Abstract
The objective of the present study is to make a comparison between various free and open-source software used for medical image processing, such as 3D Slicer (version 4.11), ITK-Snap (version 3.8), and Invesalius (version 3.1) in its application for the calculation of solitary fibrous tumor volumes. Knowing the size, shape, and volume of mesothelioma is decisive for clinical decision-making by health personnel when performing surgery; the currently used standard procedure is manual segmentation through magnetic resonance imaging (MRI). This process tends to take a long time to complete. On the other hand, automatic segmentation software is much faster and more user-friendly, so looking for software that gives us greater accuracy when doing this task is very important. This work obtained magnetic resonance imaging (MRI) of a mesothelioma patient, and the images were segmented in the 3 different programs to evaluate the concordance between the software later.
1. Introduction
Cancer is a major global public health problem because of its high disease burden. New reports show that lung cancer has been rising in men and women as leading cancer over the past decade [1]. Besides those tumors that directly affect the lungs, the pleura can also be affected by the development of fibrous tumors. A solitary fibrous tumor (SFT) are soft tissue sarcomas that usually form in the pleura, is not malignant, and causes symptoms depending on its size [2].
SFT causes Doege-Potter syndrome (DPS), a paraneoplastic disorder with symptomatic, severe, and persistent hypoglycemia), being a rare cause (2–4% of cases) of which some cases have been described [3,4,5]. To distinguish SFT from other intrathoracic tumors, computed tomography (CT) is advantageous since it presents as a homogeneous round and well-defined mass between 10 and 20 cm in diameter [6]. Surgical management of the tumor is the treatment of DPS. Chemotherapy, radiofrequency ablation, cryoablation, or chemoembolization are tools used in SFT/DPS. Additionally, management of hypoglycemia is mandatory and glucocorticoids improve insulin-related outcomes in the course of the disease. The prognosis is favorable but depends on the size of the tumor and the paraneoplastic complications, so there have been reports of cases with successful follow-up [7,8]. Given a tumor that causes a paraneoplastic syndrome, with slow growth and that adopts a large size oppressing the affected lung, it is crucial to approximate the actual size of the tumor with the use of software from the images obtained in radiology [7].
Manual segmentation of MRI slices has long been the standard for volume measurement; however, this process tends to be time-consuming and inaccurate [9]. Recently, different semi-automatic and automatic segmentation methods have simplified this task and tend to have greater accuracy in calculating these volumes [10]. We aimed to use three different open-source software used for semi-automated glioblastoma segmentation; these are 3D Slicer, ITK-Snap, and Invesalius, to compare the tumor volumes obtained later.
2. Materials and Methods
2.1. Study Design and Patient
We conducted an exploratory comparative study using Dycom CAT data from a recently reported patient with SFT [4]. A 74-year-old man with SFT and hypoglycemia (38 mg/dL) was admitted to hospital with chest pain for two weeks. CT showed a mass and left-sided pleural effusion, which was confirmed by bronchofiberoscopy. Pathological analysis found SFT to be positive for B-cell lymphoma 2 (BCL-2), focal CD34+ and CD99+. After the diagnosis of DPS in SFT, surgery was performed and the intrathoracic tumor (weight: 3.1 kg) was completely resected. Three weeks after discharge, a chest X-ray showed re-expantion of the left hemithorax, which gradually improved at follow-up.
2.2. Software Selection
The following criteria were used for selecting software: (1) The software should offer semi-automatic segmentation from an MRI (2) The software should have a graphical user interface that can be used without specialized knowledge. (3) The software must be open source and free. (4) There must be documentation of having been previously used for tumor segmentation. The chosen Software: 3D Slicer, Invesalius, and Itk-SNAP.
2.3. Segmentation Process of the Healthy Part of the Lung
2.3.1. 3D Slicer
As a first step, the area to work had to be delimited, sectioning the patient’s thorax using the “Cut Volume” tool found in “Converters”. Once the cut is established, a new segment must be created in the “Segment Editor” section. Then the “Threshold” tool must be used, with which the fill segment is delimited based on the intensity range in Hounsfield units (HU).
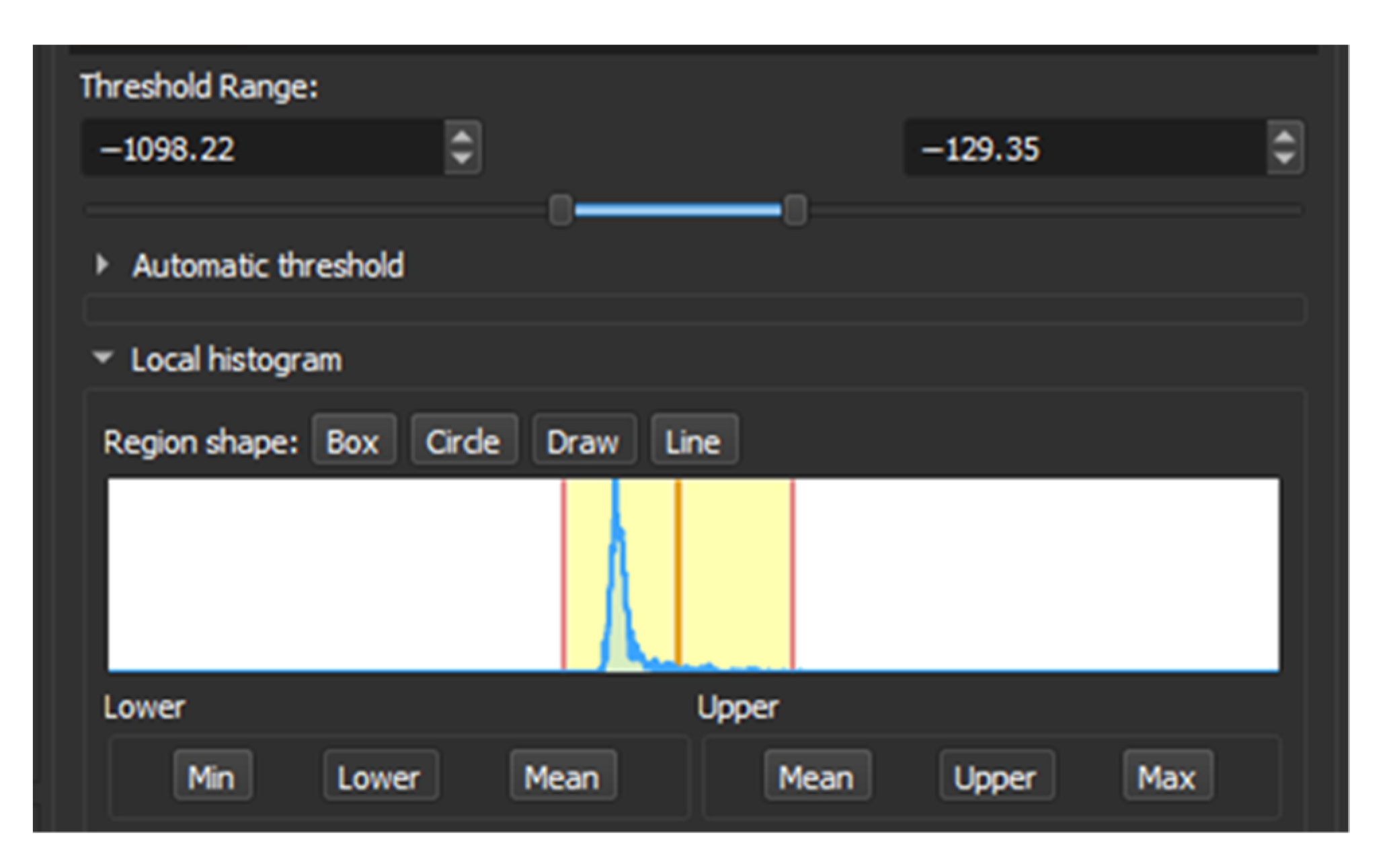
To find the appropriate range for this threshold, we can draw a line within the area to be segmented and then review the local histogram, in which we can see all the values contained in the section. Then it was found that the range is between −1098.22 to −129.35 HU (Figure 1).
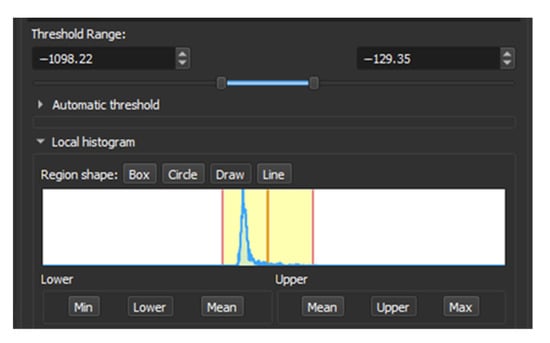
Figure 1.
Local Histogram from 3D Slicer.
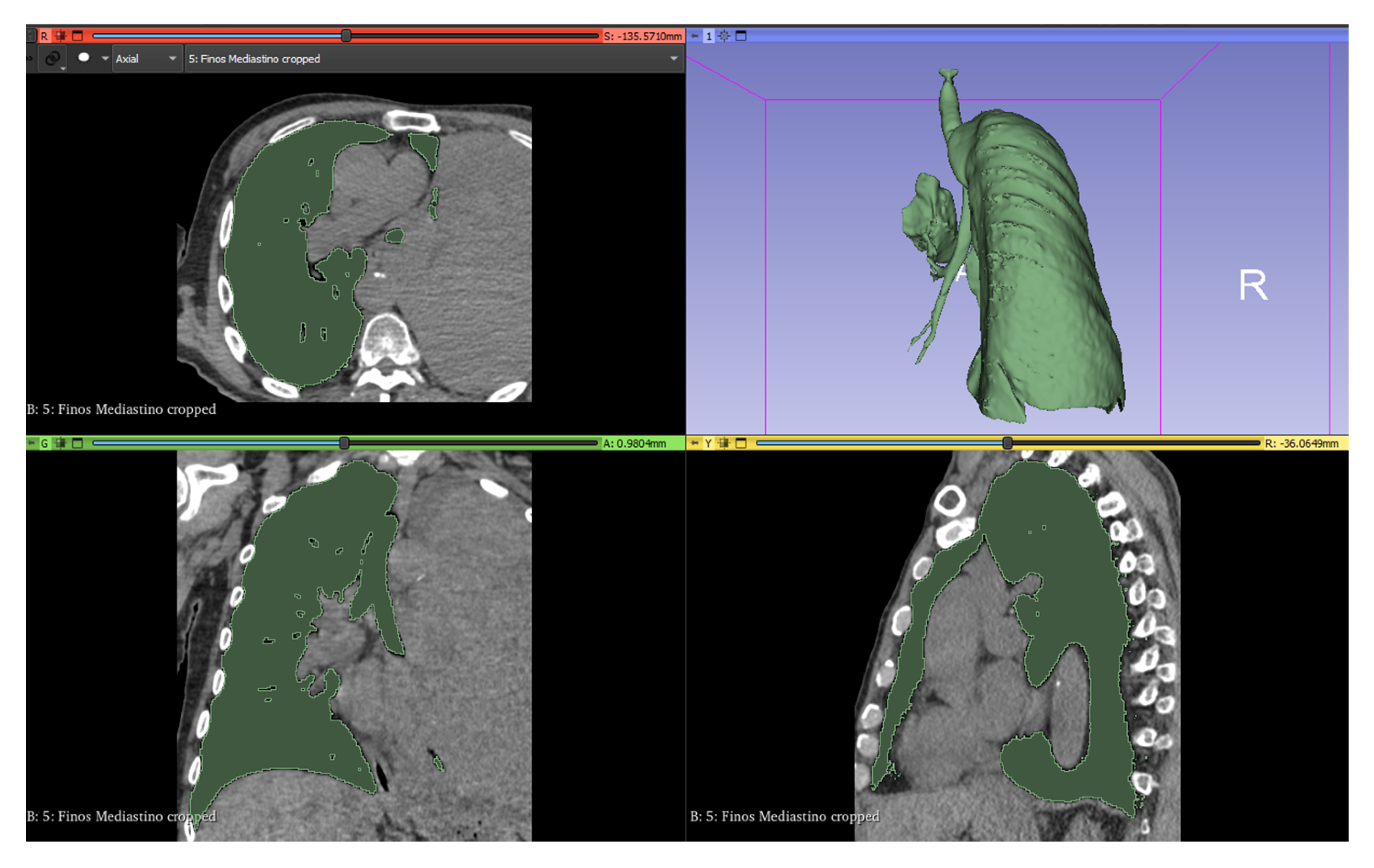
To clean those areas that have the same range of values that escape the tumor the “Scissors” tool was used in “Erase outside” mode to remove the external parts and “Erase inside” to delimit the section of interest. Finally, with the “Islands” tool in “Keep largest island” mode, we can obtain the final model of the tumor (Figure 2).
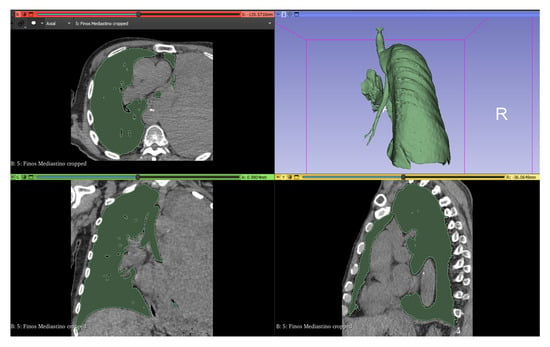
Figure 2.
Lung segmentation in 3D Slicer.
2.3.2. Invesalius
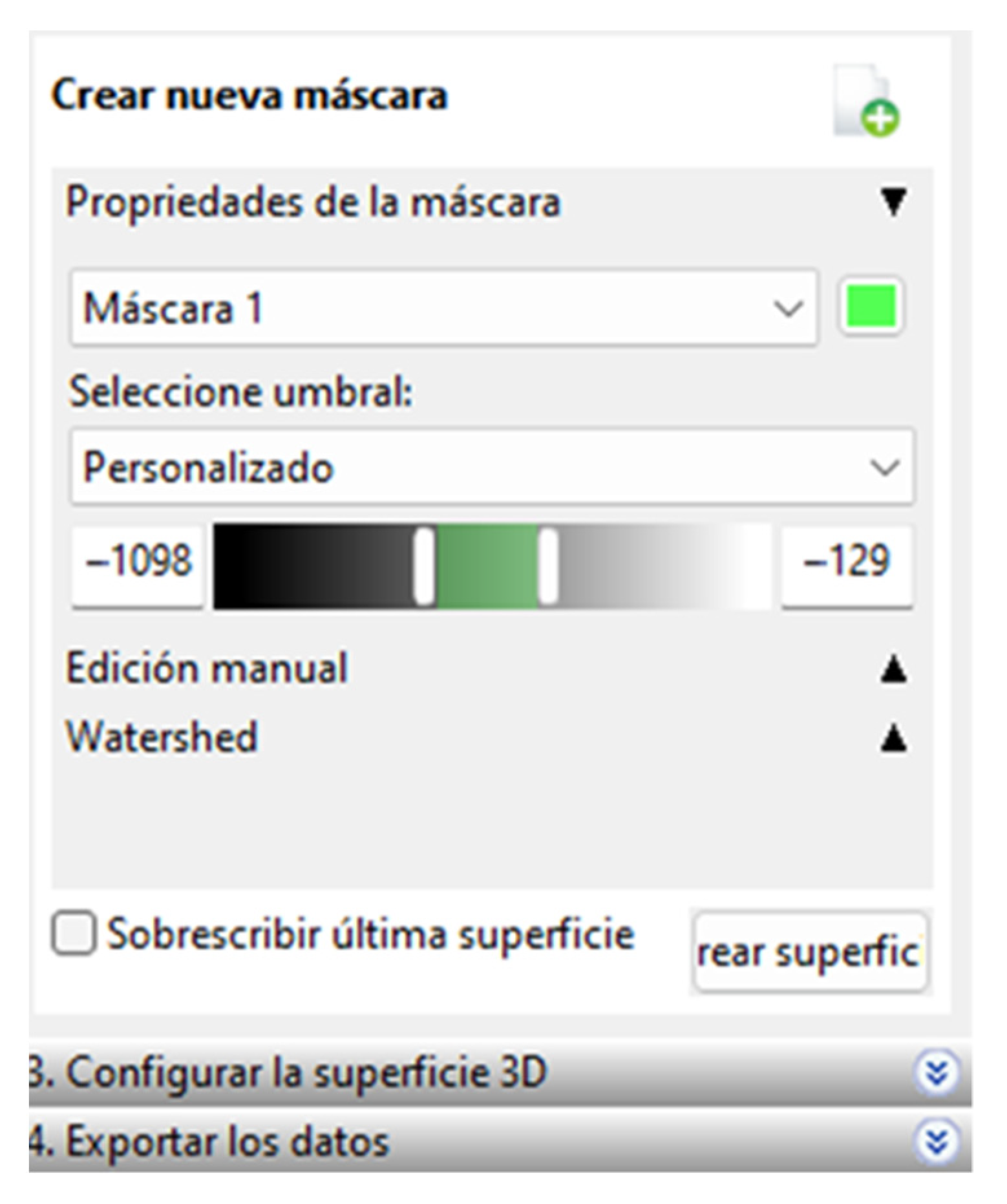
In this software, the process has a unique order of steps for segmentation; When the CT files are imported, a section of interest is cut with the “Cut” tool to create a mask by varying HU ranges depending on the tissue we want to segment. To segment this section, we must choose a threshold, and this software only allows us to visually choose the appropriate range of values for the segmentation. The values chosen can be seen in Figure 3.
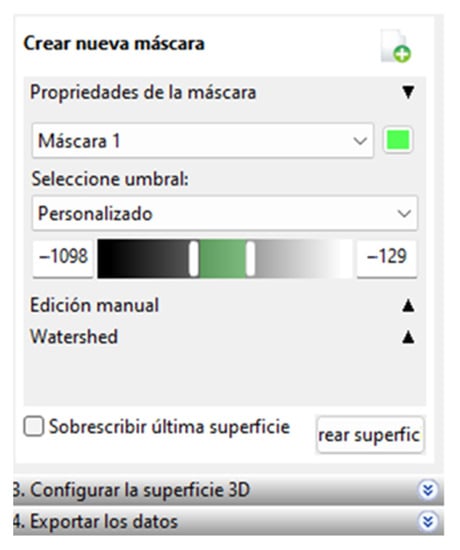
Figure 3.
Range of selected values.
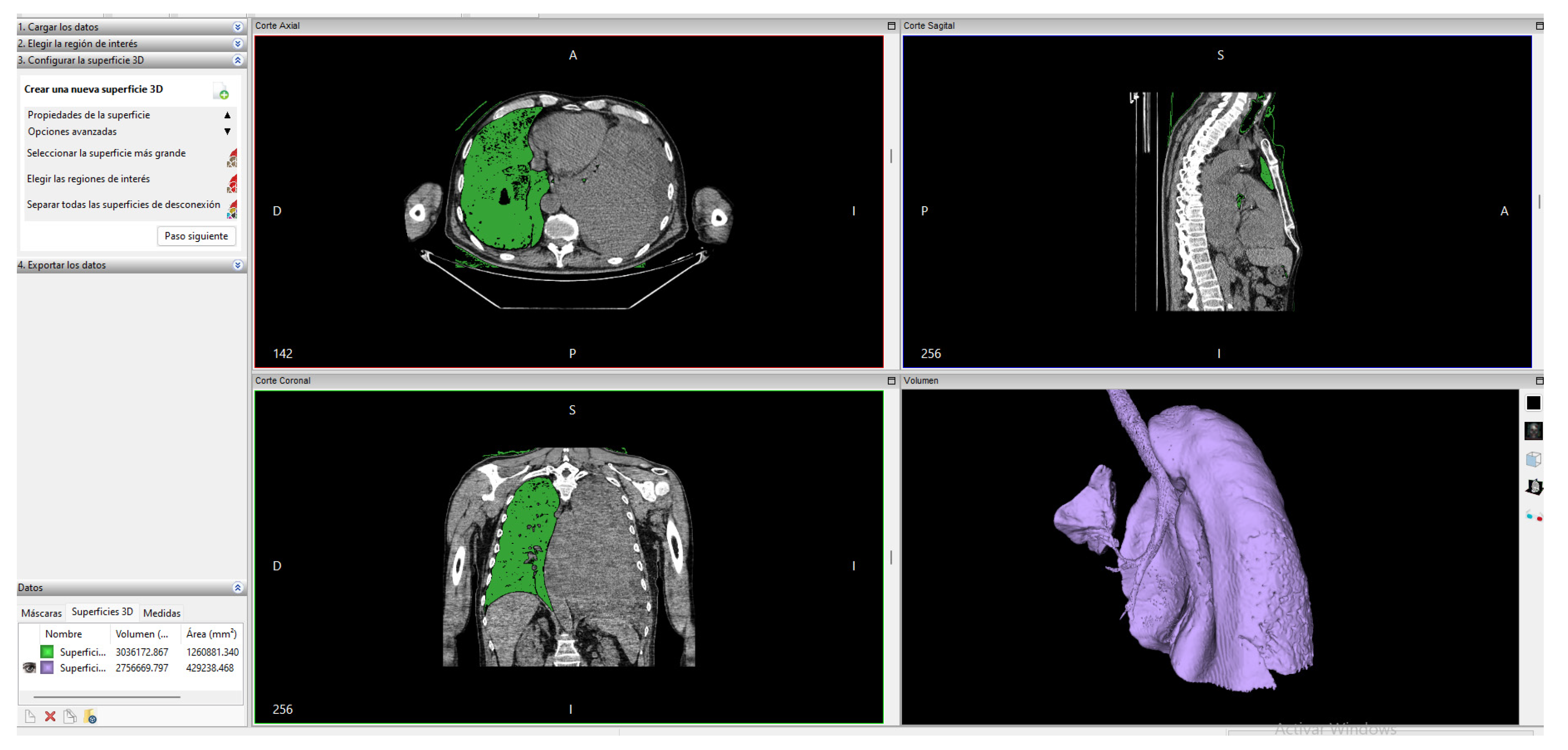
Later, within the “advanced options” we have the option to select the largest surface of the created segment; with this, we obtain the final surface that can be seen in Figure 4.
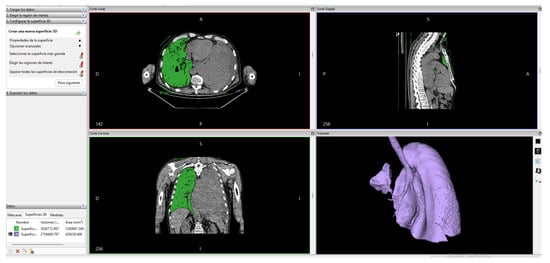
Figure 4.
Lung segmentation in Invesalius.
2.3.3. ITK-Snap
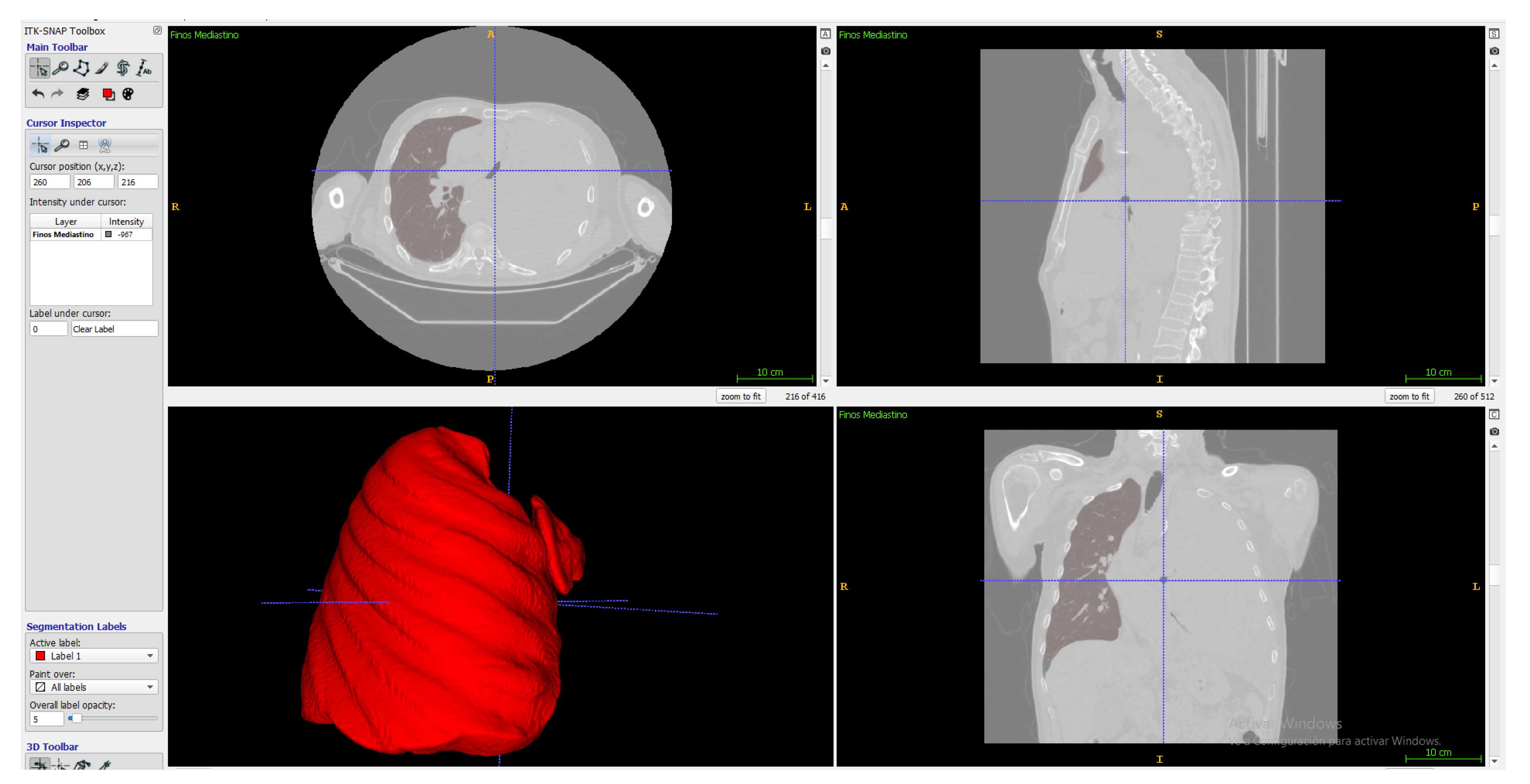
In ITK-Snap, a segmentation mode called Active Contour Segmentation Mode, aka Snake Mode, is used, which consists of several steps; in the first one, a work reference threshold is chosen, then in the second step, it is passed to create active contours based on bubbles with variable radius and different growth spots on the contrast-enhancing parts of the tumor to improve the algorithm. In the final step, an area estimate is shown graphically as a color label; the algorithm stopped when there was no more algorithm growth for 10 s when it had already made 1615 samples. (Figure 5).
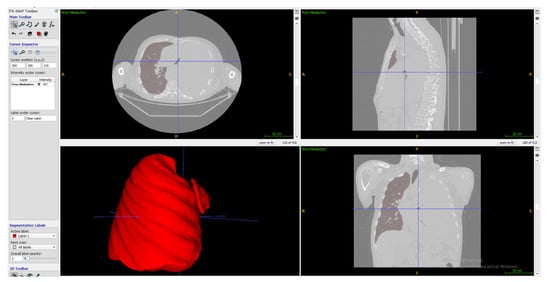
Figure 5.
Lung segmentation in ITK-Snap.
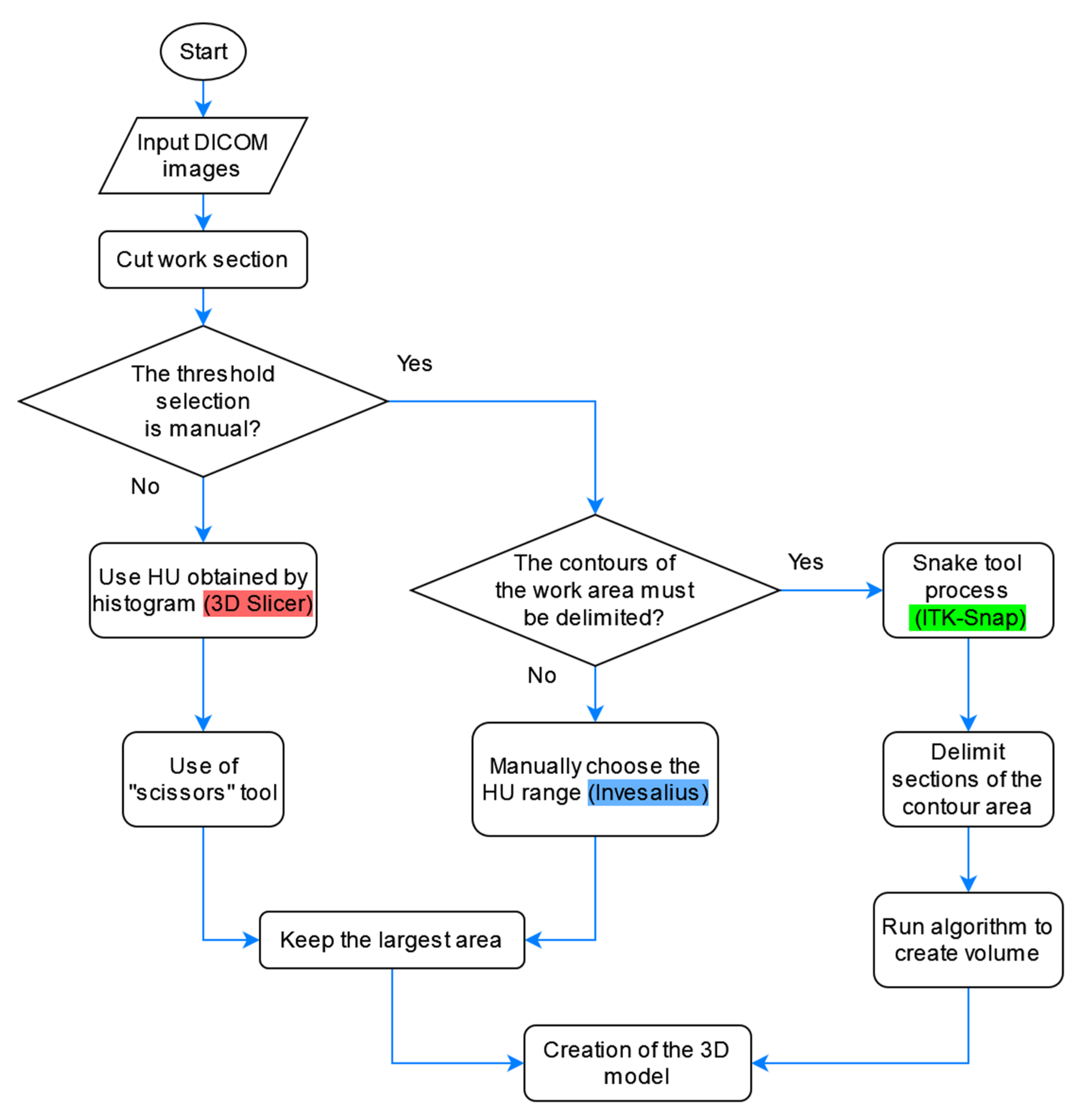
Having similarities in the segmentation process of these programs we can see a flowchart that combines the three methods in Figure 6.
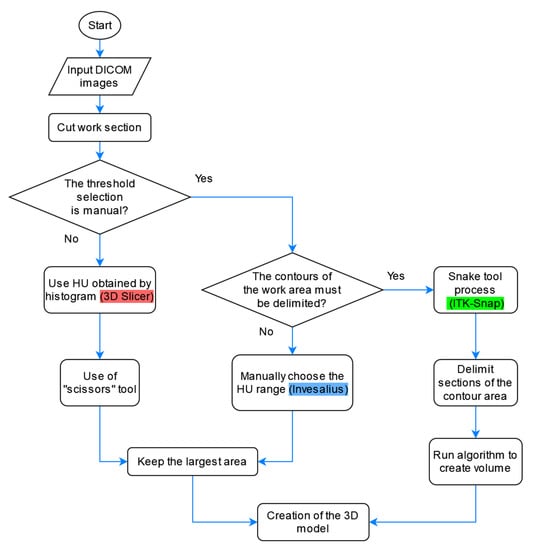
Figure 6.
Flowcharts for the segmentation of the healthy part of the lung with the three software.
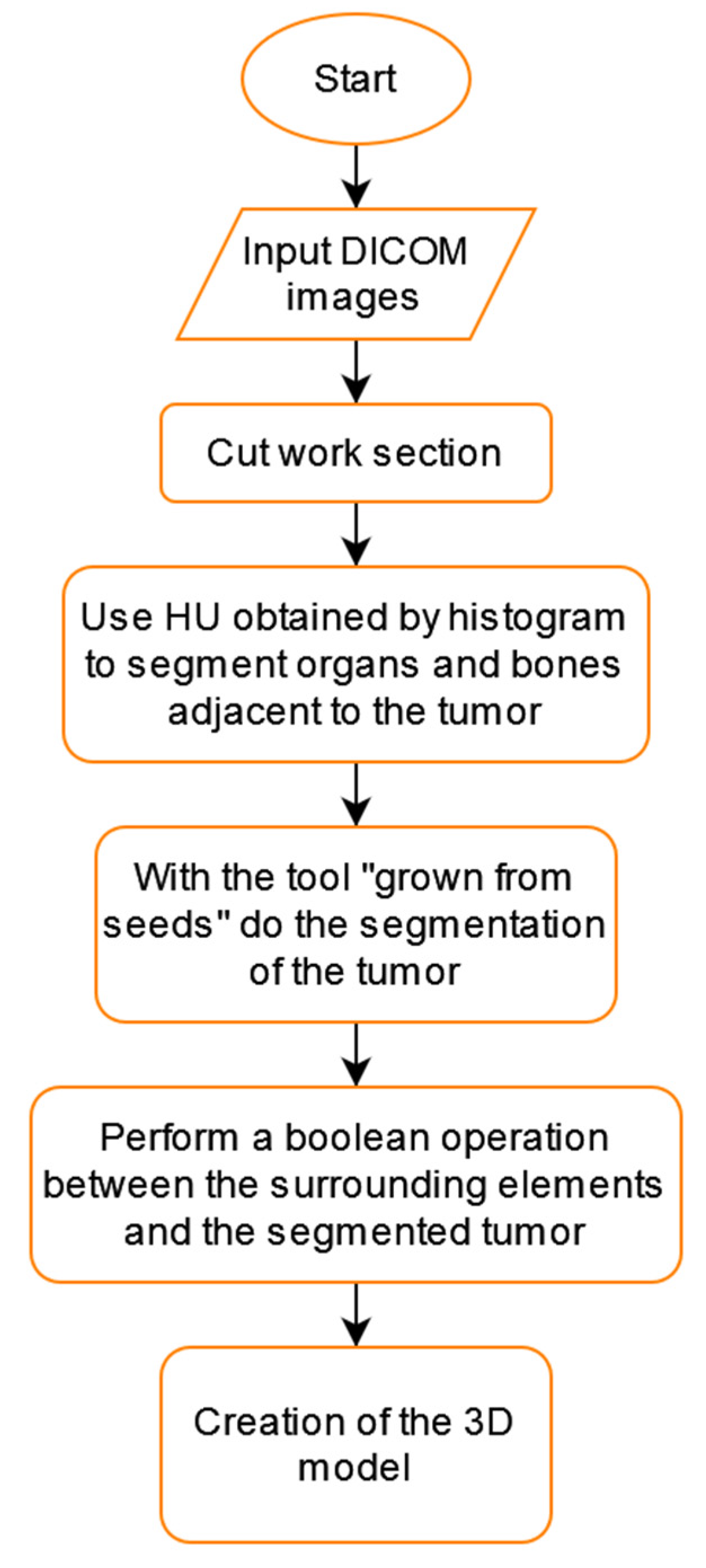
2.4. Tumor Segmentation Process
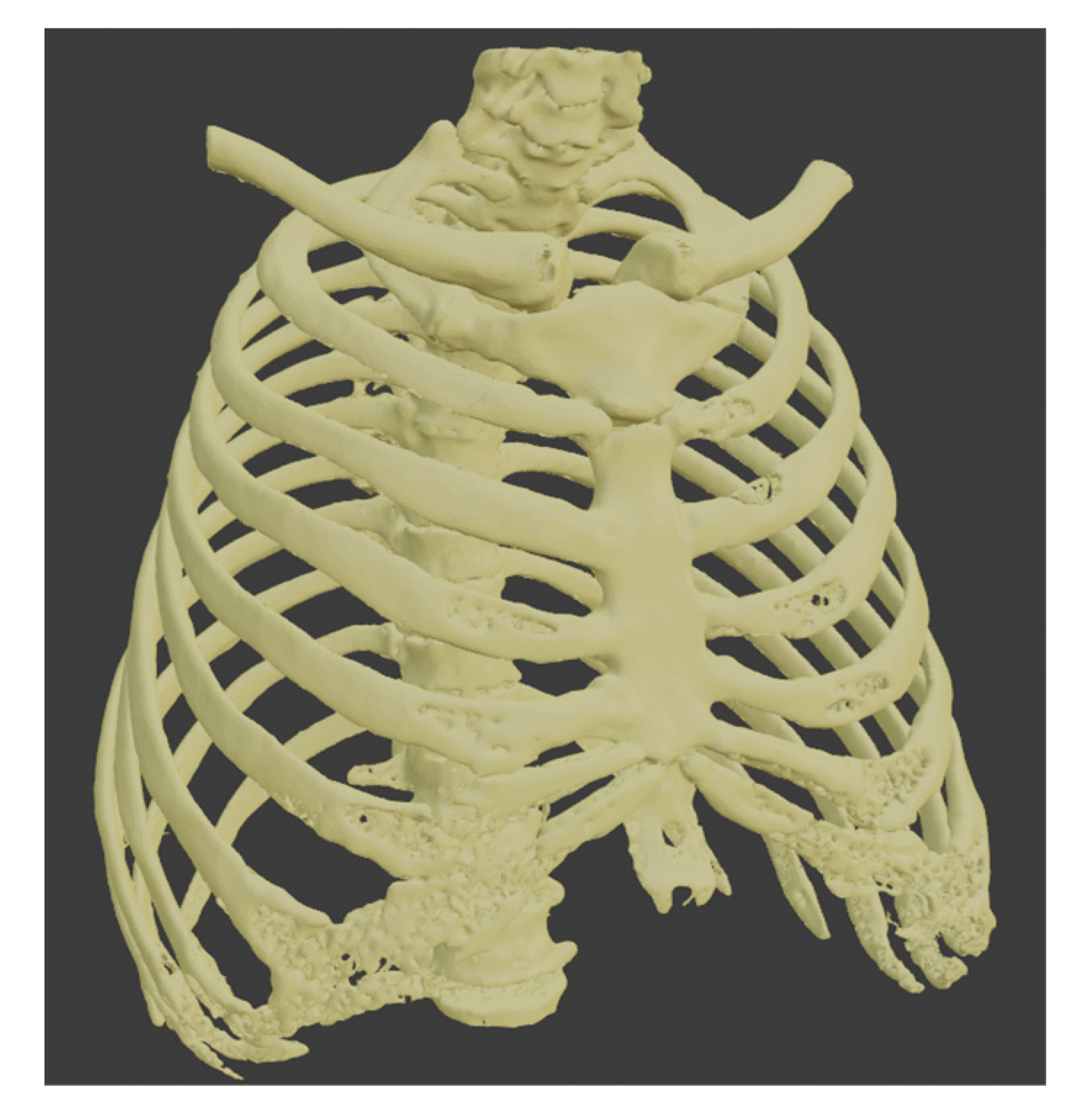
This section describes the segmentation process performed using 3D Slicer since it was the only software capable of separating the tumor from other organs using multiple tools. Since the contrast found for the tumor has similarities with different areas, such as other body tissues, segmentation was performed to create a model of the patient’s bones. Later we create a new segment in the “Segment Editor” section, and then the “Threshold” tool is used with which a fill segment is delimited based on the intensity range in Hounsfield units (HU), obtaining that the range is between −1098.22 to −129.35 HU. The result can be seen in Figure 7.
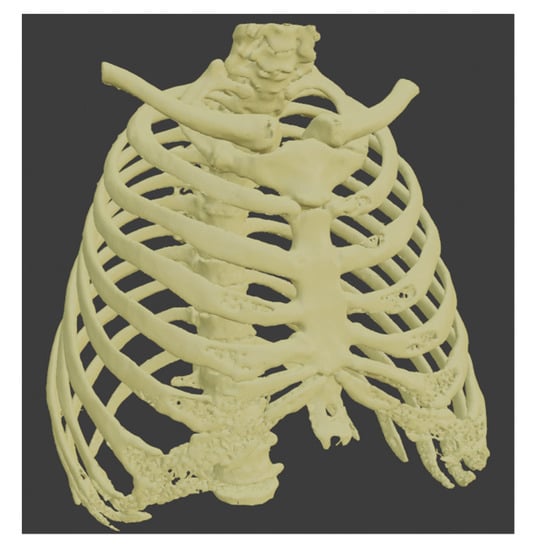
Figure 7.
Segmentation of the bones.
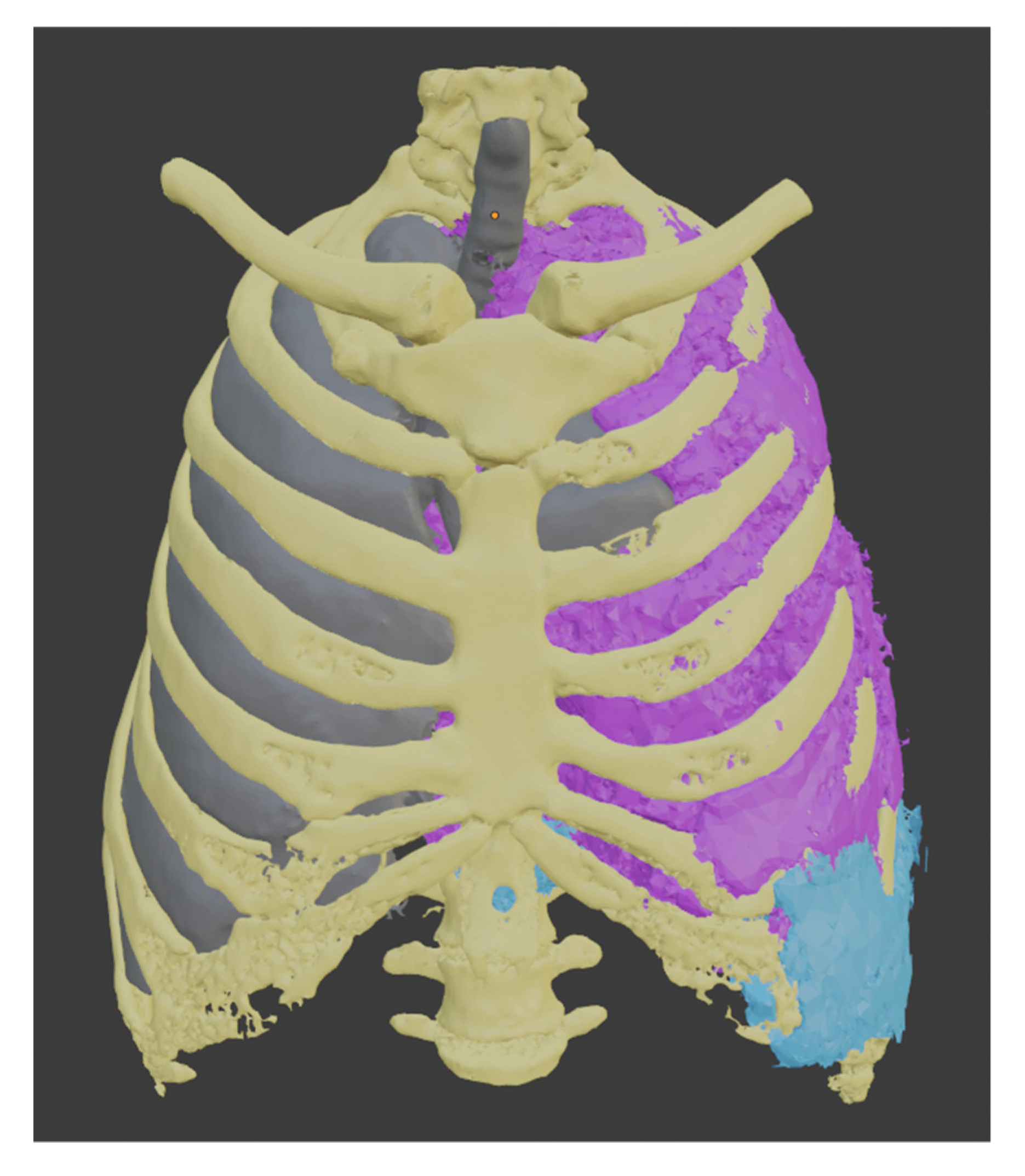
This procedure was repeated to create the liver segment, and since we have the healthy part of the lung, we used these three segments to establish the tumor’s boundaries. Each of them uses a different color, yellow for the bones, gray for the healthy part of the lung, and light blue for the liver.
To segment the tumor “Grown from seeds” tool was used, in which the internal and external edges of the tumor are delimited. This module calculates the edges of the tumor along several planes and displays them as colored areas, and these calculated borders were manually refined before proceeding to generate the tumor volume. The result was defined with purple color. We can see the union of all these segments in Figure 8.
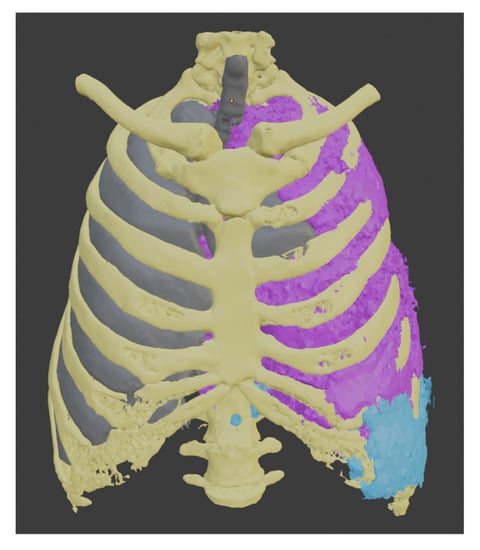
Figure 8.
Models obtained by segmentation.
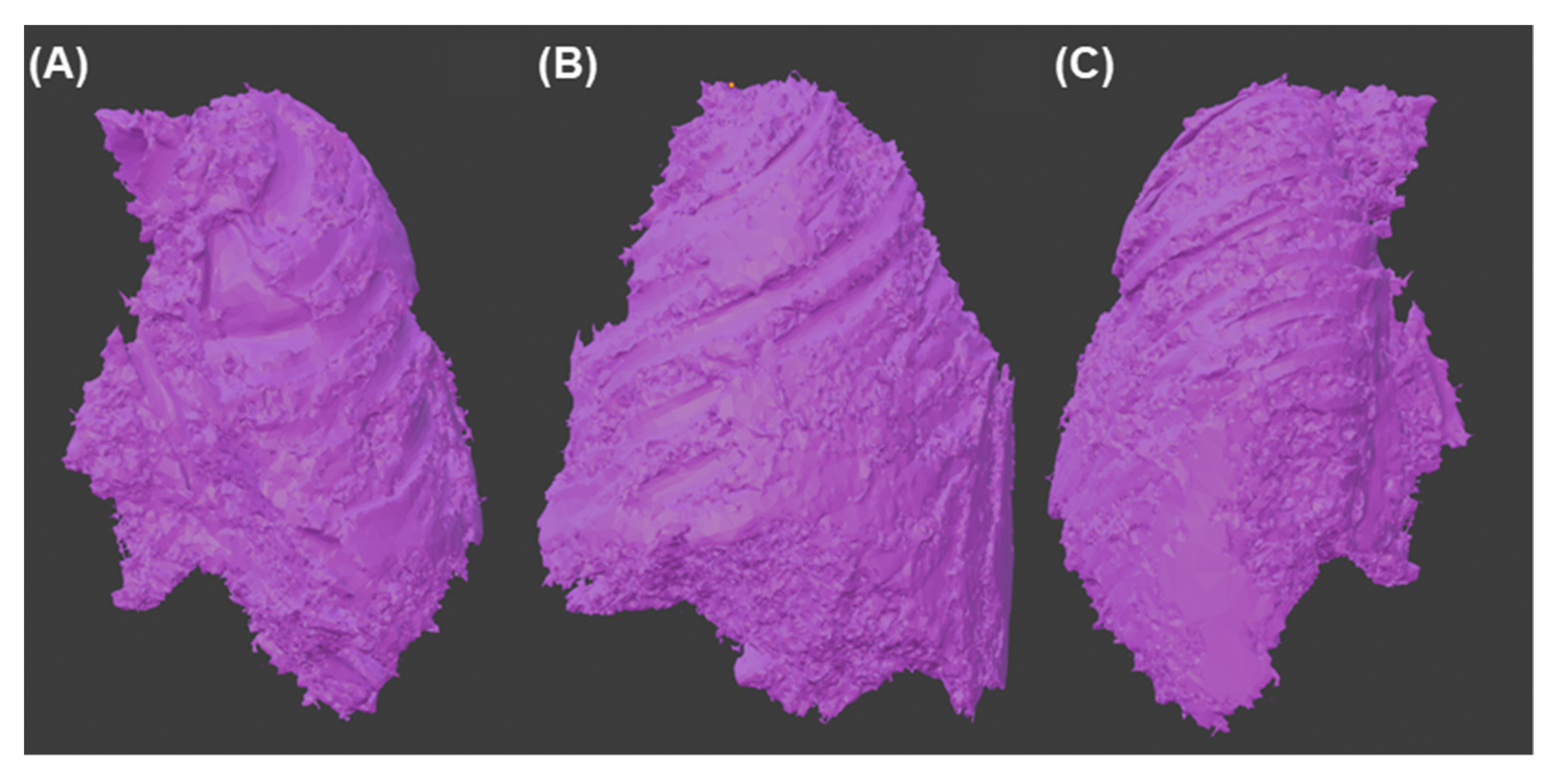
Having a visual appreciation of the segments, we know that the delimitation cannot be external to the other elements; for this reason, the “logical operator” tool was used using its “Subtract” mode of operation to remove from the tumor model all the segments that belong to other parts of the body. The resulting final model can be seen in Figure 9.
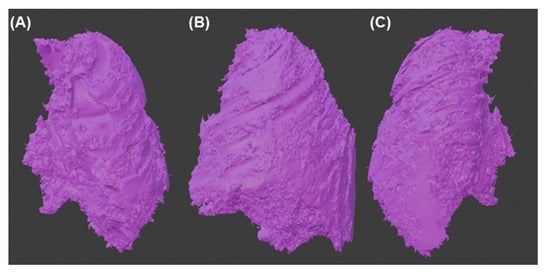
Figure 9.
Final 3d model of the tumor (A) Front view (B) Side view (C) Back view.
We can see a summary of all these steps in the flowchart shown in Figure 10.
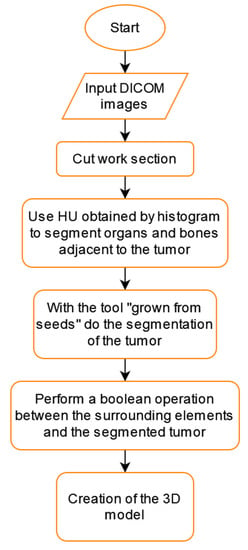
Figure 10.
Tumor segmentation flowchart using 3D Slicer.
3. Results
Comparison of the Models Obtained from the Healthy Part of the Lung
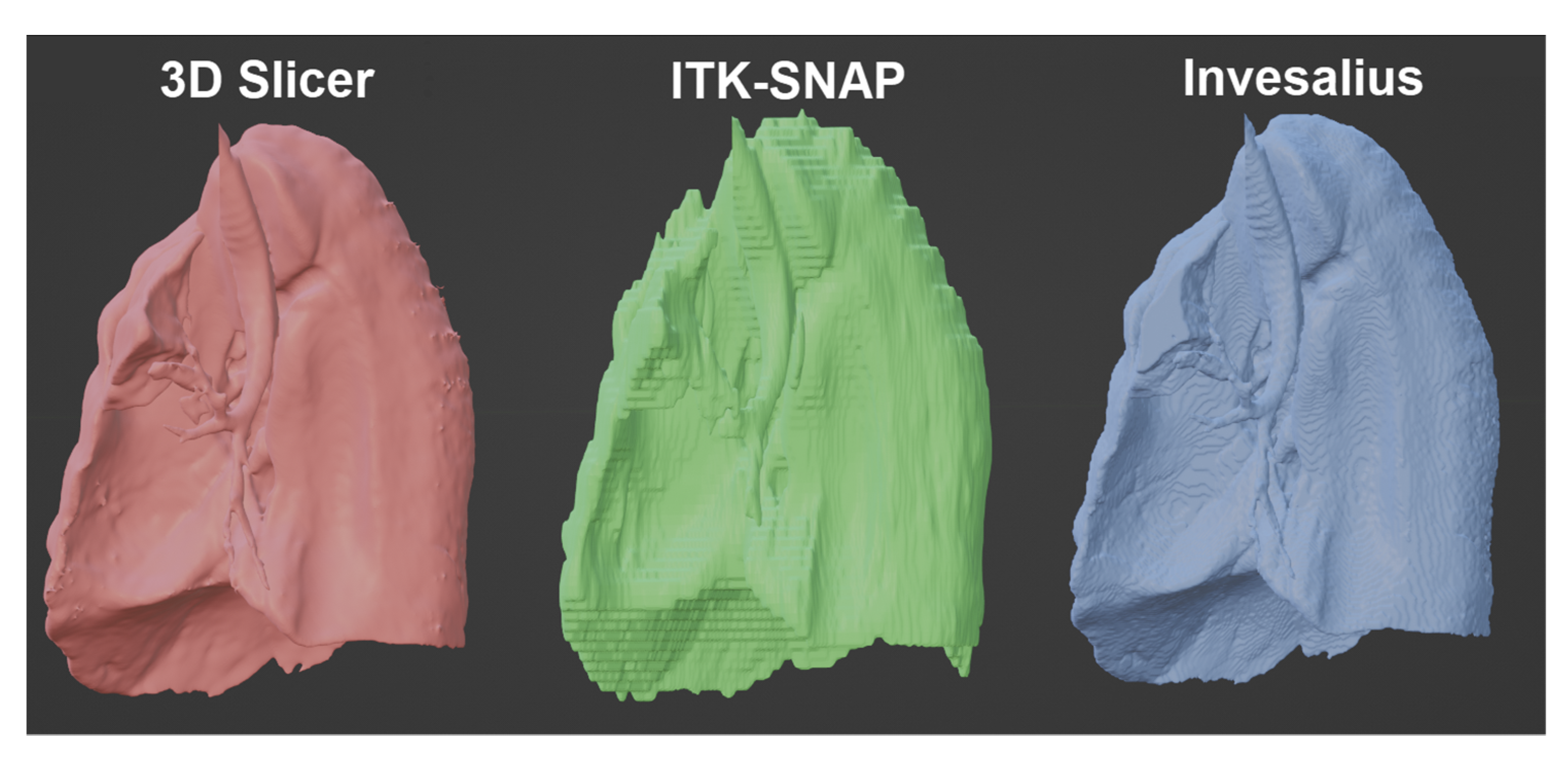
The 3 models were exported in STL format and imported into the 3D modeling software Blender in version 3.2. All renderings were made using this program. Performing an observational analysis, in Figure 11 we can see a comparison of the models obtained by the 3 software without any alteration in the rendering and using flat shadows. In it, we can see how, at first glance, the model generated by 3DSlicer has a higher resolution; therefore, the rounded parts of the organ are better appreciated. Next, in Invesalius, the spaces between layers are slightly appreciated, and finally, ITK-Snap shows quite clearly the passage between one layer and another, losing much of the detail in the tumor’s shape.

Figure 11.
Comparison of the tumor volume obtained from the 3 software.
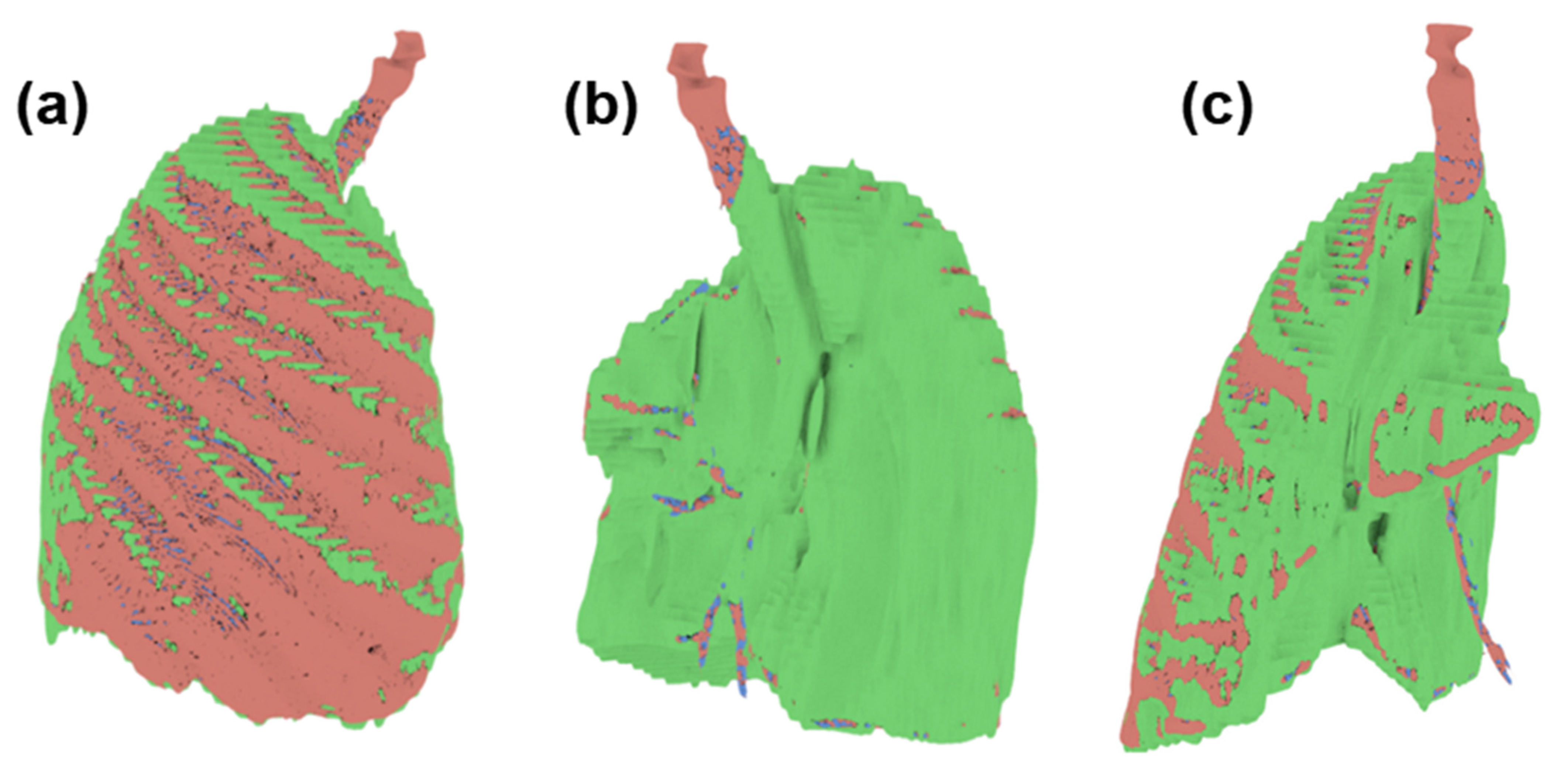
Since the three models obtained have the same reference point, it is possible to superimpose them. In Figure 12, we can see the differences between one and the other through three points of view, having a more significant presence of the green model (ITK-Snap); therefore that we can deduce that this software tends to slightly overestimate the volume of the tumor, unlike the blue model (Invesalius), being in this case that it underestimates the volume of the tumor.
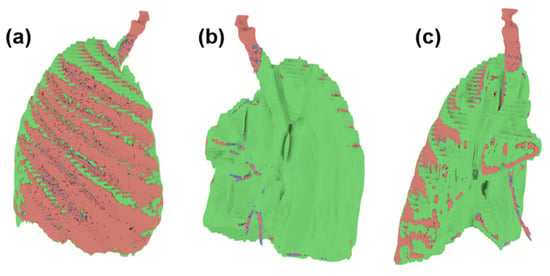
Figure 12.
Superimpose of the tumor volume obtained from the three software (a) Front view (b) Back view (c) Side view.
Regarding the quantitative analysis, relevant parameters were considered to evaluate a model, such as a volume in cubic centimeters and millimeters, the number of triangles and faces, and the model’s weight in megabytes. Additionally, the different faces or edges that tend to generate a conflict when processing the model to print it in 3D were quantified, which leads to the need to clean the mesh. In Table 1, we can observe a comparison of this; in it, we can conclude in the same way as it was appreciated in the observational analysis that taking as reference the model generated by 3D Slicer that the model obtained by ITK-Snap tends to overestimate the volume being in this case by 7.28%. In the case of Invesalius, there is an underestimation of the same parameter by 2.93%.

Table 1.
Comparison of relevant parameters in the 3D model of the segment.
Concerning the weight generated by the models, the lowest is that generated by Invesalius; since the volume generated is not far from the optimum, it can be deduced that it has the best performance in this parameter.
Regarding those parameters that indicate problems in the construction of the model, such as Intersect Faces, Zero Faces, Zero Edges, Thin Faces, and Sharp Edges, these appear mainly in the model generated by 3D Slicer, and this is relevant since these can generate conflicts when the time to be processed by laminating software used for 3D printing. With both analyses, we can conclude that the 3 software can segment the section of the healthy lung, having advantages and disadvantages.
In the case of the tumor section, it has been complicated to separate from other organs since the contrast obtained from the tomography is very similar to other sections, such as that of the heart or the liver; for this reason, it has previously been necessary to separate sections that delimit this tumor to optimize the volume measurement. The only one of the three software that had enough tools to perform all these steps was 3D Slicer; therefore, it is not feasible to compare the performance of the other programs because they did not have a comparable results.
The model obtained after the procedure performed in 3D Slicer (Figure 9) with the tumor shows a volume of 4648.544 cm3. Additionally, these values were also obtained for the following parameters:
- -
- Intersect Face: 6272
- -
- Zero Faces: 21,941
- -
- Thin Faces: 2157
- -
- Sharp Edge: 3576
4. Discussion
Our results show that 2 of the 3 software have not been able to segment the SFT of the analyzed patient. Thus, only in 3D Slicer was it possible to segment both the healthy lung and the one with the SFT that caused Dodge-Potter syndrome.
Regarding the performance of Invesalius to process the tumor section, this software works mainly by establishing a threshold to separate the different sections of the body, which is why it is widely used to separate bones since these tend to have a value in the tomography that is far from that of the organs. However, it is impossible to separate them correctly for this type of application where a tumor with contrast is very similar to other organs.
In the case of ITK-Snap, it has a seed tool very similar to the “Grown from seed” tool in 3D Slicer but with the difference that it allows you to choose how many iterations you need and how far to grow the section. But as in the case of this section of the tumor, you need to delimit its volume using other neighboring organs; additionally, you need a Boolean cutting tool like the one that 3D Slicer has, but this software lacks.
In the case of 3D Slicer, the segmentation of the healthy part of the lung and the tumor section has been able to work correctly. For the healthy part, as in the other two software, a threshold was used to generate the model; the difference lies in the fact that it has a local histogram, which allows us to more accurately delimit the Hounsfield Units in which the section is found to segment. For the segmentation of the tumoral section, it was required to use multiple tools. It starts with finding the appropriate threshold for the bones and liver, accompanied by the healthy section that was previously created, then with the segmentation of the tumor through seeds which gives us an approximate result of the tumor but without being exact. For this reason, all of them were intersected using the previously created models to perform a Boolean operation and obtain a final model.
One of the disadvantages of using this software is that the exported models have many conflicting faces and edges that, when using 3D printing laminating software, are recognized as model errors. Most laminating software can correct these errors to render the 3D model correctly. Thanks to this multiplicity of tools, a complex case such as this syndrome can be correctly segmented.
Within the selection criteria of these three software was the one that could carry out the segmentation process without having programming codes involved but instead having a graphical interface that does not require specialized knowledge. However, some of these software require an amount of more significant steps to obtain similar results as is the case with ITK-Snap since when using the Snake Mode, you must add an unknown amount of bubbles to determine estimates of the area and then wait for the software to make a set of iterations to cover the sections with similar contrast. The opposite case is Invesalius, where the steps have a direct order, making it suitable for those unfamiliar with its interface, thus optimizing the work time consumed.
The use and comparison of these open-source tools for segmentation have been used in multiple parts of the body, such as bones, as is the case of the work of Matsiushevich et al. [11], where they analyzed as points of comparison the weight and quantity of triangles of the exported models. There are also antecedents in the literature of its use in tumor sections located in the brain, such as glioblastomas; an example is an article presented by Fyllingen et al. [12], where it is concluded that the best performances at the level of time expenditure are those of 3D Slicer and ITK-Snap. But in the literature, there are no reports of using this software for a rare syndrome like the one presented in this study, where the tomography contrast is not very different from other organs.
Previous reports highlight the importance of SFT associated with Doege-Potter syndrome [4,13,14,15], showing no differences between benign and malignant cases and that all end up in surgery as an effective treatment [16]. Surgical activities use images such as CT as an aid tool and can benefit from 3D modeling by allowing us to approximate the actual size of the tumor, as seen in other medical areas [17,18]. In this sense, free-access software can give oncology a better chance at facing all stages of cancer patient care.
The use of 3D modeling is key to understanding the size of the tumor as well as its surgical margins. In addition, paraneoplastic problems can rapidly affect patient health, in the case of DPS being crucial a rapid and effective practical approach knowing the primary tumor [7,8]. The contribution of the use of the software is valuable and necessary to improve clinical practice based on open-source technologies and scientific evidence.
Current clinical practice includes a clinical approach using imaging tests (i.e., CT) that contribute to tumor management [4,5]. However, given that SFT tumors can vary considerably in size, it is also important to know the tumor distribution through pre-surgical digital approaches [6]. With this, SFT surgical practices can be improved, thus reducing the consequences of the paraneoplastic syndrome underlying the primary mesenchymal tumor, as in the case of DPS.
Within the study’s limitations, we can say that the study software’s design is focused on its use for research rather than for clinical use, declared by their programmers. Furthermore, each software has its learning curve, and the user experience makes each result have differences when used by a specialist in 3D modeling but not a recurring user of this segmentation software as in the case of this study.
5. Conclusions
In this study, we compared three open-source software for the segmentation of an SFT located in the left lung, showing easier segmentation and differentiation outcomes with the semi-automatic tools of 3D Slicer in this particular tumor, which has a contrast very close to that of the other organs. There are difficulties in data analysis with patients who have SFT due to the size of the tumor and the difficulty in distinguishing fair gray level values on a CT scan to separate a healthy organ from a tumor slice.
Open-source tools are essential in environments where economic resources are scarce, and significant communities support software such as those used in this work. Finding the limitations of each of these helps us continuously improve them and expand their use in medicine.
Author Contributions
Conceptualization, J.P.T. and J.M.-S.; methodology, J.P.T., R.S.-G. and J.M.-S.; software, J.P.T. and J.M.-S.; validation, J.P.T., B.M.-S. and H.C.-P.; formal analysis, J.P.T. and H.C.-P.; investigation, J.P.T., M.L.-H. and B.M.-S.; resources, R.S.-G. and H.C.-P.; data curation, J.P.T., M.L.-H. and R.S.-G.; writing—original draft preparation, J.P.T., J.M.-S., B.M.-S. and M.L.-H.; writing—review and editing, J.P.T., J.M.-S., R.S.-G. and H.C.-P.; visualization, J.P.T.; supervision, R.S.-G.; project administration, H.C.-P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the EsSalud Ethics and Research Committee No. 42-IETSI-ESSALUD-2020.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Acknowledgments
We thank the Nesh Hubbs team for their support in critically reviewing this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Gold, J.S.; Antonescu, C.R.; Hajdu, C.; Ferrone, C.R.; Hussain, M.; Lewis, J.J.; Brennan, M.F.; Coit, D.G. Clinicopathologic correlates of solitary fibrous tumors. Cancer 2002, 94, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Zhang, Z.; Shen, X.; Wang, K.; Zhao, Y.; He, J.; Gao, Y.; Shan, X.; Xin, G.; Li, C.; et al. Doege-Potter syndrome: A review of the literature including a new case report. Medicine 2017, 96, e7417. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Hinostroza, M.; Moya-Salazar, J.; Dávila, J.; Absencio, A.Y.; Contreras-Pulache, H. Doege-Potter syndrome due to endothoracic solitary hypoglycemic fibrous tumor. Clin. Case Rep. 2022, 10, e05611. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, T.; Ozcan, G.; Siddique, A.S.; Araneta, R.N., III; Slater, D.E.; Khan, A. Doege-Potter Syndrome with a Benign Solitary Fibrous Tumor: A Case Report and Literature Review. Case Rep. Oncol. 2021, 14, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-C.; Huang, H.-Y. Solitary fibrous tumor: An evolving and unifying entity with unsettled issues. Histol. Histopathol. 2019, 34, 313–334. [Google Scholar] [CrossRef] [PubMed]
- Badawy, M.; Nada, A.; Crim, J.; Kabeel, K.; Layfield, L.; Shaaban, A.; Elsayes, K.M.; Gaballah, A.H. Solitary fibrous tumors: Clinical and imaging features from head to toe. Eur. J. Radiol. 2022, 146, 110053. [Google Scholar] [CrossRef] [PubMed]
- Honnorat, J.; Antoine, J.-C. Paraneoplastic neurological syndromes. Orphanet J. Rare Dis. 2007, 2, 22. [Google Scholar] [CrossRef] [PubMed]
- Preim, B.; Botha, C. Chapter 4-Image Analysis for Medical Visualization. In Visual Computing for Medicine, 2nd ed.; Preim, B., Botha, C., Eds.; Morgan Kaufmann: Boston, MA, USA, 2014; pp. 111–175. ISBN 978-0-12-415873-3. [Google Scholar]
- Kim, J.J.; Nam, H.; Kaipatur, N.R.; Major, P.W.; Flores-Mir, C.; Lagravere, M.O.; Romanyk, D.L. Reliability and accuracy of segmentation of mandibular condyles from different three-dimensional imaging modalities: A systematic review. Dentomaxillofac. Radiol. 2020, 49, 20190150. [Google Scholar] [CrossRef] [PubMed]
- Matsiushevich, K.; Belvedere, C.; Leardini, A.; Durante, S. Quantitative comparison of freeware software for bone mesh from DICOM files. J. Biomech. 2019, 84, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Fyllingen, E.H.; Stensjøen, A.L.; Berntsen, E.M.; Solheim, O.; Reinertsen, I. Glioblastoma Segmentation: Comparison of Three Different Software Packages. PLoS ONE 2016, 11, e0164891. [Google Scholar] [CrossRef] [PubMed]
- Pavlíková, P.; Sochorová, L.; Snížková, O.; Malý, V.; Staněk, I. Solitary fibrous tumor of the pleura as a rare cause of severe hypoglycemia: Doege-Potter syndrome. Rozhl. V Chir. Mesic. Ceskoslovenske Chir. Spol. 2020, 99, 95–98. [Google Scholar] [CrossRef]
- Flores Cruz, G.; Aguila Gómez, M.V.; Lazo Vargas, A.; Alarcón Delgado, M.M.; Luna Catari, M.K.; Marconi Poma, E.R. Síndrome de Doege-Potter, Tumor Fibroso Solitario Endotorácico Hipoglicemiante, ¿Cuando y Cómo lo Diagnóstico? y ¿Cual es mi Conducta Final?: Presentación de un Caso y Revisión de la Literatura. Rev. Médica Paz 2019, 25, 48–57. [Google Scholar]
- Estradioto, L.; de Araujo, R.B.; Neto, N.B.; de Araújo, V.B.; Coelho, M.d.S.; da Silva, L.L.G. Doege-Potter syndrome: A rare presentation of a solitary fibrous tumor of the pleura. Braz. J. Oncol. 2021, 17, 1–4. [Google Scholar] [CrossRef]
- Meng, W.; Zhu, H.-H.; Li, H.; Wang, G.; Wei, D.; Feng, X. Solitary fibrous tumors of the pleura with Doege-Potter syndrome: A case report and three-decade review of the literature. BMC Res. Notes 2014, 7, 515. [Google Scholar] [CrossRef] [PubMed]
- Fernandes de Oliveira Santos, B.; Silva da Costa, M.D.; Centeno, R.S.; Cavalheiro, S.; Antônio de Paiva Neto, M.; Lawton, M.T.; Chaddad-Neto, F. Clinical Application of an Open-Source 3D Volume Rendering Software to Neurosurgical Approaches. World Neurosurg. 2018, 110, e864–e872. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Wang, J.; Yao, J.; Hang, F.; Lei, X.; Cao, Y. Three-dimensional image reconstruction with free open-source OsiriX software in video-assisted thoracoscopic lobectomy and segmentectomy. Int. J. Surg. Lond. Engl. 2017, 39, 16–22. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).