A Comparison Between Unimodal and Multimodal Segmentation Models for Deep Brain Structures from T1- and T2-Weighted MRI
Abstract
1. Introduction
- A heterogeneous dataset comprising 325 T1w and 325 T2w MRI scans was built by integrating data from eight public sources, enabling robust training and evaluation.
- An end-to-end multimodal framework was developed for labeling MR images, preprocessing data, and training and evaluating deep learning models for the segmentation of deep brain structures in neurosurgical settings.
- A detailed comparison between unimodal and multimodal models was conducted, highlighting the benefits and limitations of each approach.
- The developed T1w-based models were benchmarked against the state-of-the-art DBSegment tool, demonstrating clear improvements across all metrics.
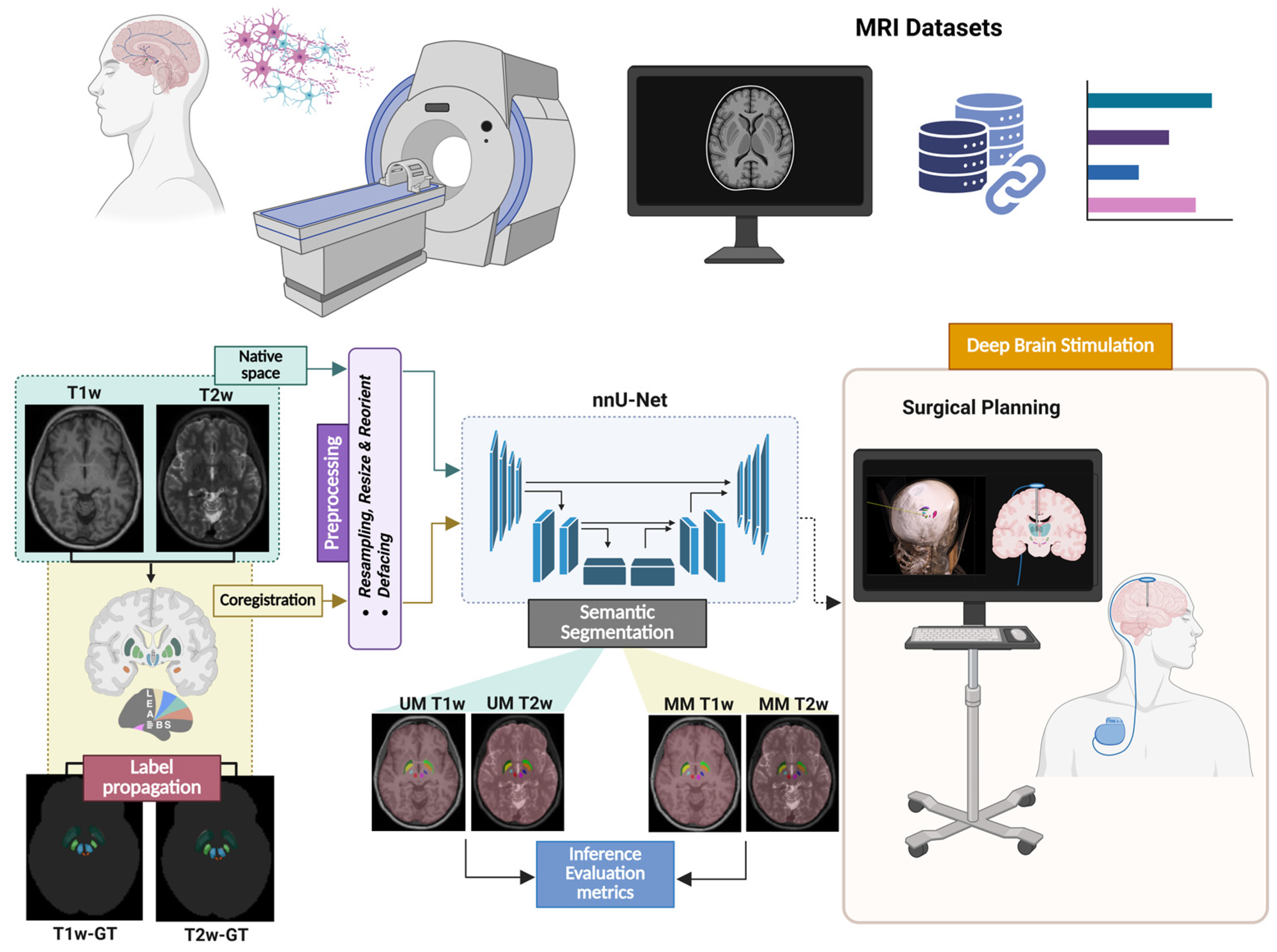
2. Materials and Methods
2.1. Dataset Collection
- HCP (Human Connectome Project) [28], which includes diffusion and anatomical neuroimaging data openly available to the scientific community for examination and exploration.
- OASIS3 (Open Access Series of Imaging Studies 3) [29], which is a retrospective compilation of data for 1378 participants with 2842 MRI sessions (encompassing T1w, T2w, and FLAIR, among others).
- ADNI (Alzheimer’s Disease Neuroimaging Initiative) [30], which is a longitudinal study started in 2004 that continuously expanded its data collection through multiple phases, contributing significantly to Alzheimer’s research. The ADNI dataset includes a variety of data types, such as clinical, biofluid, genetic, and imaging data, all of which are accessible to authorized researchers through the LONI Image and Data Archive (IDA). The latest phase of the study, ADNI4, is used in our final dataset and includes both T1w and T2w MRI scans.
- IXI (Information eXchange from Images) [31], which is a project that collected nearly 600 MR images from healthy subjects. The MR image acquisition protocols encompass T1w, T2w, and PD (Proton Density)-weighted images.
- UNC (University of North Carolina) [32], which includes paired T1-weighted and T2-weighted MRI scans acquired at both 3T and 7T from 10 healthy volunteers. The images were collected as part of a brain imaging study conducted by the University of North Carolina.
- THP (Traveling Human Phantom) [33]: This OpenNeuro dataset (accession number ds000206) was collected as part of a multi-site neuroimaging reliability study. It contains repeated multimodal MRI scans acquired from five healthy individuals across eight different imaging centers.
- NLA (Neural Correlates of Lidocaine Analgesic) [34]: This OpenNeuro dataset (accession number ds005088) includes T1w and T2w MRI scans acquired at 3T from 27 adults who participated in a single-arm, open-label study investigating the neural effects of lidocaine as an analgesic.
- neuroCOVID [35]: This OpenNeuro dataset (accession number ds005364) includes MRI data from a total of 100 participants who underwent T1w and T2w scans as part of an evaluation of the neurological effects of COVID-19.
2.2. Data Annotation
2.3. Data Preparation
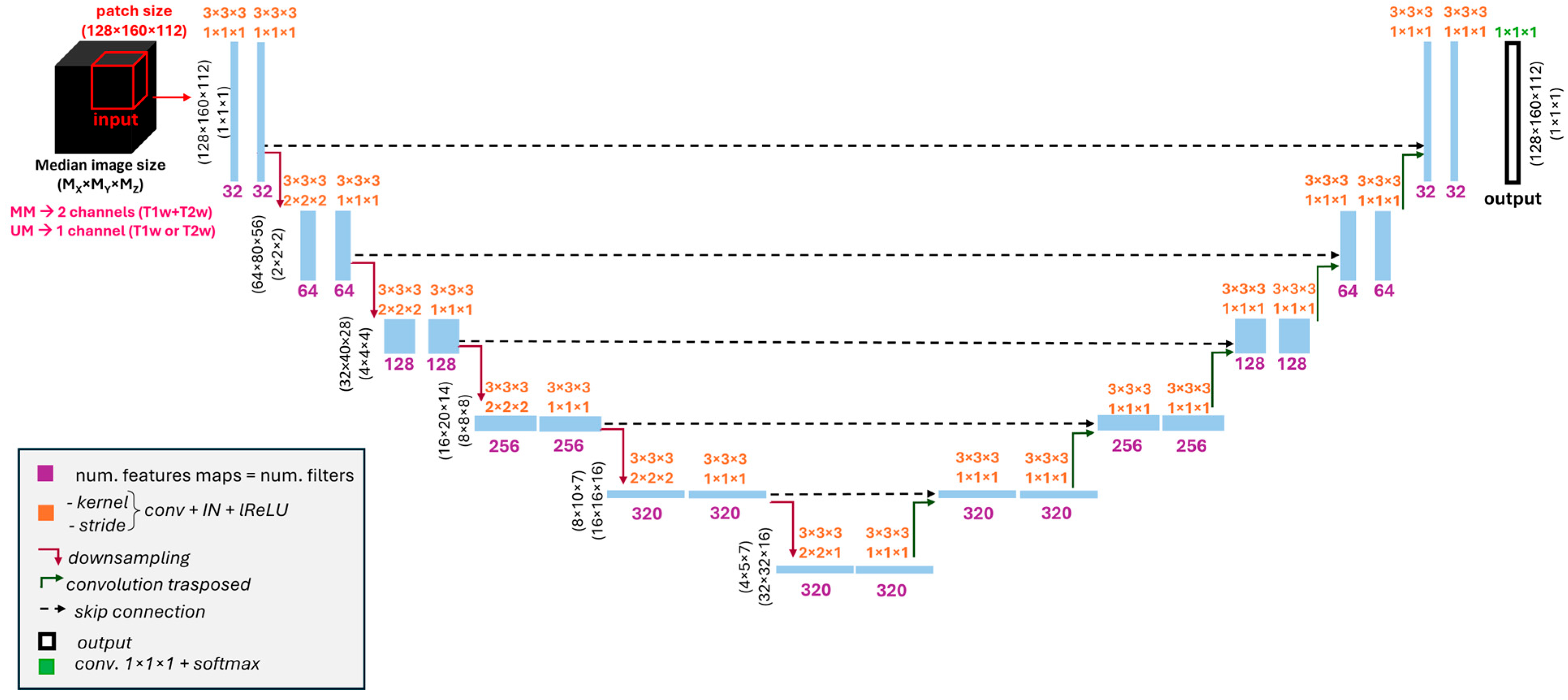
2.4. Models’ Architecture and Training
2.5. Evaluation Metrics
2.6. Statistical Evaluation
3. Results
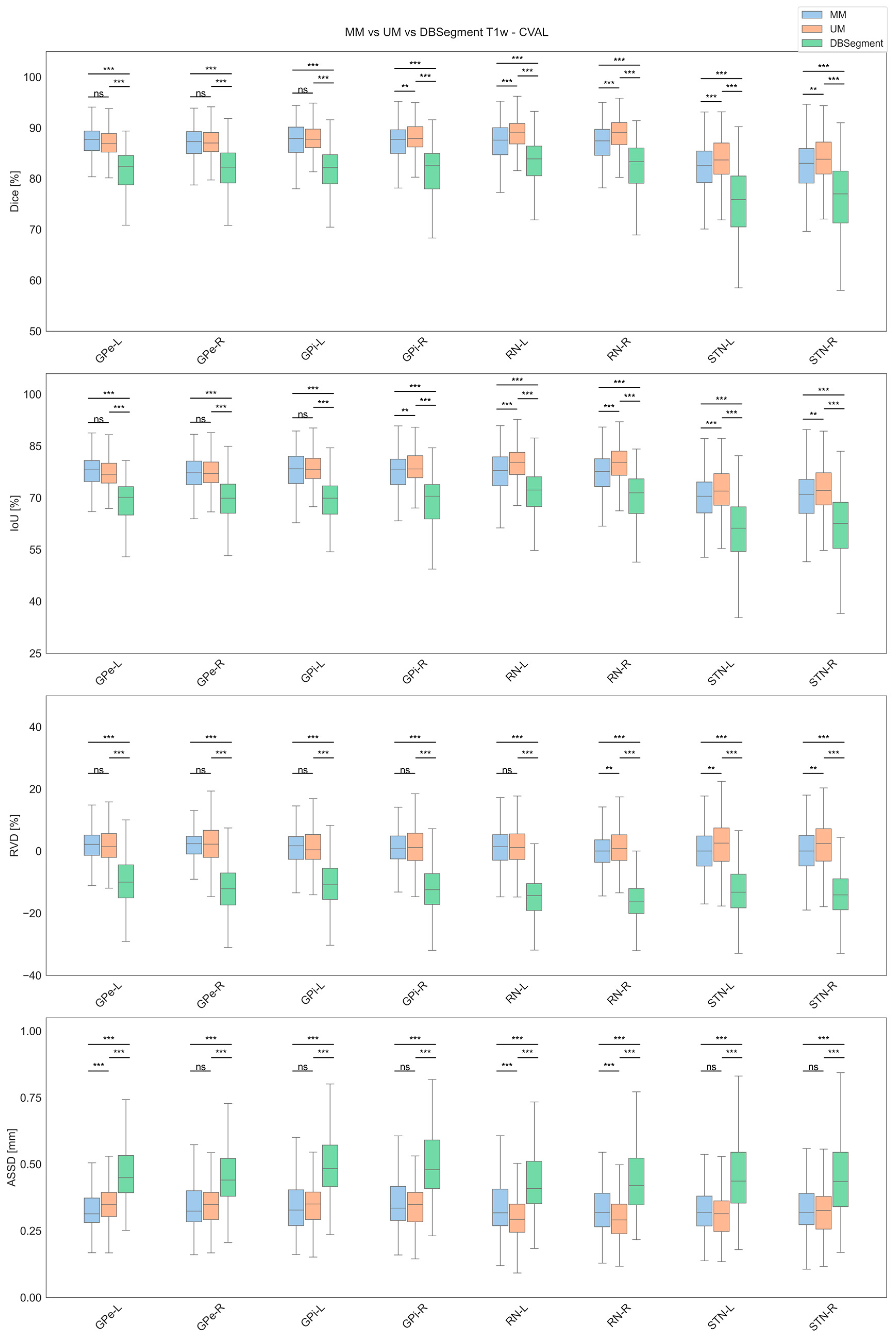
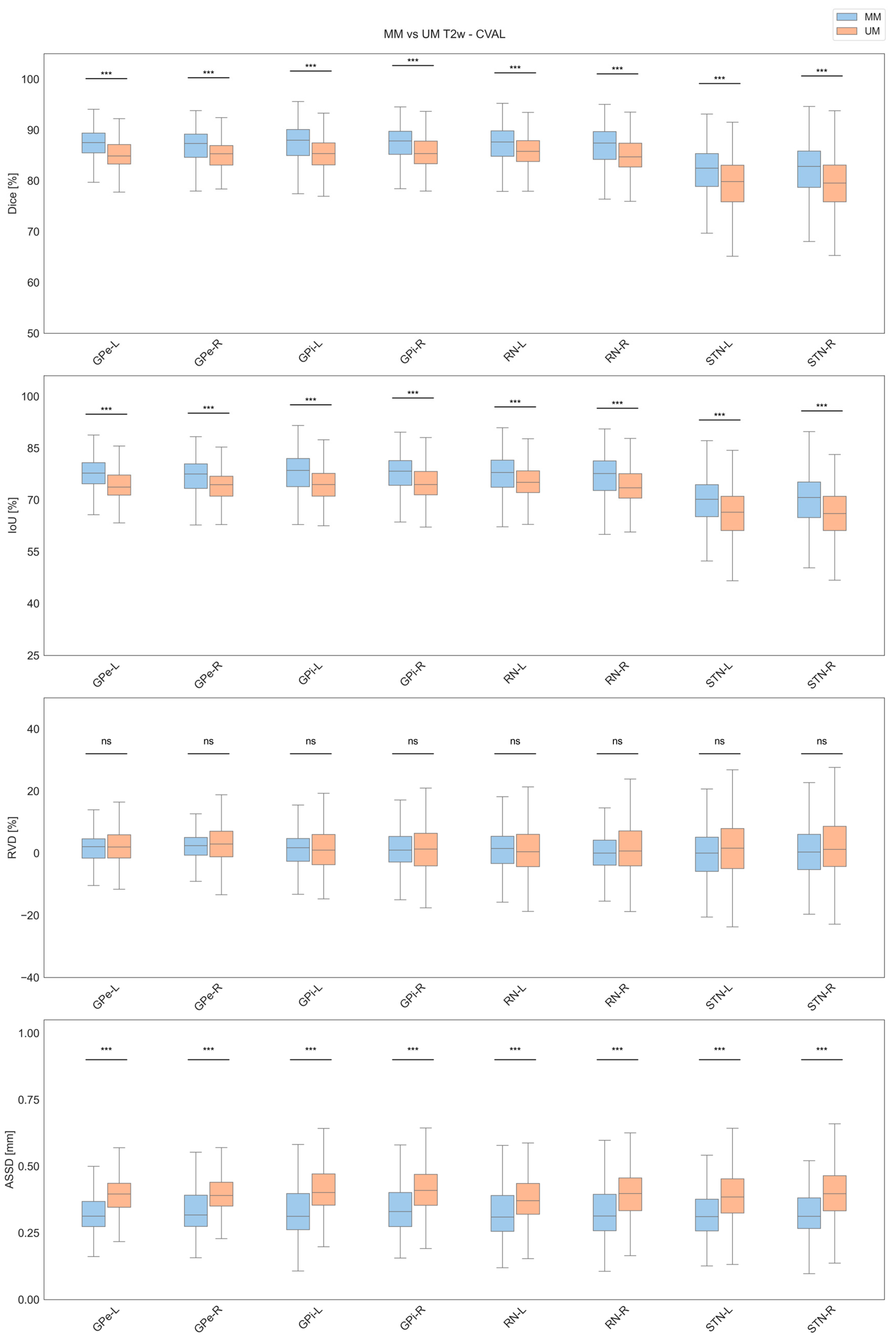
3.1. Cross-Validation Results
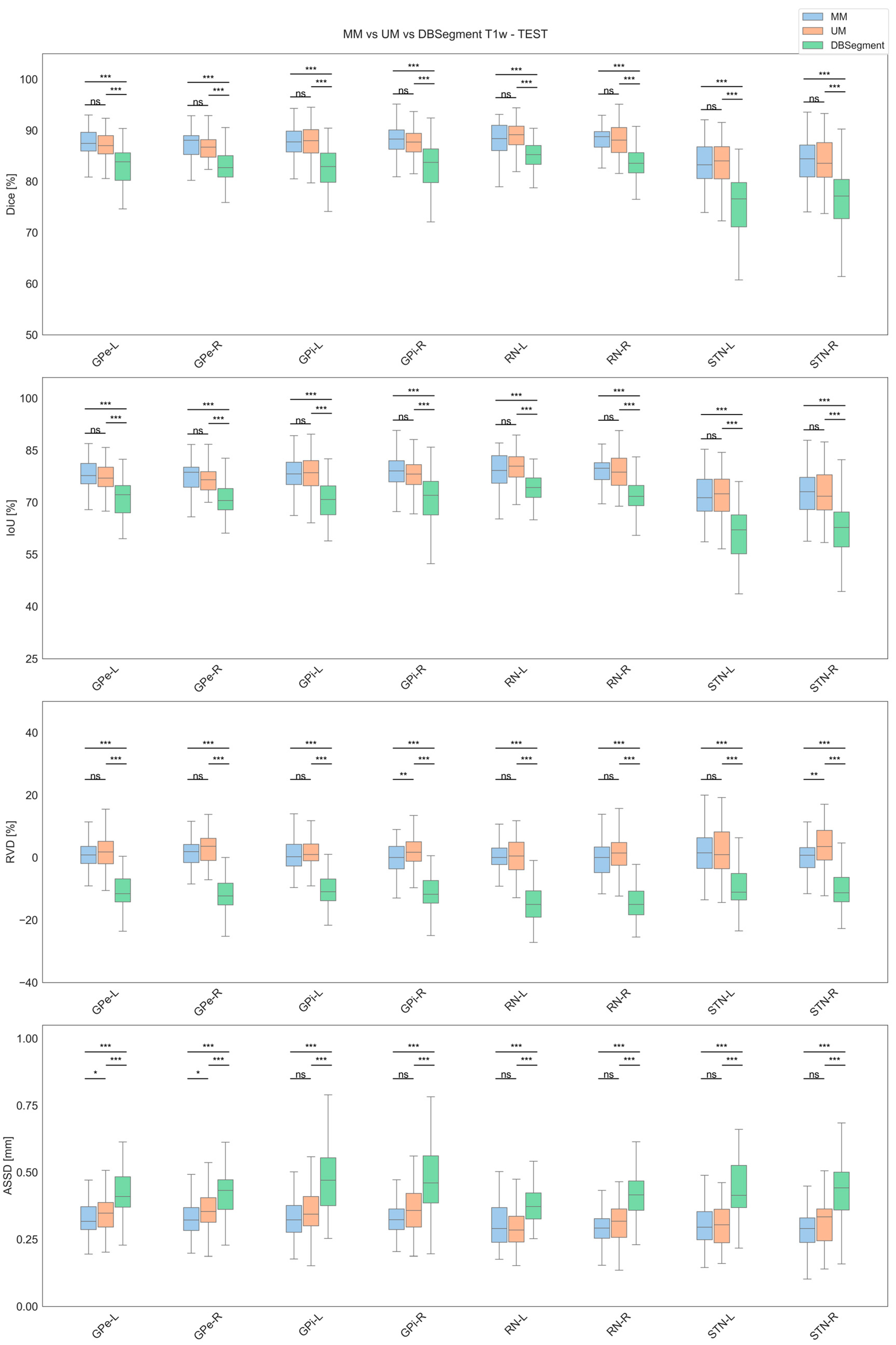
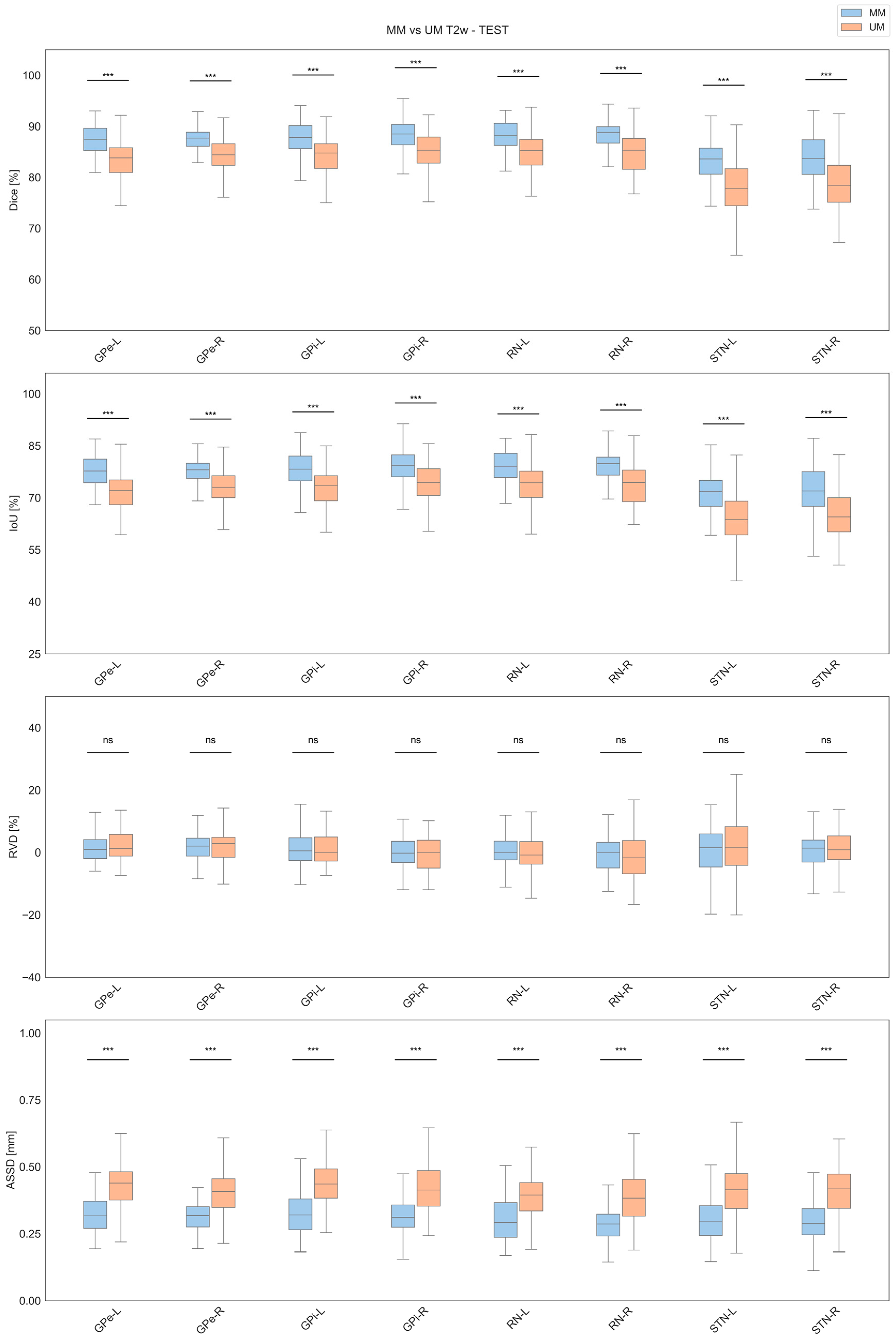
3.2. Test Results
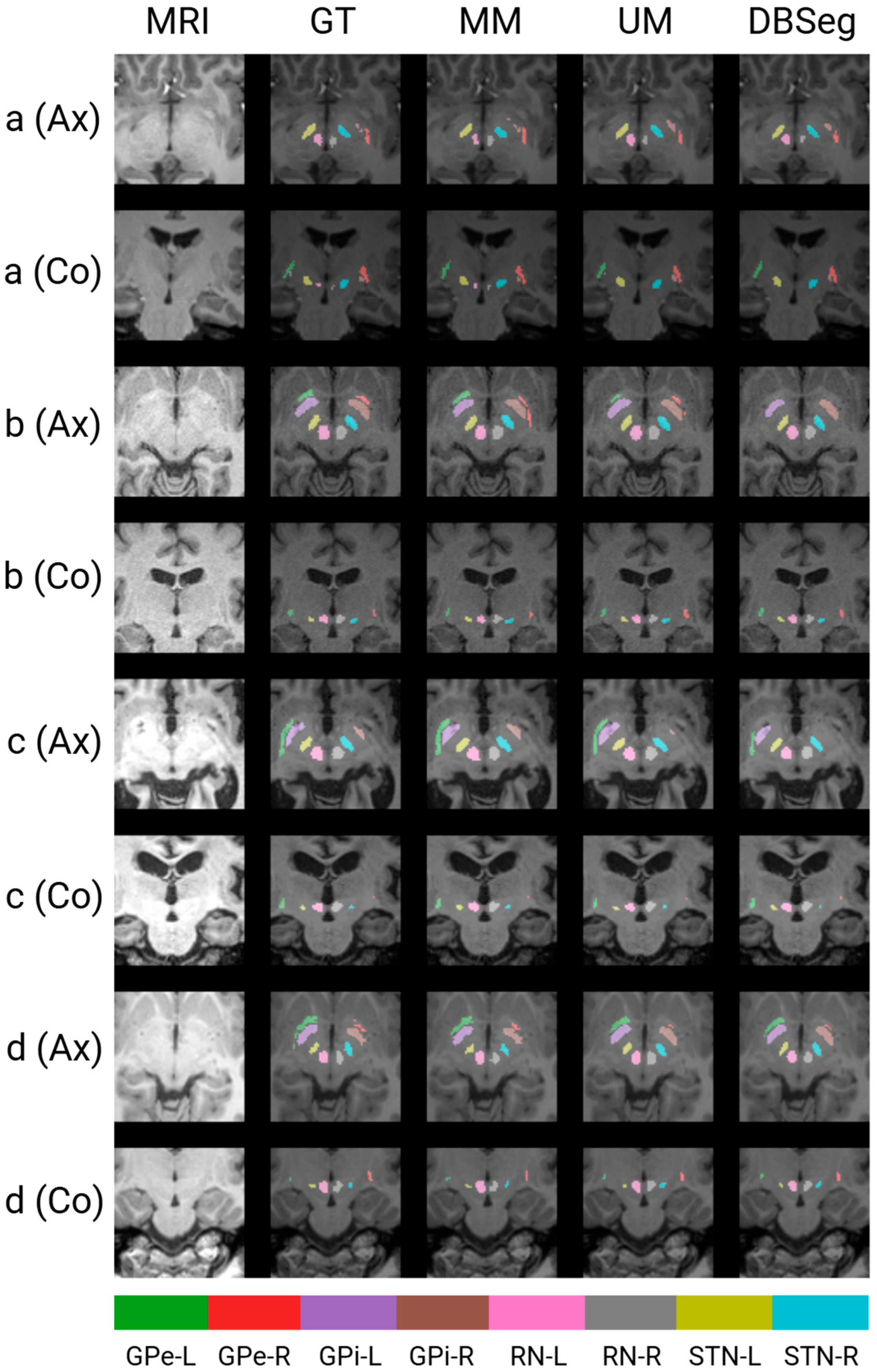
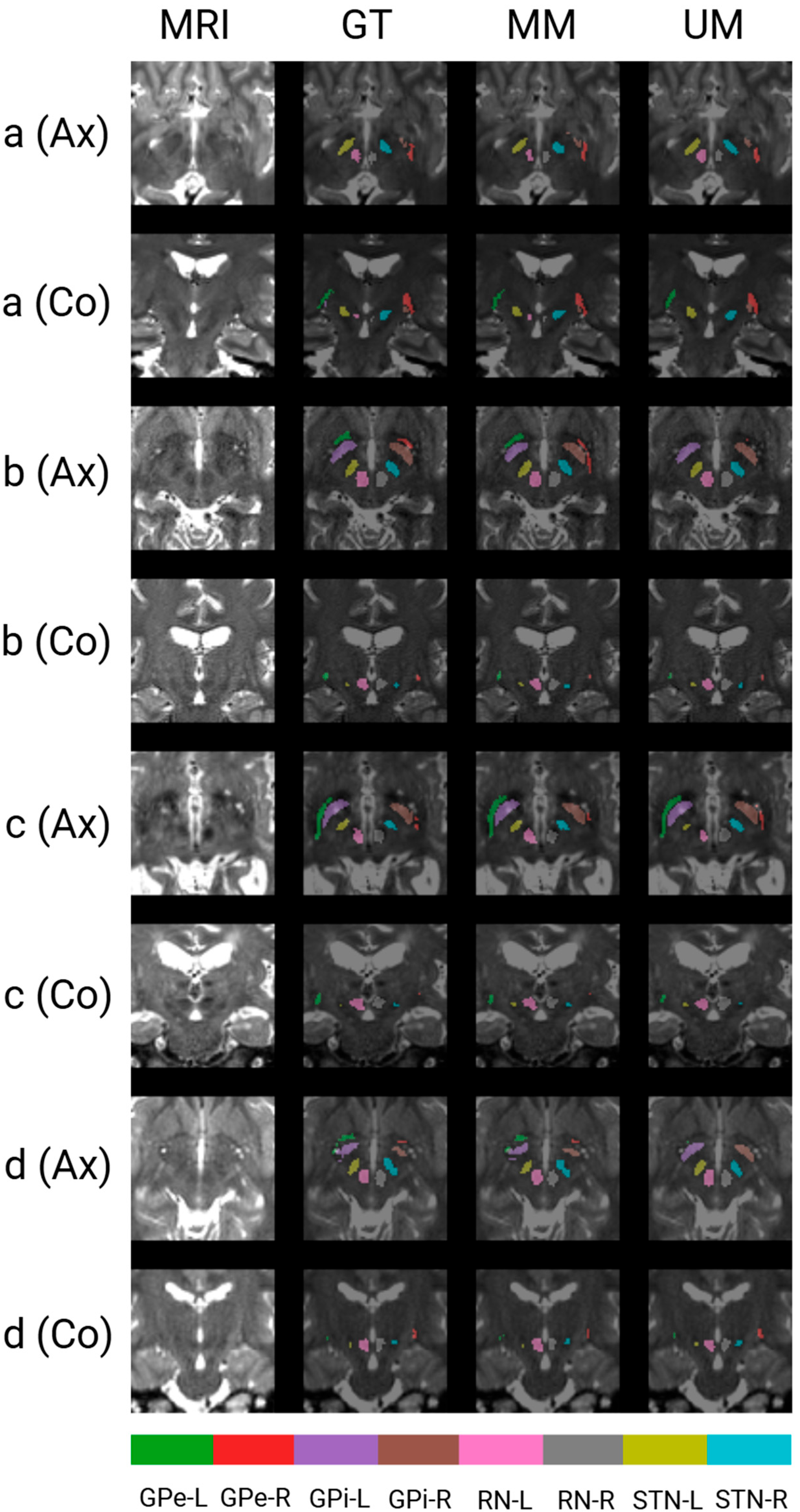
3.3. Qualitative Results
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s Disease |
| ADNI | Alzheimer’s Disease Neuroimaging Initiative |
| ANTs | Advanced Normalization Tools |
| ASSD | Average Symmetric Surface Distance |
| BM | Brain Mask |
| CVAL | Cross-Validation Set |
| DL | Deep Learning |
| DBS | Deep Brain Stimulation |
| DISTAL | DBS Intrinsic Template Atlas |
| FLAIR | Fluid Attenuated Inversion Recovery |
| GPe | Globus Pallidus Externus |
| GPi | Globus Pallidus Internus |
| HCP | Human Connectome Project |
| IoU | Intersection over Union |
| IXI | Information eXchange from Images |
| LPI | Left–Posterior–Inferior |
| MM | Multimodal |
| MNI | Montreal Neurological Institute |
| MSD | Medical Segmentation Decathlon |
| MRI | Magnetic Resonance Imaging |
| NLA | Neural Correlates of Lidocaine Analgesic |
| OASIS3 | Open Access Series of Imaging Studies 3 |
| PD | Parkinson’s Disease |
| RN | Red Nucleus |
| ROI | Region of Interest |
| RVD | Relative Volume Difference |
| SyN | Symmetric Normalization |
| STN | Subthalamic Nucleus |
| THP | Traveling Human Phantom |
| UM | Unimodal |
| UNC | University of North Carolina |
| T1w | T1-Weighted |
| T2w | T2-Weighted |
References
- Liu, Y.; D’Haese, P.-F.; Newton, A.T.; Dawant, B.M. Generation of Human Thalamus Atlases from 7 T Data and Application to Intrathalamic Nuclei Segmentation in Clinical 3 T T1-Weighted Images. Magn. Reson. Imaging 2020, 65, 114–128. [Google Scholar] [CrossRef]
- Isaacs, B.R.; Keuken, M.C.; Alkemade, A.; Temel, Y.; Bazin, P.-L.; Forstmann, B.U. Methodological Considerations for Neuroimaging in Deep Brain Stimulation of the Subthalamic Nucleus in Parkinson’s Disease Patients. J. Clin. Med. 2020, 9, 3124. [Google Scholar] [CrossRef]
- Baniasadi, M.; Petersen, M.V.; Gonçalves, J.; Horn, A.; Vlasov, V.; Hertel, F.; Husch, A. DBSegment: Fast and Robust Segmentation of Deep Brain Structures Considering Domain Generalization. Hum. Brain Mapp. 2023, 44, 762–778. [Google Scholar] [CrossRef]
- Polanski, W.H.; Zolal, A.; Sitoci-Ficici, K.H.; Hiepe, P.; Schackert, G.; Sobottka, S.B. Comparison of Automatic Segmentation Algorithms for the Subthalamic Nucleus. Stereotact. Funct. Neurosurg. 2020, 98, 256–262. [Google Scholar] [CrossRef]
- Reinacher, P.C.; Várkuti, B.; Krüger, M.T.; Piroth, T.; Egger, K.; Roelz, R.; Coenen, V.A. Automatic Segmentation of the Subthalamic Nucleus: A Viable Option to Support Planning and Visualization of Patient-Specific Targeting in Deep Brain Stimulation. Oper. Neurosurg. 2019, 17, 497. [Google Scholar] [CrossRef]
- Chen, J.; Xu, H.; Xu, B.; Wang, Y.; Shi, Y.; Xiao, L. Automatic Localization of Key Structures for Subthalamic Nucleus–Deep Brain Stimulation Surgery via Prior-Enhanced Multi-Object Magnetic Resonance Imaging Segmentation. World Neurosurg. 2023, 178, e472–e479. [Google Scholar] [CrossRef]
- Kim, J.; Duchin, Y.; Sapiro, G.; Vitek, J.; Harel, N. Clinical Deep Brain Stimulation Region Prediction Using Regression Forests from High-Field MRI. In Proceedings of the 2015 IEEE International Conference on Image Processing (ICIP), Quebec City, QC, Canada, 27–30 September 2015; pp. 2480–2484. [Google Scholar]
- Kawahara, D.; Nagata, Y. T1-Weighted and T2-Weighted MRI Image Synthesis with Convolutional Generative Adversarial Networks. Rep. Pract. Oncol. Radiother. 2021, 26, 35–42. [Google Scholar] [CrossRef]
- Haegelen, C.; Coupé, P.; Fonov, V.; Guizard, N.; Jannin, P.; Morandi, X.; Collins, D.L. Automated Segmentation of Basal Ganglia and Deep Brain Structures in MRI of Parkinson’s Disease. Int. J. CARS 2013, 8, 99–110. [Google Scholar] [CrossRef]
- Lima, T.; Varga, I.; Bakštein, E.; Novák, D.; Alves, V. Subthalamic Nucleus Segmentation in High-Field Magnetic Resonance Data. Is Space Normalization by Template Co-Registration Necessary? arXiv 2024, arXiv:2407.15485. [Google Scholar]
- Pauli, W.M.; Nili, A.N.; Tyszka, J.M. A High-Resolution Probabilistic in Vivo Atlas of Human Subcortical Brain Nuclei. Sci. Data 2018, 5, 180063. [Google Scholar] [CrossRef]
- Ewert, S.; Plettig, P.; Li, N.; Chakravarty, M.M.; Collins, D.L.; Herrington, T.M.; Kühn, A.A.; Horn, A. Toward Defining Deep Brain Stimulation Targets in MNI Space: A Subcortical Atlas Based on Multimodal MRI, Histology and Structural Connectivity. NeuroImage 2018, 170, 271–282. [Google Scholar] [CrossRef]
- Su, J.H.; Thomas, F.T.; Kasoff, W.S.; Tourdias, T.; Choi, E.Y.; Rutt, B.K.; Saranathan, M. Thalamus Optimized Multi Atlas Segmentation (THOMAS): Fast, Fully Automated Segmentation of Thalamic Nuclei from Structural MRI. NeuroImage 2019, 194, 272–282. [Google Scholar] [CrossRef]
- Brett, M.; Johnsrude, I.S.; Owen, A.M. The Problem of Functional Localization in the Human Brain. Nat. Rev. Neurosci. 2002, 3, 243–249. [Google Scholar] [CrossRef]
- Horn, A.; Kühn, A.A. Lead-DBS: A Toolbox for Deep Brain Stimulation Electrode Localizations and Visualizations. NeuroImage 2015, 107, 127–135. [Google Scholar] [CrossRef]
- Horn, A.; Li, N.; Dembek, T.A.; Kappel, A.; Boulay, C.; Ewert, S.; Tietze, A.; Husch, A.; Perera, T.; Neumann, W.-J.; et al. Lead-DBS v2: Towards a Comprehensive Pipeline for Deep Brain Stimulation Imaging. NeuroImage 2019, 184, 293–316. [Google Scholar] [CrossRef]
- Neudorfer, C.; Butenko, K.; Oxenford, S.; Rajamani, N.; Achtzehn, J.; Goede, L.; Hollunder, B.; Ríos, A.S.; Hart, L.; Tasserie, J.; et al. Lead-DBS v3.0: Mapping Deep Brain Stimulation Effects to Local Anatomy and Global Networks. NeuroImage 2023, 268, 119862. [Google Scholar] [CrossRef]
- Xiao, Y.; Fonov, V.S.; Beriault, S.; Gerard, I.; Sadikot, A.F.; Pike, G.B.; Collins, D.L. Patch-Based Label Fusion Segmentation of Brainstem Structures with Dual-Contrast MRI for Parkinson’s Disease. Int. J. CARS 2015, 10, 1029–1041. [Google Scholar] [CrossRef]
- Falahati, F.; Gustavsson, J.; Kalpouzos, G. Automated Segmentation of Midbrain Nuclei Using Deep Learning and Multisequence MRI: A Longitudinal Study on Iron Accumulation with Age. Imaging Neurosci. 2024, 2, 1–20. [Google Scholar] [CrossRef]
- Altini, N.; Rossini, M.; Turkevi-Nagy, S.; Pesce, F.; Pontrelli, P.; Prencipe, B.; Berloco, F.; Seshan, S.; Gibier, J.-B.; Pedraza Dorado, A.; et al. Performance and Limitations of a Supervised Deep Learning Approach for the Histopathological Oxford Classification of Glomeruli with IgA Nephropathy. Comput. Methods Programs Biomed. 2023, 242, 107814. [Google Scholar] [CrossRef]
- Bevilacqua, V.; Altini, N.; Prencipe, B.; Brunetti, A.; Villani, L.; Sacco, A.; Morelli, C.; Ciaccia, M.; Scardapane, A. Lung Segmentation and Characterization in COVID-19 Patients for Assessing Pulmonary Thromboembolism: An Approach Based on Deep Learning and Radiomics. Electronics 2021, 10, 2475. [Google Scholar] [CrossRef]
- Berloco, F.; Zaccaria, G.M.; Altini, N.; Colucci, S.; Bevilacqua, V. A Multimodal Framework for Assessing the Link between Pathomics, Transcriptomics, and Pancreatic Cancer Mutations. Comput. Med. Imaging Graph. 2025, 123, 102526. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A Self-Configuring Method for Deep Learning-Based Biomedical Image Segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Altini, N.; Brunetti, A.; Napoletano, V.P.; Girardi, F.; Allegretti, E.; Hussain, S.M.; Brunetti, G.; Triggiani, V.; Bevilacqua, V.; Buongiorno, D. A Fusion Biopsy Framework for Prostate Cancer Based on Deformable Superellipses and nnU-Net. Bioengineering 2022, 9, 343. [Google Scholar] [CrossRef]
- Solomon, O.; Palnitkar, T.; Patriat, R.; Braun, H.; Aman, J.; Park, M.C.; Vitek, J.; Sapiro, G.; Harel, N. Deep-learning Based Fully Automatic Segmentation of the Globus Pallidus Interna and Externa Using Ultra-high 7 Tesla MRI. Hum. Brain Mapp. 2021, 42, 2862–2879. [Google Scholar] [CrossRef]
- Beliveau, V.; Nørgaard, M.; Birkl, C.; Seppi, K.; Scherfler, C. Automated Segmentation of Deep Brain Nuclei Using Convolutional Neural Networks and Susceptibility Weighted Imaging. Hum. Brain Mapp. 2021, 42, 4809–4822. [Google Scholar] [CrossRef]
- Hanke, M.; Baumgartner, F.J.; Ibe, P.; Kaule, F.R.; Pollmann, S.; Speck, O.; Zinke, W.; Stadler, J. A High-Resolution 7-Tesla fMRI Dataset from Complex Natural Stimulation with an Audio Movie. Sci. Data 2014, 1, 140003. [Google Scholar] [CrossRef]
- Van Essen, D.C.; Ugurbil, K.; Auerbach, E.; Barch, D.; Behrens, T.E.J.; Bucholz, R.; Chang, A.; Chen, L.; Corbetta, M.; Curtiss, S.W.; et al. The Human Connectome Project: A Data Acquisition Perspective. NeuroImage 2012, 62, 2222–2231. [Google Scholar] [CrossRef]
- LaMontagne, P.J.; Benzinger, T.L.S.; Morris, J.C.; Keefe, S.; Hornbeck, R.; Xiong, C.; Grant, E.; Hassenstab, J.; Moulder, K.; Vlassenko, A.G.; et al. OASIS-3: Longitudinal Neuroimaging, Clinical, and Cognitive Dataset for Normal Aging and Alzheimer Disease. medRxiv 2019. [Google Scholar] [CrossRef]
- Mueller, S.G.; Weiner, M.W.; Thal, L.J.; Petersen, R.C.; Jack, C.R.; Jagust, W.; Trojanowski, J.Q.; Toga, A.W.; Beckett, L. Ways toward an Early Diagnosis in Alzheimer’s Disease: The Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimers Dement. 2005, 1, 55–66. [Google Scholar] [CrossRef]
- IXI Dataset—Brain Development. Available online: https://brain-development.org/ixi-dataset/ (accessed on 15 June 2025).
- Chen, X.; Qu, L.; Xie, Y.; Ahmad, S.; Yap, P.-T. A Paired Dataset of T1- and T2-Weighted MRI at 3 Tesla and 7 Tesla. Sci. Data 2023, 10, 489. [Google Scholar] [CrossRef]
- Magnotta, V.A.; Matsui, J.T.; Liu, D.; Johnson, H.J.; Long, J.D.; Bolster, B.D.; Mueller, B.A.; Lim, K.; Mori, S.; Helmer, K.G.; et al. MultiCenter Reliability of Diffusion Tensor Imaging. Brain Connect. 2012, 2, 345–355. [Google Scholar] [CrossRef]
- Vogt, K.M.; Burlew, A.C.; Simmons, M.A.; Reddy, S.N.; Kozdron, C.N.; Ibinson, J.W. Neural Correlates of Systemic Lidocaine Administration in Healthy Adults Measured by Functional MRI: A Single Arm Open Label Study. Br. J. Anaesth. 2025, 134, 414–424. [Google Scholar] [CrossRef]
- Kausel, L.; Figueroa-Vargas, A.; Zamorano, F.; Stecher, X.; Aspé-Sánchez, M.; Carvajal-Paredes, P.; Márquez-Rodríguez, V.; Martínez-Molina, M.P.; Román, C.; Soto-Fernández, P.; et al. Patients Recovering from COVID-19 Who Presented with Anosmia during Their Acute Episode Have Behavioral, Functional, and Structural Brain Alterations. Sci. Rep. 2024, 14, 19049. [Google Scholar] [CrossRef]
- Fonov, V.S.; Evans, A.C.; McKinstry, R.C.; Almli, C.R.; Collins, D.L. Unbiased Nonlinear Average Age-Appropriate Brain Templates from Birth to Adulthood. NeuroImage 2009, 47, S102. [Google Scholar] [CrossRef]
- 3D Slicer Image Computing Platform. Available online: https://slicer.org/ (accessed on 21 May 2025).
- Baek, H.-M. Diffusion Measures of Subcortical Structures Using High-Field MRI. Brain Sci. 2023, 13, 391. [Google Scholar] [CrossRef]
- Tustison, N.J.; Avants, B.B.; Cook, P.A.; Zheng, Y.; Egan, A.; Yushkevich, P.A.; Gee, J.C. N4ITK: Improved N3 Bias Correction. IEEE Trans. Med. Imaging 2010, 29, 1310–1320. [Google Scholar] [CrossRef]
- Avants, B.; Epstein, C.; Grossman, M.; Gee, J. Symmetric Diffeomorphic Image Registration with Cross-Correlation: Evaluating Automated Labeling of Elderly and Neurodegenerative Brain. Med. Image Anal. 2008, 12, 26–41. [Google Scholar] [CrossRef]
- Gulban, O.F.; Nielson, D.; Lee, J.; Poldrack, R.; Gorgolewski, C.; Vanessasaurus; Markiewicz, C. Poldracklab/Pydeface: PyDeface, version 2.0.2; Zenodo: Geneva, Switzerland, 2022.
- Altini, N.; Prencipe, B.; Cascarano, G.D.; Brunetti, A.; Brunetti, G.; Triggiani, V.; Carnimeo, L.; Marino, F.; Guerriero, A.; Villani, L.; et al. Liver, Kidney and Spleen Segmentation from CT Scans and MRI with Deep Learning: A Survey. Neurocomputing 2022, 490, 30–53. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kim, J.; Lenglet, C.; Duchin, Y.; Sapiro, G.; Harel, N. Semiautomatic Segmentation of Brain Subcortical Structures From High-Field MRI. IEEE J. Biomed. Health Inform. 2014, 18, 1678–1695. [Google Scholar] [CrossRef]
- Lee, J.-H.; Yun, J.Y.; Gregory, A.; Hogarth, P.; Hayflick, S.J. Brain MRI Pattern Recognition in Neurodegeneration With Brain Iron Accumulation. Front. Neurol. 2020, 11, 1024. [Google Scholar] [CrossRef] [PubMed]
- Hescham, S.; Lim, L.W.; Jahanshahi, A.; Blokland, A.; Temel, Y. Deep Brain Stimulation in Dementia-Related Disorders. Neurosci. Biobehav. Rev. 2013, 37, 2666–2675. [Google Scholar] [CrossRef] [PubMed]
- Links Between COVID-19 and Parkinson’s Disease/Alzheimer’s Disease: Reciprocal Impacts, Medical Care Strategies and Underlying Mechanisms | Translational Neurodegeneration | Full Text. Available online: https://translationalneurodegeneration.biomedcentral.com/articles/10.1186/s40035-023-00337-1 (accessed on 9 July 2025).
- Altini, N.; Marvulli, T.M.; Zito, F.A.; Caputo, M.; Tommasi, S.; Azzariti, A.; Brunetti, A.; Prencipe, B.; Mattioli, E.; De Summa, S.; et al. The Role of Unpaired Image-to-Image Translation for Stain Color Normalization in Colorectal Cancer Histology Classification. Comput. Methods Programs Biomed. 2023, 234, 107511. [Google Scholar] [CrossRef] [PubMed]
- Dayarathna, S.; Islam, K.T.; Zhuang, B.; Yang, G.; Cai, J.; Law, M.; Chen, Z. McCaD: Multi-Contrast MRI Conditioned, Adaptive Adversarial Diffusion Model for High-Fidelity MRI Synthesis. In Proceedings of the 2025 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Tucson, AZ, USA, 28 February–4 March 2025; pp. 670–679. [Google Scholar]
| Subset | Dataset | Scanner | FS | T1w | T2w | Disease | Age | M/F | MRI Scans |
|---|---|---|---|---|---|---|---|---|---|
| CVAL (n = 260) | HCP | SM | 3T | MPRAGE | SPACE | HT | N/A | 16/13 | 29 |
| OASIS3 | SM | 3T | MPRAGE | SPACE | HT, CI | 52–84 | 11/19 | 30 | |
| ADNI4 | PL, SM | 3T | MPRAGE | SPACE/VISTA | HT, MCI, AD | 55–85 | 13/46 | 59 | |
| IXI | N/A | N/A | N/A | N/A | HT | 21–74 | 16/13 | 29 | |
| UNC | SM | 3T, 7T | MPRAGE, MP2RAGE | SPACE, TSE | HT | 25–41 | 11/5 | 16 | |
| THP | PL, SM | 3T | MPRAGE | SPACE | HT | N/A | N/A | 40 | |
| NLA | SM | 3T | MPRAGE | SPACE | HT | 20–55 | 13/14 | 27 | |
| neuroCOVID | SM | 3T | MPRAGE | SPACE | COVID, RI | 21–64 | 17/13 | 30 | |
| TEST (n = 65) | OASIS3 | SM | 3T | MPRAGE | SPACE | HT, CI | 59–97 | 5/11 | 16 |
| ADNI4 | SM | 3T | MPRAGE | SPACE | HT, MCI, AD | 56–85 | 7/9 | 16 | |
| IXI | N/A | N/A | N/A | N/A | HT | 23–63 | 9/8 | 17 | |
| neuroCOVID | SM | 3T | MPRAGE | SPACE | COVID, RI | 19–66 | 9/7 | 16 |
| Full Name | Acronym | Label |
|---|---|---|
| Brain mask | BM | 1 |
| Globus pallidus externus (left) | GPe-L | 2 |
| Globus pallidus externus (right) | GPe-R | 3 |
| Globus pallidus internus (left) | GPi-L | 4 |
| Globus pallidus internus (right) | GPi-R | 5 |
| Red nucleus (left) | RN-L | 6 |
| Red nucleus (right) | RN-R | 7 |
| Subthalamic nucleus (left) | STN-L | 8 |
| Subthalamic nucleus (right) | STN-R | 9 |
| Label | Model | Dice [%] | IoU [%] | RVD [%] | ASSD [mm] |
|---|---|---|---|---|---|
| BM | MM | 98.18 ± 0.49 | 96.44 ± 0.92 | 0.10 ± 1.23 | 0.68 ± 0.17 |
| UM | 98.14 ± 0.58 | 96.36 ± 1.08 | 0.46 ± 1.42 | 0.72 ± 0.20 | |
| DBSegment | 96.31 ± 0.70 | 92.89 ± 1.28 | 1.21 ± 2.32 | 1.52 ± 0.77 | |
| GPe-L | MM | 86.48 ± 7.28 | 76.71 ± 8.37 | 1.77 ± 5.93 | 0.43 ± 1.44 |
| UM | 86.49 ± 6.07 | 76.56 ± 7.10 | 1.47 ± 8.72 | 0.38 ± 0.38 | |
| DBSegment | 81.30 ± 4.67 | 68.74 ± 6.37 | −9.84 ± 8.65 | 0.48 ± 0.17 | |
| GPe-R | MM | 85.91 ± 6.96 | 75.80 ± 8.57 | 2.29 ± 5.08 | 0.40 ± 0.73 |
| UM | 86.72 ± 4.55 | 76.81 ± 6.39 | 2.14 ± 6.51 | 0.35 ± 0.11 | |
| DBSegment | 81.66 ± 4.76 | 69.27 ± 6.58 | −12.17 ± 7.93 | 0.49 ± 0.47 | |
| GPi-L | MM | 86.74 ± 7.28 | 77.12 ± 8.60 | 1.30 ± 7.31 | 0.43 ± 1.34 |
| UM | 87.44 ± 4.62 | 77.95 ± 6.43 | 1.08 ± 8.03 | 0.36 ± 0.16 | |
| DBSegment | 81.13 ± 5.66 | 68.61 ± 7.56 | −10.43 ± 7.52 | 0.51 ± 0.15 | |
| GPi-R | MM | 86.39 ± 7.09 | 76.57 ± 8.82 | 1.60 ± 6.43 | 0.40 ± 0.64 |
| UM | 87.91 ± 3.75 | 78.61 ± 5.76 | 1.54 ± 7.28 | 0.35 ± 0.10 | |
| DBSegment | 81.23 ± 5.92 | 68.79 ± 8.01 | −12.05 ± 7.40 | 0.52 ± 0.21 | |
| RN-L | MM | 86.23 ± 7.24 | 76.37 ± 9.31 | 1.20 ± 9.83 | 0.36 ± 0.19 |
| UM | 87.97 ± 8.48 | 79.18 ± 8.97 | 1.70 ± 8.11 | 0.35 ± 0.59 | |
| DBSegment | 83.10 ± 4.86 | 71.38 ± 6.90 | −14.88 ± 7.52 | 0.43 ± 0.12 | |
| RN-R | MM | 86.11 ± 6.29 | 76.08 ± 8.73 | −0.01 ± 7.51 | 0.35 ± 0.15 |
| UM | 87.92 ± 8.56 | 79.10 ± 9.03 | 1.45 ± 7.82 | 0.35 ± 0.64 | |
| DBSegment | 82.34 ± 5.31 | 70.31 ± 7.41 | −15.66 ± 7.31 | 0.44 ± 0.13 | |
| STN-L | MM | 80.93 ± 8.96 | 68.74 ± 10.48 | 0.27 ± 9.14 | 0.40 ± 0.89 |
| UM | 83.15 ± 7.33 | 71.72 ± 9.12 | 2.13 ± 9.97 | 0.36 ± 0.53 | |
| DBSegment | 75.14 ± 7.61 | 60.76 ± 9.58 | −12.68 ± 8.94 | 0.45 ± 0.14 | |
| STN-R | MM | 81.27 ± 8.43 | 69.18 ± 10.32 | 0.37 ± 8.59 | 0.37 ± 0.38 |
| UM | 83.29 ± 7.42 | 71.91 ± 8.82 | 2.14 ± 9.65 | 0.39 ± 1.07 | |
| DBSegment | 75.75 ± 7.99 | 61.60 ± 9.97 | −13.64 ± 8.51 | 0.46 ± 0.15 |
| Label | Model | Dice [%] | IoU [%] | RVD [%] | ASSD [mm] |
|---|---|---|---|---|---|
| BM | MM | 98.18 ± 0.49 | 96.44 ± 0.91 | 0.09 ± 1.22 | 0.67 ± 0.17 |
| UM | 98.07 ± 0.56 | 96.22 ± 1.04 | 0.21 ± 1.47 | 0.74 ± 0.19 | |
| GPe-L | MM | 86.39 ± 7.27 | 76.56 ± 8.40 | 1.65 ± 6.08 | 0.42 ± 1.44 |
| UM | 84.36 ± 6.67 | 73.36 ± 7.40 | 1.22 ± 9.96 | 0.45 ± 0.61 | |
| GPe-R | MM | 85.90 ± 6.90 | 75.79 ± 8.55 | 2.41 ± 5.24 | 0.39 ± 0.71 |
| UM | 84.92 ± 3.36 | 73.94 ± 4.97 | 2.90 ± 6.86 | 0.40 ± 0.09 | |
| GPi-L | MM | 86.63 ± 7.34 | 76.95 ± 8.71 | 1.45 ± 7.48 | 0.43 ± 1.35 |
| UM | 84.76 ± 6.55 | 73.95 ± 7.34 | 1.11 ± 10.65 | 0.41 ± 0.11 | |
| GPi-R | MM | 86.48 ± 6.90 | 76.71 ± 8.75 | 1.56 ± 6.69 | 0.39 ± 0.63 |
| UM | 85.34 ± 3.89 | 74.62 ± 5.70 | 2.25 ± 9.56 | 0.46 ± 0.60 | |
| RN-L | MM | 86.20 ± 7.61 | 76.37 ± 9.60 | 1.09 ± 9.75 | 0.35 ± 0.21 |
| UM | 85.54 ± 4.34 | 74.97 ± 6.26 | 1.40 ± 9.97 | 0.38 ± 0.10 | |
| RN-R | MM | 86.09 ± 6.17 | 76.04 ± 8.64 | 0.13 ± 8.02 | 0.34 ± 0.15 |
| UM | 84.73 ± 3.98 | 73.70 ± 5.89 | 1.81 ± 9.59 | 0.40 ± 0.10 | |
| STN-L | MM | 80.85 ± 8.91 | 68.62 ± 10.48 | 0.14 ± 9.79 | 0.38 ± 0.86 |
| UM | 78.92 ± 6.63 | 65.63 ± 8.32 | 1.88 ± 12.59 | 0.40 ± 0.16 | |
| STN-R | MM | 81.13 ± 8.63 | 69.00 ± 10.60 | 0.67 ± 8.73 | 0.36 ± 0.37 |
| UM | 79.45 ± 5.74 | 66.29 ± 7.90 | 2.54 ± 11.18 | 0.40 ± 0.11 |
| Label | Model | Dice [%] | IoU [%] | RVD [%] | ASSD [mm] |
|---|---|---|---|---|---|
| BM | MM | 98.24 ± 0.41 | 96.54 ± 0.79 | −0.25 ± 1.20 | 0.67 ± 0.15 |
| UM | 98.19 ± 0.54 | 96.45 ± 1.02 | 0.34 ± 1.56 | 0.70 ± 0.20 | |
| DBSegment | 96.31 ± 0.59 | 92.88 ± 1.08 | −0.00 ± 2.02 | 1.42 ± 0.21 | |
| GPe-L | MM | 86.76 ± 4.01 | 76.83 ± 5.87 | 1.81 ± 6.13 | 0.34 ± 0.10 |
| UM | 86.93 ± 2.96 | 77.00 ± 4.61 | 1.48 ± 6.17 | 0.35 ± 0.07 | |
| DBSegment | 82.44 ± 4.86 | 70.40 ± 6.62 | −10.84 ± 5.69 | 0.44 ± 0.12 | |
| GPe-R | MM | 86.88 ± 3.95 | 77.01 ± 5.91 | 1.83 ± 5.19 | 0.34 ± 0.10 |
| UM | 86.48 ± 3.54 | 76.34 ± 5.39 | 2.42 ± 5.46 | 0.36 ± 0.09 | |
| DBSegment | 82.04 ± 5.43 | 69.88 ± 7.44 | −11.44 ± 7.47 | 0.52 ± 0.66 | |
| GPi-L | MM | 87.39 ± 3.80 | 77.80 ± 5.82 | 1.70 ± 6.15 | 0.34 ± 0.10 |
| UM | 87.53 ± 3.54 | 78.00 ± 5.55 | 1.51 ± 5.12 | 0.35 ± 0.09 | |
| DBSegment | 82.52 ± 4.31 | 70.46 ± 6.16 | −10.10 ± 5.31 | 0.47 ± 0.11 | |
| GPi-R | MM | 88.01 ± 3.43 | 78.75 ± 5.40 | 0.23 ± 5.36 | 0.33 ± 0.09 |
| UM | 87.68 ± 3.12 | 78.20 ± 4.94 | 1.84 ± 5.44 | 0.36 ± 0.09 | |
| DBSegment | 83.04 ± 4.89 | 71.29 ± 7.05 | −10.98 ± 5.75 | 0.48 ± 0.20 | |
| RN-L | MM | 88.17 ± 3.21 | 78.98 ± 5.05 | 0.06 ± 5.55 | 0.31 ± 0.08 |
| UM | 88.91 ± 3.12 | 80.18 ± 5.02 | 0.25 ± 6.07 | 0.29 ± 0.08 | |
| DBSegment | 84.69 ± 3.57 | 73.61 ± 5.16 | −14.61 ± 6.14 | 0.39 ± 0.09 | |
| RN-R | MM | 88.29 ± 2.59 | 79.12 ± 4.12 | −0.46 ± 5.13 | 0.30 ± 0.07 |
| UM | 88.08 ± 3.40 | 78.87 ± 5.44 | 1.13 ± 5.89 | 0.31 ± 0.08 | |
| DBSegment | 83.42 ± 3.25 | 71.69 ± 4.77 | −14.63 ± 5.29 | 0.42 ± 0.08 | |
| STN-L | MM | 82.79 ± 5.55 | 70.99 ± 7.63 | 2.09 ± 8.18 | 0.31 ± 0.09 |
| UM | 83.37 ± 4.53 | 71.73 ± 6.62 | 2.21 ± 7.24 | 0.31 ± 0.08 | |
| DBSegment | 75.44 ± 6.20 | 60.95 ± 7.86 | −10.03 ± 6.81 | 0.44 ± 0.11 | |
| STN-R | MM | 83.86 ± 4.95 | 72.50 ± 7.19 | 0.60 ± 6.94 | 0.30 ± 0.09 |
| UM | 84.01 ± 4.58 | 72.69 ± 6.83 | 3.45 ± 6.64 | 0.31 ± 0.09 | |
| DBSegment | 76.45 ± 6.72 | 62.34 ± 8.68 | −10.13 ± 6.20 | 0.44 ± 0.12 |
| Label | Model | Dice [%] | IoU [%] | RVD [%] | ASSD [mm] |
|---|---|---|---|---|---|
| BM | MM | 98.24 ± 0.41 | 96.54 ± 0.78 | −0.25 ± 1.20 | 0.66 ± 0.15 |
| UM | 98.10 ± 0.51 | 96.27 ± 0.97 | −0.10 ± 1.48 | 0.73 ± 0.19 | |
| GPe-L | MM | 86.74 ± 4.09 | 76.79 ± 5.97 | 2.01 ± 6.19 | 0.34 ± 0.10 |
| UM | 82.49 ± 8.16 | 70.83 ± 9.32 | 0.70 ± 12.61 | 0.52 ± 0.64 | |
| GPe-R | MM | 86.93 ± 3.87 | 77.08 ± 5.78 | 1.96 ± 5.27 | 0.34 ± 0.10 |
| UM | 84.38 ± 3.69 | 73.16 ± 5.49 | 1.86 ± 5.08 | 0.41 ± 0.09 | |
| GPi-L | MM | 87.30 ± 3.89 | 77.66 ± 5.94 | 1.83 ± 5.97 | 0.34 ± 0.10 |
| UM | 83.24 ± 7.28 | 71.83 ± 8.96 | −0.21 ± 11.18 | 0.48 ± 0.27 | |
| GPi-R | MM | 88.06 ± 3.51 | 78.83 ± 5.51 | 0.19 ± 5.61 | 0.33 ± 0.09 |
| UM | 84.74 ± 4.61 | 73.78 ± 6.60 | 0.18 ± 7.80 | 0.52 ± 0.75 | |
| RN-L | MM | 88.16 ± 3.20 | 78.97 ± 5.04 | 0.37 ± 6.06 | 0.30 ± 0.08 |
| UM | 84.81 ± 4.85 | 73.92 ± 7.13 | −0.18 ± 7.14 | 0.40 ± 0.12 | |
| RN-R | MM | 88.47 ± 2.71 | 79.43 ± 4.33 | −0.95 ± 5.20 | 0.29 ± 0.07 |
| UM | 85.06 ± 4.19 | 74.23 ± 6.38 | −1.43 ± 7.62 | 0.38 ± 0.10 | |
| STN-L | MM | 82.72 ± 5.69 | 70.89 ± 7.76 | 1.26 ± 8.32 | 0.30 ± 0.09 |
| UM | 77.75 ± 5.82 | 63.97 ± 7.75 | 1.80 ± 10.76 | 0.41 ± 0.10 | |
| STN-R | MM | 83.59 ± 5.12 | 72.13 ± 7.45 | 0.72 ± 7.31 | 0.30 ± 0.09 |
| UM | 78.85 ± 5.89 | 65.48 ± 8.12 | 2.29 ± 8.88 | 0.40 ± 0.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altini, N.; Lasaracina, E.; Galeone, F.; Prunella, M.; Suglia, V.; Carnimeo, L.; Triggiani, V.; Ranieri, D.; Brunetti, G.; Bevilacqua, V. A Comparison Between Unimodal and Multimodal Segmentation Models for Deep Brain Structures from T1- and T2-Weighted MRI. Mach. Learn. Knowl. Extr. 2025, 7, 84. https://doi.org/10.3390/make7030084
Altini N, Lasaracina E, Galeone F, Prunella M, Suglia V, Carnimeo L, Triggiani V, Ranieri D, Brunetti G, Bevilacqua V. A Comparison Between Unimodal and Multimodal Segmentation Models for Deep Brain Structures from T1- and T2-Weighted MRI. Machine Learning and Knowledge Extraction. 2025; 7(3):84. https://doi.org/10.3390/make7030084
Chicago/Turabian StyleAltini, Nicola, Erica Lasaracina, Francesca Galeone, Michela Prunella, Vladimiro Suglia, Leonarda Carnimeo, Vito Triggiani, Daniele Ranieri, Gioacchino Brunetti, and Vitoantonio Bevilacqua. 2025. "A Comparison Between Unimodal and Multimodal Segmentation Models for Deep Brain Structures from T1- and T2-Weighted MRI" Machine Learning and Knowledge Extraction 7, no. 3: 84. https://doi.org/10.3390/make7030084
APA StyleAltini, N., Lasaracina, E., Galeone, F., Prunella, M., Suglia, V., Carnimeo, L., Triggiani, V., Ranieri, D., Brunetti, G., & Bevilacqua, V. (2025). A Comparison Between Unimodal and Multimodal Segmentation Models for Deep Brain Structures from T1- and T2-Weighted MRI. Machine Learning and Knowledge Extraction, 7(3), 84. https://doi.org/10.3390/make7030084







