Drug-Drug Interaction Extraction from Biomedical Text Using Relation BioBERT with BLSTM
Abstract
1. Introduction
- Our study proposes a novel approach that leverages the power of integrating BLSTM and Relation BioBERT to accurately extract drug-drug interactions (DDIs) and classify their respective types of relationships.
- To evaluate the efficacy of our proposed model, we conducted experiments on three distinct datasets: SemEval 2013, TAC 2018, and TAC 2019 DDIs Extraction, all of which involve drug-drug interactions (DDIs) extraction tasks. Our experimental results demonstrate that our proposed method (is R-BioBERT with BLSTM) outperforms the baseline model.
2. Related Works
3. Literature Review
3.1. BERT Language Model
3.2. BioBERT
3.3. Relation BERT
3.4. Bi-Directional Long Short-Term Memory Network
4. Materials and Methods
4.1. Datasets
4.1.1. SemEval 2013 DDIs Extraction
- Advice: Advice is a type of DDI that refers to recommendations or cautions given in a document about the concurrent use of two drugs. For instance, an example of advice could be “Extreme caution should be exercised when taking alosetron and ketoconazole together”.
- Effect: This type in the DDIs corpus refers to the resulting effect or pharmacodynamic mechanism of interaction between two drugs. For instance, an example sentence for this type could be: “After a single administration of oxytocin, PGF2alpha caused significantly increased vasoconstriction”.
- Int: This refers to an interaction between drugs without providing any further information. An example of this would be “Possible interaction between atorvastatin and cyclosporine”.
- Mechanism: This type of DDI refers to a description of the pharmacokinetic mechanism, as in the example, “Withdrawal of rifampin decreased the warfarin requirement by 50%”.
- Negative: This refers to drug entity pairs that do not have any interaction. For example, “Ibogaine, but not 18-MC, decreases heart rate at high doses”.
4.1.2. TAC 2018 and TAC 2019 DDIs Extraction
- Pharmacokinetic (PK)
- Pharmacodynamic (PD)
- Unspecified (U)
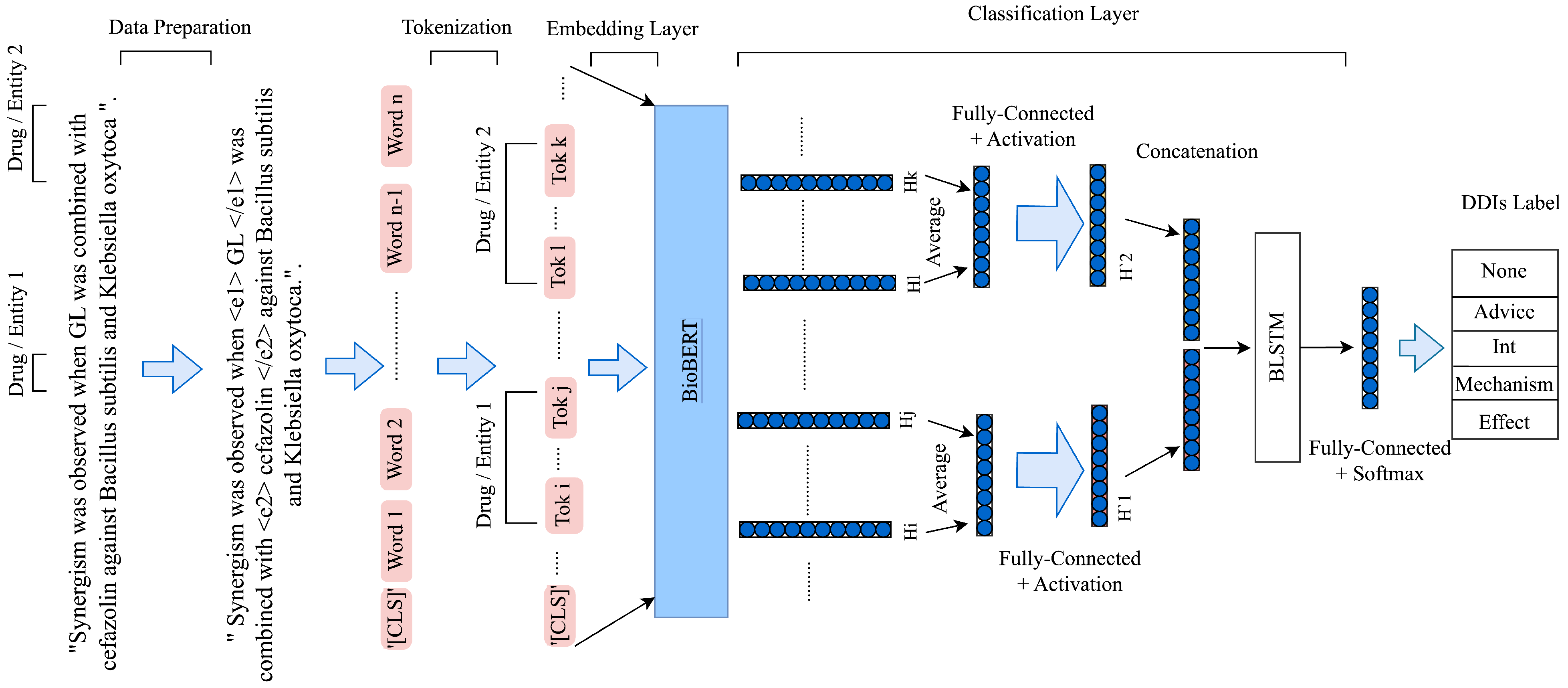
4.2. Data Preprocessing
- Firstly, instances with the same drug names in a pair were removed, as a drug cannot interact with itself. In addition, instances with only one drug in a sentence were eliminated.
- Secondly, to identify the location of two drugs in a pair, a special token was added before and was added after the first drug, and was added before and was added after the second drug. Unlike many other related studies, the original drug names were retained.
4.3. Model Architecture
5. Experimental Evaluation
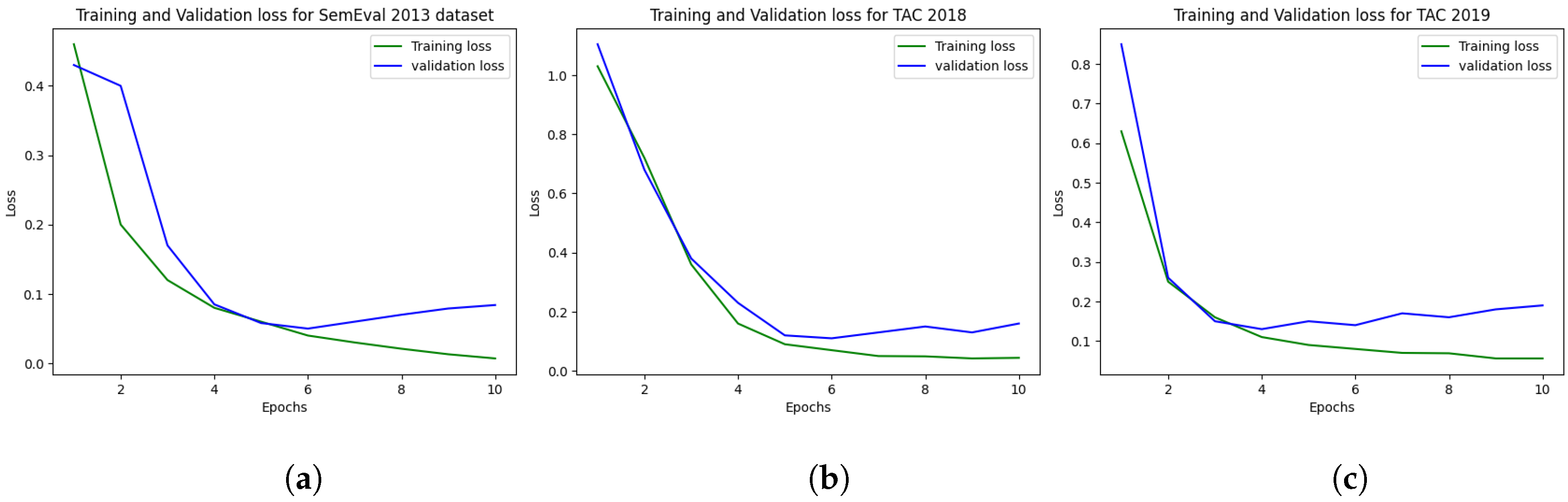
5.1. Experimental Setup
5.2. Evaluation Metrics
6. Results
6.1. Results on SemEval 2013 DDIs Extraction
6.2. Results on TAC 2018
6.3. Results on TAC 2019
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Miranda, V.; Fede, A.; Nobuo, M.; Ayres, V.; Giglio, A.; Miranda, M.; Riechelmann, R.P. Adverse drug reactions and drug interactions as causes of hospital admission in oncology. J. Pain Symptom Manag. 2011, 42, 342–353. [Google Scholar] [CrossRef]
- Lazarou, J.; Pomeranz, B.H.; Corey, P.N. Incidence of adverse drug reactions in hospitalized patients: A meta-analysis of prospective studies. JAMA 1998, 279, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.L.; Kallewaard, M.; Caspers, P.W.; Visser, L.E.; Leufkens, H.G.; Stricker, B.H. Hospitalisations and emergency department visits due to drug–drug interactions: A literature review. Pharmacoepidemiol. Drug Saf. 2007, 16, 641–651. [Google Scholar] [CrossRef]
- Businaro, R. Why we need an efficient and careful pharmacovigilance? J. Pharmacovigil. 2013. [Google Scholar] [CrossRef]
- Hohl, C.M.; Dankoff, J.; Colacone, A.; Afilalo, M. Polypharmacy, adverse drug-related events, and potential adverse drug interactions in elderly patients presenting to an emergency department. Ann. Emerg. Med. 2001, 38, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Paczynski, R.P.; Alexander, G.C.; Chinchilli, V.M.; Kruszewski, S.P. Quality of evidence in drug compendia supporting off-label use of typical and atypical antipsychotic medications. Int. J. Risk Saf. Med. 2012, 24, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Terol, A.; Caraballo, M.; Palma, D.; Santos-Ramos, B.; Molina, T.; Desongles, T.; Aguilar, A. Quality of interaction database management systems. Farm. Hosp. (Engl. Ed.) 2009, 33, 134–146. [Google Scholar] [CrossRef]
- Ammar, W.; Groeneveld, D.; Bhagavatula, C.; Beltagy, I.; Crawford, M.; Downey, D.; Dunkelberger, J.; Elgohary, A.; Feldman, S.; Ha, V.; et al. Construction of the literature graph in semantic scholar. arXiv 2018, arXiv:1805.02262. [Google Scholar]
- Asada, M.; Miwa, M.; Sasaki, Y. Enhancing drug-drug interaction extraction from texts by molecular structure information. arXiv 2018, arXiv:1805.05593. [Google Scholar]
- Beltagy, I.; Lo, K.; Cohan, A. SciBERT: A pretrained language model for scientific text. arXiv 2019, arXiv:1903.10676. [Google Scholar]
- Zhang, T.; Leng, J.; Liu, Y. Deep learning for drug–drug interaction extraction from the literature: A review. Brief. Bioinform. 2020, 21, 1609–1627. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Lauriola, I.; Lavelli, A.; Aiolli, F. An introduction to deep learning in natural language processing: Models, techniques, and tools. Neurocomputing 2022, 470, 443–456. [Google Scholar] [CrossRef]
- Pennington, J.; Socher, R.; Manning, C.D. Glove: Global vectors for word representation. In Proceedings of the 2014 Conference On Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [Google Scholar]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Shen, S.; Liu, J.; Lin, L.; Huang, Y.; Zhang, L.; Liu, C.; Feng, Y.; Wang, D. SsciBERT: A pre-trained language model for social science texts. Scientometrics 2023, 128, 1241–1263. [Google Scholar] [CrossRef]
- Hong, L.; Lin, J.; Li, S.; Wan, F.; Yang, H.; Jiang, T.; Zhao, D.; Zeng, J. A novel machine learning framework for automated biomedical relation extraction from large-scale literature repositories. Nat. Mach. Intell. 2020, 2, 347–355. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, Z.; Luo, L.; Lin, H.; Wang, J. Drug drug interaction extraction from biomedical literature using syntax convolutional neural network. Bioinformatics 2016, 32, 3444–3453. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.K.; Anand, A. Drug-drug interaction extraction from biomedical texts using long short-term memory network. J. Biomed. Inform. 2018, 86, 15–24. [Google Scholar] [CrossRef]
- Williams, R.J.; Zipser, D. A learning algorithm for continually running fully recurrent neural networks. Neural Comput. 1989, 1, 270–280. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, L.; Zhang, H.; Xiao, X. Breast cancer histopathological image classification using convolutional neural networks with small SE-ResNet module. PLoS ONE 2019, 14, e0214587. [Google Scholar] [CrossRef]
- Dai, X.; Chen, Y.; Xiao, B.; Chen, D.; Liu, M.; Yuan, L.; Zhang, L. Dynamic head: Unifying object detection heads with attentions. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 7373–7382. [Google Scholar]
- Behzadi, M.M.; Ilieş, H.T. Real-time topology optimization in 3d via deep transfer learning. Comput.-Aided Des. 2021, 135, 103014. [Google Scholar] [CrossRef]
- Basiri, M.E.; Nemati, S.; Abdar, M.; Cambria, E.; Acharya, U.R. ABCDM: An attention-based bidirectional CNN-RNN deep model for sentiment analysis. Future Gener. Comput. Syst. 2021, 115, 279–294. [Google Scholar] [CrossRef]
- Shen, Y.; He, X.; Gao, J.; Deng, L.; Mesnil, G. Learning semantic representations using convolutional neural networks for web search. In Proceedings of the 23rd International Conference on World Wide Web, Seoul, Republic of Korea, 7–11 April 2014; pp. 373–374. [Google Scholar]
- Yih, W.T.; He, X.; Meek, C. Semantic parsing for single-relation question answering. In Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers), Baltimore, MD, USA, 22–27 June 2014; pp. 643–648. [Google Scholar]
- Liu, S.; Tang, B.; Chen, Q.; Wang, X. Drug-drug interaction extraction via convolutional neural networks. Comput. Math. Methods Med. 2016, 2016, 6918381. [Google Scholar] [CrossRef]
- Asada, M.; Miwa, M.; Sasaki, Y. Extracting Drug-Drug Interactions with Attention CNNs. In BioNLP 2017; Association for Computational Linguistics: Vancouver, BC, Canada, 2017; pp. 9–18. [Google Scholar] [CrossRef]
- Dewi, I.N.; Dong, S.; Hu, J. Drug-drug interaction relation extraction with deep convolutional neural networks. In Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, USA, 13–16 November 2017; IEEE Computer Society: Washington, DC, USA, 2017; pp. 1795–1802. [Google Scholar] [CrossRef]
- Sun, X.; Ma, L.; Du, X.; Feng, J.; Dong, K. Deep convolution neural networks for drug-drug interaction extraction. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (Bibm), Madrid, Spain, 3–6 December 2018; IEEE: New York, NY, USA, 2018; pp. 1662–1668. [Google Scholar] [CrossRef]
- Quan, C.; Hua, L.; Sun, X.; Bai, W. Multichannel convolutional neural network for biological relation extraction. BioMed Res. Int. 2016, 2016, 1850404. [Google Scholar] [CrossRef]
- Sun, X.; Dong, K.; Ma, L.; Sutcliffe, R.; He, F.; Chen, S.; Feng, J. Drug-drug interaction extraction via recurrent hybrid convolutional neural networks with an improved focal loss. Entropy 2019, 21, 37. [Google Scholar] [CrossRef] [PubMed]
- Kavuluru, R.; Rios, A.; Tran, T. Extracting drug-drug interactions with word and character-level recurrent neural networks. In Proceedings of the 2017 IEEE International Conference on Healthcare Informatics (ICHI), Park City, UT, USA, 23–26 August 2017; IEEE: New York, NY, USA, 2017; pp. 5–12. [Google Scholar] [CrossRef]
- Huang, D.; Jiang, Z.; Zou, L.; Li, L. Drug-drug interaction extraction from biomedical literature using support vector machine and long short term memory networks. Inf. Sci. 2017, 415, 100–109. [Google Scholar] [CrossRef]
- Yi, Z.; Li, S.; Yu, J.; Tan, Y.; Wu, Q.; Yuan, H.; Wang, T. Drug-drug interaction extraction via recurrent neural network with multiple attention layers. In Advanced Data Mining and Applications, Proceedings of the 13th International Conference, ADMA 2017, Singapore, 5–6 November 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 554–566. [Google Scholar]
- Zhou, D.; Miao, L.; He, Y. Position-aware deep multi-task learning for drug–drug interaction extraction. Artif. Intell. Med. 2018, 87, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yan, S.; Lu, Z. Transfer learning in biomedical natural language processing: An evaluation of BERT and ELMo on ten benchmarking datasets. arXiv 2019, arXiv:1906.05474. [Google Scholar]
- Datta, T.T.; Shill, P.C.; Al Nazi, Z. BERT-D2: Drug-Drug Interaction Extraction using BERT. In Proceedings of the 2022 International Conference for Advancement in Technology (ICONAT), Goa, India, 21–22 January 2022; IEEE: New York, NY, USA, 2022; pp. 1–6. [Google Scholar]
- Zaikis, D.; Vlahavas, I. TP-DDI: Transformer-based pipeline for the extraction of Drug-Drug Interactions. Artif. Intell. Med. 2021, 119, 102153. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Rasmy, L.; Xiang, Y.; Xie, Z.; Tao, C.; Zhi, D. Med-BERT: Pretrained contextualized embeddings on large-scale structured electronic health records for disease prediction. NPJ Digit. Med. 2021, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mondal, I. Bertchem-ddi: Improved drug-drug interaction prediction from text using chemical structure information. arXiv 2020, arXiv:2012.11599. [Google Scholar]
- Asada, M.; Miwa, M.; Sasaki, Y. Using drug descriptions and molecular structures for drug–drug interaction extraction from literature. Bioinformatics 2021, 37, 1739–1746. [Google Scholar] [CrossRef]
- Dou, M.; Ding, J.; Chen, G.; Duan, J.; Guo, F.; Tang, J. IK-DDI: A novel framework based on instance position embedding and key external text for DDI extraction. Brief. Bioinform. 2023, 24, bbad099. [Google Scholar] [CrossRef]
- Huang, Z.; An, N.; Liu, J.; Ren, F. EMSI-BERT: Asymmetrical Entity-Mask Strategy and Symbol-Insert Structure for Drug–Drug Interaction Extraction Based on BERT. Symmetry 2023, 15, 398. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, L.; Lu, H.; Zhou, A.; Qin, X. Extracting drug-drug interactions from texts with BioBERT and multiple entity-aware attentions. J. Biomed. Inform. 2020, 106, 103451. [Google Scholar] [CrossRef]
- Peters, M.E.; Neumann, M.; Iyyer, M.; Gardner, M.; Clark, C.; Lee, K.; Zettlemoyer, L. Deep Contextualized Word Representations. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers), New Orleans, LA, USA, 1–6 June 2018; pp. 2227–2237. [Google Scholar] [CrossRef]
- McCann, B.; Bradbury, J.; Xiong, C.; Socher, R. Learned in translation: Contextualized word vectors. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar] [CrossRef]
- Radford, A.; Narasimhan, K.; Salimans, T.; Sutskever, I. Improving Language Understanding by Generative Pre-Training. 2018. Available online: https://s3-us-west-2.amazonaws.com/openai-assets/research-covers/language-unsupervised/language_understanding_paper.pdf (accessed on 16 May 2023).
- Hendrickx, I.; Kim, S.N.; Kozareva, Z.; Nakov, P.; Séaghdha, D.O.; Padó, S.; Pennacchiotti, M.; Romano, L.; Szpakowicz, S. Semeval-2010 task 8: Multi-way classification of semantic relations between pairs of nominals. arXiv 2019, arXiv:1911.10422. [Google Scholar]
- Nogueira dos Santos, C.; Xiang, B.; Zhou, B. Classifying relations by ranking with convolutional neural networks. arXiv 2015, arXiv:1504.06580. [Google Scholar]
- Lee, J.; Seo, S.; Choi, Y.S. Semantic relation classification via bidirectional lstm networks with entity-aware attention using latent entity typing. Symmetry 2019, 11, 785. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Zhang, J.; Lopyrev, K.; Liang, P. Squad: 100,000+ questions for machine comprehension of text. arXiv 2016, arXiv:1606.05250. [Google Scholar]
- Wu, S.; He, Y. Enriching pre-trained language model with entity information for relation classification. In Proceedings of the 28th ACM International Conference on Information and Knowledge Management, Beijing, China, 3–7 November 2019; pp. 2361–2364. [Google Scholar]
- Wu, Y.; Yuan, M.; Dong, S.; Lin, L.; Liu, Y. Remaining useful life estimation of engineered systems using vanilla LSTM neural networks. Neurocomputing 2018, 275, 167–179. [Google Scholar] [CrossRef]
- Zhou, P.; Qi, Z.; Zheng, S.; Xu, J.; Bao, H.; Xu, B. Text classification improved by integrating bidirectional LSTM with two-dimensional max pooling. arXiv 2016, arXiv:1611.06639. [Google Scholar]
- Herrero-Zazo, M.; Segura-Bedmar, I.; Martínez, P.; Declerck, T. The DDI corpus: An annotated corpus with pharmacological substances and drug–drug interactions. J. Biomed. Inform. 2013, 46, 914–920. [Google Scholar] [CrossRef]
- Demner-Fushman, D.; Fung, K.W.; Do, P.; Boyce, R.D.; Goodwin, T.R. Overview of the TAC 2018 Drug-Drug Interaction Extraction from Drug Labels Track. In Proceedings of the TAC, Gaithersburg, MD, USA, 13–14 November 2018. [Google Scholar]
- Goodwin, T.R.; Demner-Fushman, D.; Fung, K.W.; Do, P. Overview of the TAC 2019 Track on Drug-Drug Interaction Extraction from Drug Labels. In Proceedings of the TAC, Gaithersburg, MD, USA, 12–13 November 2019. [Google Scholar]
- Segura-Bedmar, I.; Martínez Fernández, P.; Sánchez Cisneros, D. The 1st DDIExtraction-2011 Challenge Task: Extraction of Drug-Drug Interactions from Biomedical Texts. In Proceedings of the 1st Challenge Task on Drug-Drug Interaction Extraction, Huelva, Spain, 11 September 2011. [Google Scholar]
- Rai, N.; Kumar, D.; Kaushik, N.; Raj, C.; Ali, A. Fake News Classification using transformer based enhanced LSTM and BERT. Int. J. Cogn. Comput. Eng. 2022, 3, 98–105. [Google Scholar] [CrossRef]
- Deepak, S.; Chitturi, B. Deep neural approach to Fake-News identification. Procedia Comput. Sci. 2020, 167, 2236–2243. [Google Scholar]
- Nguyen, D.P.; Ho, T.B. Drug-drug interaction extraction from biomedical texts via relation BERT. In Proceedings of the 2020 RIVF International Conference on Computing and Communication Technologies (RIVF), Ho Chi Minh City, Vietnam, 14–15 October 2020; IEEE: New York, NY, USA, 2020; pp. 1–7. [Google Scholar] [CrossRef]
- Tang, S.; Zhang, Q.; Zheng, T.; Zhou, M.; Chen, Z.; Shen, L.; Ren, X.; Zhuang, Y.; Pu, S.; Wu, F. Two step joint model for drug drug interaction extraction. arXiv 2020, arXiv:2008.12704. [Google Scholar]
- Tran, T.; Kavuluru, R.; Kilicoglu, H. A multi-task learning framework for extracting drugs and their interactions from drug labels. arXiv 2019, arXiv:1905.07464. [Google Scholar]
- Mahajan, D.; Poddar, A.; Lin, Y.T. A hybrid model for drug-drug interaction extraction from structured product labeling documents. In Proceedings of the TAC, Gaithersburg, MD, USA, 12–13 November 2019. [Google Scholar]
| Train | Test | |||
|---|---|---|---|---|
| DDIs | TAC 2018 | TAC 2019 | TAC 2018 | TAC 2019 |
| Pharmacodynamic (PD) | 47 | 553 | 335 | 292 |
| Pharmacokinetic (PK) | 60 | 494 | 296 | 118 |
| Unspecified | 62 | 665 | 440 | 202 |
| Training | Test | |||
|---|---|---|---|---|
| DDI Samples | Original | Filtered | Original | Filtered |
| Positive | 4020 | 3840 | 979 | 971 |
| Negative | 23,772 | 8989 | 4782 | 2084 |
| Total | 27,792 | 12,829 | 5761 | 3055 |
| Ratio | 1:5.9 | 1:2.3 | 1:4.9 | 1:2.2 |
| Batch size | 8 |
| Max sentence length | 400 |
| Adam learning rate | 2 × 10−5 |
| Number of epochs | 10 |
| Dropout rate | 0.1 |
| F1-Score (F) | Overall Performance | ||||||
|---|---|---|---|---|---|---|---|
| Model | Negative | Mechanism | Effect | Advice | Int | F1-Score | F1-Macro |
| Joint AB-Lstm [20] | - | 72.26 | 65.46 | 80.26 | 44.11 | 69.39 | 65.52 |
| MCCNN [33] | - | 72.2 | 68.2 | 78.0 | 51.0 | 70.21 | - |
| RHCNN [34] | - | 78.3 | 73.5 | 80.5 | 58.9 | 75.5 | - |
| BioBERT [48] | - | 84.6 | 80.1 | 86 | 56.6 | 80.09 (micro-averaged) | - |
| BERT-D2 [40] | - | - | - | - | - | 81.97 | - |
| EMSI-BERT [47] | - | 86.6 | 80.07 | 86.8 | 56 | 82 (micro-averaged) | - |
| TP-DDI [41] | - | - | - | - | - | 82.4 | - |
| BERTChem [44] | 87 | 80 | 88 | 58 | 83 | - | |
| IK-DDI [46] | - | - | - | - | - | - | 79.04 |
| R-BioBERT (Baseline) [65] | - | 97.42 | 77.80 | 87.32 | 57.31 | - | 80.89 |
| R-BioBERT with BLSTM (Our method) | 95.70 | 86.47 | 82.5 | 90.79 | 61.12 | 91.79 (weighted) | 83.32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
KafiKang, M.; Hendawi, A. Drug-Drug Interaction Extraction from Biomedical Text Using Relation BioBERT with BLSTM. Mach. Learn. Knowl. Extr. 2023, 5, 669-683. https://doi.org/10.3390/make5020036
KafiKang M, Hendawi A. Drug-Drug Interaction Extraction from Biomedical Text Using Relation BioBERT with BLSTM. Machine Learning and Knowledge Extraction. 2023; 5(2):669-683. https://doi.org/10.3390/make5020036
Chicago/Turabian StyleKafiKang, Maryam, and Abdeltawab Hendawi. 2023. "Drug-Drug Interaction Extraction from Biomedical Text Using Relation BioBERT with BLSTM" Machine Learning and Knowledge Extraction 5, no. 2: 669-683. https://doi.org/10.3390/make5020036
APA StyleKafiKang, M., & Hendawi, A. (2023). Drug-Drug Interaction Extraction from Biomedical Text Using Relation BioBERT with BLSTM. Machine Learning and Knowledge Extraction, 5(2), 669-683. https://doi.org/10.3390/make5020036







