.P.; Xu, L.; Shahbaz, S.; Sligl, W.; Osman, M.; Kutsogiannis, D.J.; Macintyre, E.; O’neil, C.R.; et al. Galectin-9, a player in cytokine release syndrome and a surrogate diagnostic biomarker in SARS-CoV-2 infection. MBio 2021, 12, e00384-21. [Google Scholar] [CrossRef]
- Iwasaki-Hozumi, H.; Maeda, Y.; Niki, T.; Chagan-Yasutan, H.; Bai, G.; Matsuba, T.; Furushima, D.; Ashino, Y.; Hattori, T. Plasma N-Cleaved Galectin-9 Is a Surrogate Marker for Determining the Severity of COVID-19 and Monitoring the Therapeutic Effects of Tocilizumab. Int. J. Mol. Sci. 2023, 24, 3591. [Google Scholar] [CrossRef]
- Al-Nesf, M.A.Y.; Abdesselem, H.B.; Bensmail, I.; Ibrahim, S.; Saeed, W.A.H.; Mohammed, S.S.I.; Razok, A.; Alhussain, H.; Aly, R.M.A.; Al Maslamani, M.; et al. Prognostic tools and candidate drugs based on plasma proteomics of patients with severe COVID-19 complications. Nat. Commun. 2022, 13, 946. [Google Scholar] [CrossRef]
- Toro-Huamanchumo, C.J.; Benites-Meza, J.K.; Mamani-García, C.S.; Bustamante-Paytan, D.; Gracia-Ramos, A.E.; Diaz-Vélez, C.; Barboza, J.J. Efficacy of Colchicine in the Treatment of COVID-19 Patients: A Systematic Review and Meta-Analysis. J. Clin. Med. 2022, 11, 2615. [Google Scholar] [CrossRef]
- Bafadhel, M.; Faner, R.; Taillé, C.; Russell, R.E.K.; Barnes, P.J.; Agustí, A.; Welte, T. Inhaled corticosteroids for the treatment of COVID-19. Eur. Respir. Rev. 2022, 31, 220099. [Google Scholar] [CrossRef]
- Wu, P.; Chen, D.; Ding, W.; Wu, P.; Hou, H.; Bai, Y.; Zhou, Y.; Li, K.; Xiang, S.; Liu, P.; et al. The trans-omics landscape of COVID-19. Nat. Commun. 2021, 12, 4543. [Google Scholar] [CrossRef] [PubMed]
- Saheb Sharif-Askari, F.; Saheb Sharif-Askari, N.; Hafezi, S.; Goel, S.; Ali Hussain Alsayed, H.; Ansari, A.W.; Mahboub, B.; Al-Muhsen, S.; Temsah, M.H.; Hamid, Q.; et al. Upregulation of interleukin-19 in saliva of patients with COVID-19. Sci. Rep. 2022, 12, 16019. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.Y.; Kim, S.C.; Ahn, D.h.; Lee, S.; Chang, S.H.; Jung, S.Y.; Kim, Y.J.; Kim, E.; Kim, J.E.; Kim, Y.S.; et al. Establishment of the large-scale longitudinal multi-omics dataset in COVID-19 patients: Data profile and biospecimen. BMB Rep. 2022, 55, 465–472. [Google Scholar] [CrossRef]
- Schultheiß, C.; Willscher, E.; Paschold, L.; Gottschick, C.; Klee, B.; Henkes, S.S.; Bosurgi, L.; Dutzmann, J.; Sedding, D.; Frese, T.; et al. The IL-1β, IL-6, and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep. Med. 2022, 3, 100663. [Google Scholar] [CrossRef] [PubMed]
- Pathak, G.A.; Singh, K.; Miller-Fleming, T.W.; Wendt, F.R.; Ehsan, N.; Hou, K.; Johnson, R.; Lu, Z.; Gopalan, S.; Yengo, L.; et al. Integrative genomic analyses identify susceptibility genes underlying COVID-19 hospitalization. Nat. Commun. 2021, 12, 4569. [Google Scholar] [CrossRef]
- Kutky, M.; Cross, E.; Treleaven, D.J.; Alam, A.; Lanktree, M.B. The Impact of COVID-19 on Patients With ADPKD. Can. J. Kidney Health Dis. 2021, 8, 20543581211056479. [Google Scholar] [CrossRef]
- Hu, Y.; Rehawi, G.; Moyon, L.; Gerstner, N.; Ogris, C.; Knauer-Arloth, J.; Bittner, F.; Marsico, A.; Mueller, N.S. Network Embedding Across Multiple Tissues and Data Modalities Elucidates the Context of Host Factors Important for COVID-19 Infection. Front. Genet. 2022, 13, 909714. [Google Scholar] [CrossRef]
- Wang, D.; Kumar, V.; Burnham, K.L.; Mentzer, A.J.; Marsden, B.D.; Knight, J.C. COMBATdb: A database for the COVID-19 Multi-Omics Blood ATlas. Nucleic Acids Res. 2023, 51, D896–D905. [Google Scholar] [CrossRef]
- Bhattarai, S.; Gupta, A.; Ali, E.; Ali, M.; Riad, M.; Adhikari, P.; Mostafa, J.A. Can Big Data and Machine Learning Improve Our Understanding of Acute Respiratory Distress Syndrome? Cureus 2021, 13, e13529. [Google Scholar] [CrossRef]
- Lipman, D.; Safo, S.E.; Chekouo, T. Multi-omic analysis reveals enriched pathways associated with COVID-19 and COVID-19 severity. PLoS ONE 2022, 17, e0267047. [Google Scholar] [CrossRef]
- Richard, V.R.; Gaither, C.; Popp, R.; Chaplygina, D.; Brzhozovskiy, A.; Kononikhin, A.; Mohammed, Y.; Zahedi, R.P.; Nikolaev, E.N.; Borchers, C.H. Early Prediction of COVID-19 Patient Survival by Targeted Plasma Multi-Omics and Machine Learning. Mol. Cell. Proteom. 2022, 21, 100277. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hasan, M.R.; Ahmed, K.A.; Hossain, M.Z. Machine learning to analyse omic-data for COVID-19 diagnosis and prognosis. BMC Bioinform. 2023, 24, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Chen, Z.; Xiao, Y.; Lai, D.; Wang, X.; Fang, X.; Shu, Q. Multi-omics and immune cells’ profiling of COVID-19 patients for ICU admission prediction: In silico analysis and an integrated machine learning-based approach in the framework of Predictive, Preventive, and Personalized Medicine. EPMA J. 2023, 14, 101–117. [Google Scholar] [CrossRef]
- de Andrés-Galiana, E.J.; Fernández-Martínez, J.L.; Álvarez-Machancoses, Ó.; Bea, G.; Galmarini, C.M.; Kloczkowski, A. Analysis of transcriptomic responses to SARS-CoV-2 reveals plausible defective pathways responsible for increased susceptibility to infection and complications and helps to develop fast-track repositioning of drugs against COVID-19. Comput. Biol. Med. 2022, 149, 106029. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Bhattacharya, M.; Agoramoorthy, G.; Lee, S.S. The Drug Repurposing for COVID-19 Clinical Trials Provide Very Effective Therapeutic Combinations: Lessons Learned From Major Clinical Studies. Front. Pharmacol. 2021, 12, 704205. [Google Scholar] [CrossRef]
- Mousavi, S.; Zare, S.; Mirzaei, M.; Feizi, A. Novel Drug Design for Treatment of COVID-19: A Systematic Review of Preclinical Studies. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 2044282. [Google Scholar] [CrossRef]
- Abbasi, J. COVID-19 and mRNA vaccines—First large test for a new approach. JAMA—J. Am. Med. Assoc. 2020, 324, 1125–1127. [Google Scholar] [CrossRef]
- Akinosoglou, K.; Rigopoulos, E.A.; Schinas, G.; Kaiafa, G.; Polyzou, E.; Tsoupra, S.; Tzouvelekis, A.; Gogos, C.; Savopoulos, C. Remdesivir Use in the Real-World Setting: An Overview of Available Evidence. Viruses 2023, 15, 1167. [Google Scholar] [CrossRef]
- Chera, A.; Tanca, A. Remdesivir: The first FDA-approved anti-COVID-19 Treatment for Young Children. Discoveries 2022, 10, e151. [Google Scholar] [CrossRef]
- Kiraz, A. SARS-CoV-2 infection, hypercoagulability and hereditary thrombophilia factors. Gazi Med. J. 2022, 33, P1–P2. [Google Scholar]
- Jayk Bernal, A.; Gomes da Silva, M.M.; Musungaie, D.B.; Kovalchuk, E.; Gonzalez, A.; Delos Reyes, V.; Martín-Quirós, A.; Caraco, Y.; Williams-Diaz, A.; Brown, M.L.; et al. Molnupiravir for Oral Treatment of Covid-19 in Nonhospitalized Patients. N. Engl. J. Med. 2022, 386, 509–520. [Google Scholar] [CrossRef]
- Stephenson, J. FDA Authorizes Pharmacists to Prescribe Oral Antiviral Medication for COVID-19. JAMA Health Forum 2022, 3, e222968. [Google Scholar] [CrossRef] [PubMed]
- Chourasia, P.; Maringanti, B.S.; Edwards-Fligner, M.; Gangu, K.; Bobba, A.; Sheikh, A.B.; Shekhar, R. Paxlovid (Nirmatrelvir and Ritonavir) Use in Pregnant and Lactating Woman: Current Evidence and Practice Guidelines—A Scoping Review. Vaccines 2023, 11, 107. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, H.; Peluso, M.J.; Rodgers, K.; Aberg, J.A.; Patterson, T.F.; Tamburro, R.; Baizer, L.; Goldman, J.D.; Rouphael, N.; Deitchman, A.; et al. Therapeutic trials for long COVID-19: A call to action from the interventions taskforce of the RECOVER initiative. Front. Immunol. 2023, 14, 1129459. [Google Scholar] [CrossRef] [PubMed]
- Henao-Restrepo, A.M.; Pan, H.; Peto, R.; Preziosi, M.P.; Sathiyamoorthy, V.; Abdool Karim, Q.; Alejandria, M.; Hernández García, C.; Marie-Paule, K.; Malekzadeh, R.; et al. Remdesivir and three other drugs for hospitalised patients with COVID-19: Final results of the WHO Solidarity randomised trial and updated meta-analyses. Lancet 2022, 399, 1941–1953. [Google Scholar] [CrossRef]
- Louis, I.V.; Abadie, J. Prophylactic Ribonucleic Acid Vaccines to Combat RNA Viral Infections in Humans. In RNA Therapeutics—History, Design, Manufacturing, and Applications; IntechOpen: London, UK, 2022; pp. 14–38. ISBN 978-1-80355-658-1. [Google Scholar]
- Aggarwal, N.R.; Beaty, L.E.; Bennett, T.D.; Carlson, N.E.; Davis, C.B.; Kwan, B.M.; Mayer, D.A.; Ong, T.C.; Russell, S.; Steele, J.; et al. Real-World Evidence of the Neutralizing Monoclonal Antibody Sotrovimab for Preventing Hospitalization and Mortality in COVID-19 Outpatients. J. Infect. Dis. 2022, 226, 2129–2136. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, R.L.; Nirula, A.; Chen, P.; Boscia, J.; Heller, B.; Morris, J.; Huhn, G.; Cardona, J.; Mocherla, B.; Stosor, V.; et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: A randomized clinical trial. JAMA—J. Am. Med. Assoc. 2021, 325, 632–644. [Google Scholar] [CrossRef]
- Tamura, T.; Ito, J.; Uriu, K.; Zahradnik, J.; Kida, I.; Nasser, H.; Shofa, M.; Oda, Y.; Lytras, S.; Nao, N.; et al. Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants. Nat. Commun. 2023, 14, 2800. [Google Scholar] [CrossRef]
- Reis, G.; Moreira Silva, E.A.S.; Medeiros Silva, D.C.; Thabane, L.; Campos, V.H.S.; Ferreira, T.S.; Santos, C.V.Q.; Nogueira, A.M.R.; Almeida, A.P.F.G.; Savassi, L.C.M.; et al. Early Treatment with Pegylated Interferon Lambda for Covid-19. N. Engl. J. Med. 2023, 388, 518–528. [Google Scholar] [CrossRef]
- Siemieniuk, R.A.C.; Bartoszko, J.J.; Ge, L.; Zeraatkar, D.; Izcovich, A.; Pardo-Hernandez, H.; Rochwerg, B.; Lamontagne, F.; Han, M.A.; Kum, E.; et al. Drug treatments for covid-19: Living systematic review and network meta-Analysis. BMJ 2020, 370, m2980. [Google Scholar] [CrossRef]
- Marconi, V.C.; Ramanan, A.V.; de Bono, S.; Kartman, C.E.; Krishnan, V.; Liao, R.; Piruzeli, M.L.B.; Goldman, J.D.; Alatorre-Alexander, J.; de Cassia Pellegrini, R.; et al. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): A randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir. Med. 2021, 9, 1407–1418. [Google Scholar] [CrossRef]
- Yaqinuddin, A.; Kashir, J. Novel therapeutic targets for SARS-CoV-2-induced acute lung injury: Targeting a potential IL-1β/neutrophil extracellular traps feedback loop. Med. Hypotheses 2020, 143, 109906. [Google Scholar] [CrossRef] [PubMed]
- NCT04482699. RAPA-501-Allo Off-the-Shelf Therapy of COVID-19. Clinicaltrials 2020. Available online: https://clinicaltrials.gov/show/NCT04482699 (accessed on 4 June 2023).
- Peterson, J.H.; Paranjape, N.S.; Grundlingh, N.; Priestley, J.L. Outcomes and Adverse Effects of Baricitinib Versus Tocilizumab in the Management of Severe COVID-19. Crit. Care Med. 2023, 51, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Aydin, S.I.; Derespina, K.R.; Bansal, P.B.; Kowalsky, S.; Trachtman, R.; Gillen, J.K.; Perez, M.M.; Soshnick, S.H.; Conway, E.E.; et al. Multisystem Inflammatory Syndrome in Children Associated with Severe Acute Respiratory Syndrome Coronavirus 2 Infection (MIS-C): A Multi-institutional Study from New York City. J. Pediatr. 2020, 224, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Alavi Darazam, I.; Shokouhi, S.; Pourhoseingholi, M.A.; Naghibi Irvani, S.S.; Mokhtari, M.; Shabani, M.; Amirdosara, M.; Torabinavid, P.; Golmohammadi, M.; Hashemi, S.P.; et al. Role of interferon therapy in severe COVID-19: The COVIFERON randomized controlled trial. Sci. Rep. 2021, 11, 8059. [Google Scholar] [CrossRef]
- Amer, M.; Kamel, A.M.; Bawazeer, M.; Maghrabi, K.; Butt, A.; Dahhan, T.; Kseibi, E.; Khurshid, S.M.; Abujazar, M.; Alghunaim, R.; et al. Clinical characteristics and outcomes of critically ill mechanically ventilated COVID-19 patients receiving interleukin-6 receptor antagonists and corticosteroid therapy: A preliminary report from a multinational registry. Eur. J. Med. Res. 2021, 26, 117. [Google Scholar] [CrossRef]
- Abani, O.; Abbas, A.; Abbas, F.; Abbas, M.; Abbasi, S.; Abbass, H.; Abbott, A.; Abdallah, N.; Abdelaziz, A.; Abdelfattah, M.; et al. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet 2021, 397, 1637–1645. [Google Scholar] [CrossRef]
- Malekzadeh, R.; Abedini, A.; Mohsenpour, B.; Sharifipour, E.; Ghasemian, R.; Javad-Mousavi, S.A.; Khodashahi, R.; Darban, M.; Kalantari, S.; Abdollahi, N.; et al. Subcutaneous tocilizumab in adults with severe and critical COVID-19: A prospective open-label uncontrolled multicenter trial. Int. Immunopharmacol. 2020, 89, 107102. [Google Scholar] [CrossRef]
- Al-Mistarehi, A.H.; Al Sbihi, A.L.; Ata, E.M.; Alomari, S.; Khassawneh, A.B.; Kheirallah, K.A.; Allowh, S.H.; Ata, R.H.; Mashina, M.M.; Alzoubi, A.; et al. The added clinical value of tocilizymab in treating adults hospitalized with COVID-19: A retrospective cohort study. Chest 2022, 162, A655–A656. [Google Scholar] [CrossRef]
- Lescure, F.X.; Honda, H.; Fowler, R.A.; Lazar, J.S.; Shi, G.; Wung, P.; Patel, N.; Hagino, O.; Bazzalo, I.J.; Casas, M.M.; et al. Sarilumab in patients admitted to hospital with severe or critical COVID-19: A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Respir. Med. 2021, 9, 522–532. [Google Scholar] [CrossRef]
- Gritti, G.; Raimondi, F.; Bottazzi, B.; Ripamonti, D.; Riva, I.; Landi, F.; Alborghetti, L.; Frigeni, M.; Damiani, M.; Micò, C.; et al. Siltuximab downregulates interleukin-8 and pentraxin 3 to improve ventilatory status and survival in severe COVID-19. Leukemia 2021, 35, 2710–2714. [Google Scholar] [CrossRef] [PubMed]
- Monk, P.D.; Brookes, J.L.; Tear, V.J.; Batten, T.N.; Mankowski, M.; Adzic-Vukicevic, T.; Crooks, M.G.; Dosanjh, D.P.S.; Kraft, M.; Brightling, C.E.; et al. Nebulised interferon-β1a (SNG001) in hospitalised COVID-19: SPRINTER phase III study. ERJ Open Res. 2023, 9, 00605–02022. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhan, Y.; Zhu, L.; Hou, Z.; Liu, F.; Song, P.; Qiu, F.; Wang, X.; Zou, X.; Wan, D.; et al. Retrospective Multicenter Cohort Study Shows Early Interferon Therapy Is Associated with Favorable Clinical Responses in COVID-19 Patients. Cell Host Microbe 2020, 28, 455–464.e2. [Google Scholar] [CrossRef] [PubMed]
- Saeedi-Boroujeni, A.; Nashibi, R.; Ghadiri, A.A.; Nakajima, M.; Salmanzadeh, S.; Mahmoudian-Sani, M.R.; Hanafi, M.G.; Sharhani, A.; Khodadadi, A. Tranilast as an Adjunctive Therapy in Hospitalized Patients with Severe COVID-19: A Randomized Controlled Trial. Arch. Med. Res. 2022, 53, 368–377. [Google Scholar] [CrossRef]
- Generali, D.; Bosio, G.; Malberti, F.; Cuzzoli, A.; Testa, S.; Romanini, L.; Fioravanti, A.; Morandini, A.; Pianta, L.; Giannotti, G.; et al. Canakinumab as treatment for COVID-19-related pneumonia: A prospective case-control study. Int. J. Infect. Dis. 2021, 104, 433–440. [Google Scholar] [CrossRef]
- Cavalli, G.; De Luca, G.; Campochiaro, C.; Della-Torre, E.; Ripa, M.; Canetti, D.; Oltolini, C.; Castiglioni, B.; Tassan Din, C.; Boffini, N.; et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: A retrospective cohort study. Lancet Rheumatol. 2020, 2, e325–e331. [Google Scholar] [CrossRef]
- Kyriazopoulou, E.; Panagopoulos, P.; Metallidis, S.; Dalekos, G.N.; Poulakou, G.; Gatselis, N.; Karakike, E.; Saridaki, M.; Loli, G.; Stefos, A.; et al. An open label trial of anakinra to prevent respiratory failure in COVID-19. Elife 2021, 10, e66125. [Google Scholar] [CrossRef]
- Abani, O.; Abbas, A.; Abbas, F.; Abbas, J.; Abbas, K.; Abbas, M.; Abbasi, S.; Abbass, H.; Abbott, A.; Abbott, A.; et al. Baricitinib in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial and updated meta-analysis. Lancet 2022, 400, 1102. [Google Scholar] [CrossRef]
- Kalil, A.C.; Patterson, T.F.; Mehta, A.K.; Tomashek, K.M.; Wolfe, C.R.; Ghazaryan, V.; Marconi, V.C.; Ruiz-Palacios, G.M.; Hsieh, L.; Kline, S.; et al. Baricitinib plus Remdesivir for Hospitalized Adults with Covid-19. N. Engl. J. Med. 2021, 384, 795–807. [Google Scholar] [CrossRef]
- NCT04970719. Baricitinib in Hospitalized Covid-19 Patients With Diabetes Mellitus. Available online: https://clinicaltrials.gov/show/NCT04970719 (accessed on 6 June 2023).
- Guimarães, P.O.; Quirk, D.; Furtado, R.H.; Maia, L.N.; Saraiva, J.F.; Antunes, M.O.; Kalil Filho, R.; Junior, V.M.; Soeiro, A.M.; Tognon, A.P.; et al. Tofacitinib in Patients Hospitalized with Covid-19 Pneumonia. N. Engl. J. Med. 2021, 385, 406–415. [Google Scholar] [CrossRef]
- Gianfrancesco, M.; Hyrich, K.L.; Al-Adely, S.; Al-Adely, S.; Carmona, L.; Danila, M.I.; Gossec, L.; Gossec, L.; Izadi, Z.; Jacobsohn, L.; et al. Characteristics associated with hospitalisation for COVID-19 in people with rheumatic disease: Data from the COVID-19 Global Rheumatology Alliance physician-reported registry. Ann. Rheum. Dis. 2020, 79, 859–866. [Google Scholar] [CrossRef]
- Izadi, Z.; Brenner, E.J.; Mahil, S.K.; Dand, N.; Yiu, Z.Z.N.; Yates, M.; Ungaro, R.C.; Zhang, X.; Agrawal, M.; Colombel, J.F.; et al. Association between Tumor Necrosis Factor Inhibitors and the Risk of Hospitalization or Death among Patients with Immune-Mediated Inflammatory Disease and COVID-19. JAMA Netw. Open 2021, 4, e2129639. [Google Scholar] [CrossRef]
- Fisher, B.A.; Veenith, T.; Slade, D.; Gaskell, C.; Rowland, M.; Whitehouse, T.; Scriven, J.; Parekh, D.; Balasubramaniam, M.S.; Cooke, G.; et al. Namilumab or infliximab compared with standard of care in hospitalised patients with COVID-19 (CATALYST): A randomised, multicentre, multi-arm, multistage, open-label, adaptive, phase 2, proof-of-concept trial. Lancet Respir. Med. 2022, 10, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Temesgen, Z.; Burger, C.D.; Baker, J.; Polk, C.; Libertin, C.R.; Kelley, C.F.; Marconi, V.C.; Orenstein, R.; Catterson, V.M.; Aronstein, W.S.; et al. Lenzilumab in hospitalised patients with COVID-19 pneumonia (LIVE-AIR): A phase 3, randomised, placebo-controlled trial. Lancet Respir. Med. 2022, 10, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Umemura, Y.; Mitsuyama, Y.; Minami, K.; Nishida, T.; Watanabe, A.; Okada, N.; Yamakawa, K.; Nochioka, K.; Fujimi, S. Efficacy and safety of nintedanib for pulmonary fibrosis in severe pneumonia induced by COVID-19: An interventional study. Int. J. Infect. Dis. 2021, 108, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; De Souza, R.; Nadkar, M.; Guleria, R.; Trikha, A.; Joshi, S.R.; Loganathan, S.; Vaidyanathan, S.; Marwah, A.; Athalye, S.N. A two-arm, randomized, controlled, multi-centric, open-label phase-2 study to evaluate the efficacy and safety of Itolizumab in moderate to severe ARDS patients due to COVID-19. Expert. Opin. Biol. Ther. 2021, 21, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Maslennikov, R.; Ivashkin, V.; Vasilieva, E.; Chipurik, M.; Semikova, P.; Semenets, V.; Russkova, T.; Levshina, A.; Grigoriadis, D.; Magomedov, S.; et al. Interleukin 17 antagonist netakimab is effective and safe in the new coronavirus infection (COVID-19). Eur. Cytokine Netw. 2021, 32, 8–14. [Google Scholar] [CrossRef]
- Declercq, J.; Van Damme, K.F.A.; De Leeuw, E.; Maes, B.; Bosteels, C.; Tavernier, S.J.; De Buyser, S.; Colman, R.; Hites, M.; Verschelden, G.; et al. Effect of anti-interleukin drugs in patients with COVID-19 and signs of cytokine release syndrome (COV-AID): A factorial, randomised, controlled trial. Lancet Respir. Med. 2021, 9, 1427–1438. [Google Scholar] [CrossRef]
- Arish, M.; Naz, F. Personalized therapy: Can it tame the COVID-19 monster? Per. Med. 2021, 18, 583–593. [Google Scholar] [CrossRef]
- Dearlove, B.; Lewitus, E.; Bai, H.; Li, Y.; Reeves, D.B.; Joyce, M.G.; Scott, P.T.; Amare, M.F.; Vasan, S.; Michael, N.L.; et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. USA 2020, 117, 23652–23662. [Google Scholar] [CrossRef]
- Lisewski, A.M. Interim estimates in null models of COVID-19 vaccine effectiveness. Int. J. Infect. Dis. 2021, 106, 169–170. [Google Scholar] [CrossRef] [PubMed]
- Haas, E.J.; Angulo, F.J.; McLaughlin, J.M.; Anis, E.; Singer, S.R.; Khan, F.; Brooks, N.; Smaja, M.; Mircus, G.; Pan, K.; et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: An observational study using national surveillance data. Lancet 2021, 397, 1819–1829. [Google Scholar] [CrossRef]
- Iwasaki, A. What reinfections mean for COVID-19. Lancet Infect. Dis. 2021, 21, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Tillett, R.L.; Sevinsky, J.R.; Hartley, P.D.; Kerwin, H.; Crawford, N.; Gorzalski, A.; Laverdure, C.; Verma, S.C.; Rossetto, C.C.; Jackson, D.; et al. Genomic evidence for reinfection with SARS-CoV-2: A case study. Lancet Infect. Dis. 2021, 21, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Anderegg, N.; Schwab, T.; Borcard, L.; Mugglin, C.; Keune-Dübi, B.; Ramette, A.; Fenner, L. Population-Based Severe Acute Respiratory Syndrome Coronavirus 2 Whole-Genome Sequencing and Contact Tracing During the Coronavirus Disease 2019 Pandemic in Switzerland. J. Infect. Dis. 2023, 228, 251–260. [Google Scholar] [CrossRef]
- To, K.K.W.; Hung, I.F.N.; Ip, J.D.; Chu, A.W.H.; Chan, W.M.; Tam, A.R.; Fong, C.H.Y.; Yuan, S.; Tsoi, H.W.; Ng, A.C.K.; et al. Coronavirus Disease 2019 (COVID-19) Re-infection by a Phylogenetically Distinct Severe Acute Respiratory Syndrome Coronavirus 2 Strain Confirmed by Whole Genome Sequencing. Clin. Infect. Dis. 2021, 73, e2946–e2951. [Google Scholar] [CrossRef]
- Satpati, P.; Sarangi, S.; Gantait, K.; Endow, S.; Mandal, N.; Panchanan, K.; Subhadip, B.; Soham, S. Sero-surveillance (IgG) of SARS-CoV-2 among Asymptomatic General population of Paschim Medinipur, West Bengal, India. medRxiv 2020. [Google Scholar] [CrossRef]
- Deshmukh, K.; Khanna, A.; Talwar, D. “COVID Vaccine” is not the excuse to delay adaptation to the “New-Normal”. J. Fam. Med. Prim. Care 2020, 9, 5076–5078. [Google Scholar] [CrossRef]
- Chirico, F.; Nucera, G.; Ilesanmi, O.; Afolabi, A.; Pruc, M.; Szarpak, L. Identifying asymptomatic cases during the mass COVID-19 vaccination campaign: Insights and implications for policy makers. Future Virol. 2022, 17, 141–144. [Google Scholar] [CrossRef]
- Rambo, A.P.S.; Gonçalves, L.F.; Gonzáles, A.I.; Rech, C.R.; de Paiva, K.M.; Haas, P. Impact of super-spreaders on COVID-19: Systematic review. Sao Paulo Med. J. 2021, 139, 163–169. [Google Scholar] [CrossRef]
- WHO. Tracking SARS-CoV-2 Variants. 2021. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 17 June 2023).
- ClinicalTrials. A Study of SARS CoV-2 Infection and Potential Transmission in Individuals Immunized with Moderna COVID-19 Vaccine (CoVPN 3006). NCT04811664. 2021. Available online: https://www.clinicaltrials.gov/ct2/show/study/NCT04811664 (accessed on 30 May 2023).
- Teijaro, J.R.; Farber, D.L. COVID-19 vaccines: Modes of immune activation and future challenges. Nat. Rev. Immunol. 2021, 21, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Keehner, J.; Horton, L.E.; Pfeffer, M.A.; Longhurst, C.A.; Schooley, R.T.; Currier, J.S.; Abeles, S.R.; Torriani, F.J. SARS-CoV-2 Infection after Vaccination in Health Care Workers in California. N. Engl. J. Med. 2021, 384, 1774–1775. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, J.; Hozumi, Y.; Yin, C.; Wei, G.W. Emerging Vaccine-Breakthrough SARS-CoV-2 Variants. ACS Infect. Dis. 2022, 8, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, M.; Penfold, R.S.; Merino, J.; Sudre, C.H.; Molteni, E.; Berry, S.; Canas, L.S.; Graham, M.S.; Klaser, K.; Modat, M.; et al. Risk factors and disease profile of post-vaccination SARS-CoV-2 infection in UK users of the COVID Symptom Study app: A prospective, community-based, nested, case-control study. Lancet Infect. Dis. 2022, 22, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Iwagami, M.; Yasuhara, J.; Takagi, H.; Kuno, T. Protective effect of COVID-19 vaccination against long COVID syndrome: A systematic review and meta-analysis. Vaccine 2023, 41, 1783–1790. [Google Scholar] [CrossRef]
- Marks, P. Coronavirus (COVID-19) Update: FDA Recommends Inclusion of Omicron BA.4/5 Component for COVID-19 Vaccine Booster Doses. 2022. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-recommends-inclusion-omicron-ba45-component-covid-19-vaccine-booster (accessed on 29 June 2023).
- Pfizer & BioNTech. Pfizer and BioNTech Submit Application to U.S. FDA for Emergency Use Authorization of Omicron BA.4/BA.5-Adapted Bivalent COVID-19 Vaccine. 2022. Available online: https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-submit-application-us-fda-emergency-use (accessed on 29 May 2023).
- Aguiar, M.; Van-Dierdonck, J.B.; Mar, J.; Stollenwerk, N. The role of mild and asymptomatic infections on COVID-19 vaccines performance: A modeling study. J. Adv. Res. 2022, 39, 157–166. [Google Scholar] [CrossRef]
- Liu, Y.-t.; Wang, S.-s. Ixekizumab successfully treated severe pityriasis rubra pilaris after COVID-19 vaccination. Ski. Health Dis. 2023, 3, e139. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vlasova-St. Louis, I.; Fang, D.; Amer, Y.; Mohei, H. COVID-19-Omics Report: From Individual Omics Approaches to Precision Medicine. Reports 2023, 6, 45. https://doi.org/10.3390/reports6040045
Vlasova-St. Louis I, Fang D, Amer Y, Mohei H. COVID-19-Omics Report: From Individual Omics Approaches to Precision Medicine. Reports. 2023; 6(4):45. https://doi.org/10.3390/reports6040045
Chicago/Turabian StyleVlasova-St. Louis, Irina, Daniel Fang, Yara Amer, and Hesham Mohei. 2023. "COVID-19-Omics Report: From Individual Omics Approaches to Precision Medicine" Reports 6, no. 4: 45. https://doi.org/10.3390/reports6040045
APA StyleVlasova-St. Louis, I., Fang, D., Amer, Y., & Mohei, H. (2023). COVID-19-Omics Report: From Individual Omics Approaches to Precision Medicine. Reports, 6(4), 45. https://doi.org/10.3390/reports6040045








