Abstract
Cancer is the second leading cause of death in the world and is often untreatable. Protein-based therapeutics, such as immunotherapeutics, show promising results in the fight against cancer, resulting in their market share increasing every year. Unfortunately, most protein-based therapeutics suffer from fast degradation in the blood, making effective treatment expensive, causing more off-target effects (due to the high doses necessary), and often require repeated injections to stay within the correct therapeutic range. Encapsulation of these proteins inside nanocarriers is prompted to overcome these problems by enhancing targeted drug delivery and, thus, leading to a less frequent administration and lower required dose. However, most current protein encapsulation methods show very low loading capacities (LC). This leads to even more expensive treatments and might pose a further risk for the patient caused by systemic toxicity against high concentrations of the carrier material. We investigated and optimized protein nanoprecipitation as a method to obtain a high protein LC and encapsulation efficiency (EE) inside poly(lactic-co-glycolic acid; PLGA) nanoparticles via a simple two-step process. In this work, we used model proteins to investigate the influence of various parameters such as precipitation solvent, addition speed, and protein concentration on protein activity. Our work is a critical step towards the high-loading encapsulation of immunotherapeutics.
1. Introduction
Cancer is the second leading cause of death in the world and is often untreatable [1,2]. However, protein therapeutics have revolutionized the oncology field, showing promising results in the fight against cancer [3]. Unfortunately, while performing well in vitro, protein-based active pharmaceutical ingredients (APIs) are often rapidly cleared from the blood [4] and are less effective in vivo. To overcome these problems, high doses are administered in short intervals, thus increasing the costs and invasiveness of the procedure. Encapsulation of these protein-based APIs inside nanoparticles could prevent protein degradation, promote passive or active targeting and reduce side effects [4,5]. However, the nanoparticles should remain below 200 nm in size in order to prevent quick opsonization by the reticuloendothelial system, prevent particle filtration by the spleen, and still make use of the enhanced permeability and retention effect [6,7]. Moreover, most current protein encapsulation methods show very low loading capacities (LC) or remaining protein activity after encapsulation. In this paper, protein nanoprecipitation was investigated as a method for encapsulation of high concentrations of protein inside poly(lactic-co-glycolic acid; PLGA) nanoparticles.
Nanoprecipitation, also known as desolvation, was first described by Fessi et al. [8] in 1989 and involves two miscible solvents, one of which is a good solvent for the polymer and API (e.g., acetone), while the other is a bad solvent (e.g., water). When this good solvent containing polymer and API is added to a large volume of the bad solvent, API encapsulated nanoparticles are formed (for a detailed review of the mechanism, see [9]). This method works well for hydrophobic drugs that can be dissolved together with the polymer. However, most proteins are not directly soluble or lose their activity in these organic solvents, making this original method unsuitable for protein encapsulation. Recently, Morales-Cruz et al. [10] introduced a modified, two-step nanoprecipitation method for encapsulation of proteins inside PLGA nanoparticles. In this case, the protein was precipitated into protein nanoparticles, mixed with PLGA, and then precipitated, creating protein-loaded PLGA nanoparticles. However, this method only encapsulated smaller proteins (12–21 kDa), and it remains uncertain if this method can be used for encapsulation of larger immunotherapeutics (~150 kDa). In this work, two model proteins, bovine serum albumin (BSA; 66.5 kDa) and amylase (51–54 kDa), which are two times larger than previously reported, were precipitated by the non-solvent (NS) acetonitrile (ACN) and encapsulated inside PLGA nanoparticles. We tested various parameters that may affect the protein and polymer precipitation and demonstrated that the activity of amylase was preserved during the process.
2. Materials and Methods
2.1. Chemicals
Poly(lactic-co-glycolic acid), PLGA 5002 (50:50), and PLGA 5002A (50:50) were kindly provided as a gift by Corbion (Amsterdam, The Netherlands). All solvents were purchased from VWR (Radnor, United States). Bovine serum albumin (BSA; K35-011) was purchased from PAA laboratories(Pasching, Austria). Amylase (10044725) was purchased from MP biomedicals (Irvine, CA, USA).
2.2. Protein Precipitation
The protocol was adapted from [10] with few modifications. 200 µL of protein of various concentrations, amylase, or BSA was precipitated by dropwise (0.5 mL/min) addition of NS (ACN, ethanol, acetone) at various NS:water ratios while stirring (500 rpm). Used concentrations, solvents, and ratios will be further indicated in the text. After addition, the samples were incubated for 5 min at room temperature, and their size and distribution were characterized using DLS (NanoPhox, Sympatec, Clausthal-Zellerfeld, Germany).
2.3. PLGA Nanoprecipitation
The protocol was adapted from [10] with few modifications. 100 µL of PLGA (5002 or 5002A) in ACN:water (5.5:1 ratio) at various concentrations (3.3, 6.8, 10.2, 13.4 mg/mL, final concentration) was slowly added to the precipitated protein mixture (described above) under stirring (500 rpm). Once PLGA was mixed, the samples were immediately precipitated. The precipitation was done by dropwise addition at 0.1 mL/min of 1 mL of PLGA/protein mixture to 9, 19, or 39 mL of water containing 10 mg/mL of F127 or F68 under stirring (700 rpm). Blank nanoparticles without protein were also created. The PLGA concentrations and water volume are further indicated in the text.
2.4. PLGA Solubility
The solubility of PLGA (5002, 5002A) in ACN/water mixtures was tested by dropwise addition of 10 µL of water to 19 mg of PLGA in ACN for various concentrations, till visible precipitation was observed. The maximum water percentage was recorded as the last addition before precipitation.
2.5. Dynamic Light Scattering
The protein and PLGA nanoparticles were analyzed by dynamic light scattering (DLS; NanoPhox, Sympatec, Clausthal-Zellerfeld, Germany) at 25 °C. Protein nanoparticles were measured directly as a suspension in their NS:water mixture, and PLGA particles were measured as a suspension in water with surfactant. Samples were diluted if the observed kilocounts per second (KCPS) succeeded 500 KCPS. Further data analysis was performed using Python 3.0.
2.6. Encapsulation Efficiency and Protein Activity
The encapsulation efficiency (EE) was measured indirectly by spinning down the protein particles for 20 min at 22,000 RCF (Centurion Scientific Benchtop Centrifuge, Stoughton, United Kingdom) and measuring the protein concentration in the supernatant, following standard microBCA assay (ThermoFisher, Waltham, United States). Absorbance was measured at 562 nm (microplate reader, SpectraMax, Molecular Devices, San Jose, United States). The EE was calculated using the following equation:
For activity measurements, 2 mg/mL of amylase was precipitated as described above. After precipitation, 1 mL of precipitated protein was directly added to 14 mL of PBS. The protein was then incubated at 4 °C for at least 2 h to let the protein fully redissolve. 2 mg/mL stock amylase was diluted (100×) in PBS, and the activity of both samples was measured following the standard amylase activity colorimetric assay kit (BioVision, K711, Milpitas, United States) protocol at 405 nm (microplate reader, SpectraMax). The measured activity of redissolved protein was then normalized against the activity of stock amylase.
3. Results and Discussion
Protein nanoencapsulation by nanoprecipitation required two steps: first, nanoprecipitation of the protein; second, nanoencapsulation of this precipitated protein by PLGA during a second nanoprecipitation step. In order to preserve the activity of the protein throughout the steps and obtain nanoparticles below 200 nm with high EE, each step had to be carefully optimized. In the following sections, we discuss factors affecting each process: protein nanoprecipitation, PLGA nanoprecipitation, and encapsulation.
3.1. Protein Precipitation
Protein precipitation was the first step that may lead to the loss of protein activity. In order to get a better understanding of protein nanoprecipitation, three different parameters were investigated: NS, addition speed, and initial protein concentration.
The NS in protein nanoprecipitation could have a big influence on protein particle size, polydispersity index (pdi), and stability. Ethanol was most commonly used as an NS in protein nanoprecipitation of BSA [11,12]. However, PLGA was needed in the second precipitation step and was only soluble in ACN or acetone. When comparing ACN and acetone for protein precipitation of lysozyme and alpha-chemotrypsin, Morales-Cruz et al. [10] observed that ACN induced the least amount of non-soluble aggregates and retained the highest protein activity. To find the best precipitation conditions for proteins of interest, we tested three water-miscible solvents for BSA precipitation: acetone, ACN, and ethanol.
As shown in Figure 1A, only precipitation with ACN led to a homogeneous distribution of BSA nanoparticles below 200 nm. Furthermore, ACN led to the highest particle count on DLS (+500 KCPS), indicating a large number of protein particles. According to Von Storp et al. [13], the higher the dielectric constant of the non-solvent (ACN > ethanol > acetone), the smaller the formed protein particles should be. However, due to the broad distribution of BSA particles after ethanol and acetone precipitation, these findings could not be compared. Furthermore, even though ACN was not often used for protein precipitation in the literature, it showed a homogeneous distribution and high concentration of protein particles, making it a good candidate for two-step precipitation. ACN was therefore selected as a solvent for all further experiments.
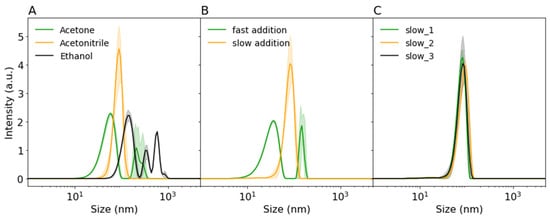
Figure 1.
Dynamic light scattering (DLS) intensity distributions of (A) precipitated 12.5 mg/mL bovine serum albumin (BSA) by various non-solvents (1:4 water:acetonitirile (ACN)), (B) the influence of addition speed of ACN (fast = instant pipetting, slow = 0.5 mL/min) on nanoprecipitation of 25 mg/mL BSA, (C) repeatability of precipitation of 25 mg/mL BSA by slow addition (0.5 mL/min) of ACN. Shaded areas indicate standard deviation.
One of the possible explanations of the broad particle size distributions observed in Figure 1A is a too fast addition speed. Paik et al. [12] also observed larger BSA protein particles at fast addition of ethanol and showed that slow controlled addition to BSA led to robust size control. They suggested that slower addition stabilized particle formation by allowing for more equilibrium time. As shown in Figure 1B, instant addition of ACN to BSA solution led to smaller particles, but the resulting particle size distribution was larger. Furthermore, some larger aggregates were observed in the same solution (~200 nm). On the other hand, slow and controlled addition (controlled by a syringe pump) of ACN led to repeatable, larger, and more homogeneously distributed particles (see Figure 1C). Therefore, slow addition of ACN was used to further investigate the effect of initial BSA concentration on protein particle size and distribution.
Another factor that may influence the size of nanoprecipitated proteins was their concentration [11,14]. Only minor differences in protein size were found by Tarhini et al., with the lowest concentration leading to the smallest nanoparticles. Rahimnejad et al. [14] showed that, initially, the protein particle size decreased with increasing concentration, but beyond a certain point, the particle size remained the same. Here, BSA was precipitated in a range between 10 and 25 mg/mL at 5.5:1 ACN:water ratio. As shown in Figure 2A and Table 1, in our case, the size of the BSA protein particles increased with increasing protein concentration, from 35.2 ± 0.2 nm to 67.0 ± 4.1 nm for 10 mg/mL and 25 mg/mL, respectively. The increase appeared to be linear within this given range.
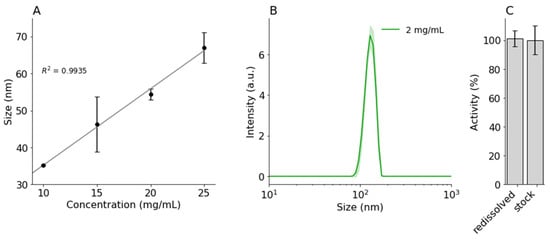
Figure 2.
DLS intensity distributions of (A) nanoprecipitated BSA (10, 15, 20, 25 mg/mL) and (B) amylase (2 mg/mL) with ACN (5.5:1; 0.1 mL/min). (C) Normalized activity of redissolved nanoprecipitated amylase in PBS, compared to amylase stock. Error bars and shaded areas indicate standard deviation.

Table 1.
Overview of particle size (z-average) and polydispersity index (pdi) of acetonitrile precipitated BSA and amylase at various concentrations.
As a model protein for activity measurement, amylase was precipitated. Due to lower solubility, a lower initial concentration (2 mg/mL) of amylase was used. Interestingly, the particle size of nanoprecipitated amylase was a lot larger than for BSA, 133.7 ± 4.7 nm (see Figure 2B and Table 1). This suggested that the precipitation process may be influenced by certain unique protein properties (hydrophobicity, molecular weight, or charge), and optimized precipitation parameters for one protein might not lead to similar results for others. Therefore, the initial protein concentration might be used to alter the protein particle size of specific proteins but is not a universal method to control particle size.
There are many publications where precipitated and cross-linked protein nanoparticles were used for encapsulation of other APIs [11,13,15,16,17]. However, in order to use this method for protein encapsulation of protein immunotherapeutics, the proteins should be able to redissolve in an aqueous solution and retain their activity. Morales-Cruz et al. [10] showed that lysozyme and alpha-chemotrypsin remained active after protein nanoprecipitation using ACN. To evaluate the influence of the nanoprecipitation on the activity of large proteins, amylase nanoparticles were redissolved in PBS, and their activity was quantified. As seen in Figure 2C, redissolved amylase retained 101.1 ± 5.4% of the activity compared to the stock solution. This seems to suggest that nanoprecipitation of amylase did not lead to irreversible denaturation.
In conclusion, our results suggested that the first protein nanoprecipitation step was not detrimental to the protein structure and might be applicable for a variety of proteins.
3.2. PLGA Precipitation
The second step of protein nanoencapsulation was the encapsulation of the obtained protein nanoparticle inside PLGA. For that, PLGA should be dissolved into the protein suspension, and the whole solution precipitated out in an NS (water) for PLGA. One of the most critical parameters affecting this nanoprecipitation step was the polymer solubility in the solvent mixture. That is, if PLGA already precipitates out of solution upon addition to the protein mixture, the protein will not be encapsulated when the PLGA/protein mixture is precipitated in water. In our preliminary experiments, the proteins were precipitated by ACN addition to a final 4:1 ACN:water ratio since this ratio led to sufficient protein precipitation. However, the addition of PLGA in any concentration to this 4:1 ACN:water ratio led to immediate nanoprecipitation of PLGA. The water concentration was too high to keep the PLGA dissolved. As shown in Table 2, PLGA nanoprecipitated out of ACN when it contained a water concentration between 16 and 18%. Therefore, the initial 4:1 ratio was raised to 5.5:1 and used to investigate other factors that may influence PLGA nanoprecipitation.

Table 2.
Highest acceptable water percentage in acetonitrile/water mixtures before poly(lactic-co-glycolic acid; PLGA) precipitates out of solution. Increments of 10 µL of water were added to 19 mg of PLGA (5002, 5002A) in acetonitrile at various concentrations till visible precipitation occurred.
The ratio of NS:water used for protein precipitation in our experiments was higher than previously reported. To validate the published observations on PLGA nanoprecipitation in our setup, the effects of PLGA concentration, surfactants, and organic:aqueous phase ratio were investigated without the protein particle suspension. The surfactants in the water phase were supposed to stabilize the particles during the nucleation and growth process and were necessary for obtaining stable sub 200 nm particles [9]. In agreement with Lebouille et al., the PLGA particles aggregated into bigger microparticles upon the addition to water if surfactants were not used (see Figure 3A). Another factor that may affect PLGA precipitation was the ratio of the organic to aqueous phase. The organic phase needed to be precipitated in an excess of water; however, larger excess would lead to a lower final concentration of particles, potentially requiring large volumes or up-concentration to be effective in vivo. In our case, a decrease in water phase ratio from 1:39 to 1:9 did not notably affect the particle size or size distribution (see Figure 3B). Therefore, in our setup a 1:9 ratio already provided enough excess volume for successful precipitation leading to higher particle concentrations. Finally, we also tested the influence of PLGA concentration on particle size. As shown in Figure 3C, PLGA particle size decreased from 181.3 ± 0.34 nm to 80.5 ± 0.75 nm with a decreasing PLGA concentration from 13.4 mg/mL to 3.3 mg/mL, respectively. Our results suggested that the final particle size could be controlled by adjusting the PLGA concentration. However, the addition of protein particles inside this PLGA mixture might influence the precipitation outcome. Amylase encapsulation inside PLGA is discussed in the following section.
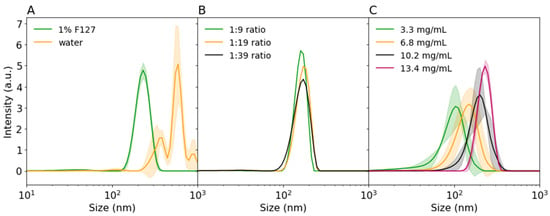
Figure 3.
DLS intensity distributions of (A) precipitated 5002A PLGA in water of 1% F127 (1:9 ratio), (B) precipitated 5002 PLGA in different 1% F127 ACN:water ratios, (C) precipitated 5002A PLGA at various PLGA concentrations in 1% F127 (1:9 ratio). Shaded areas indicate standard deviation.
3.3. Encapuslation
Finally, for practical applications, therapeutical proteins should be encapsulated for drug delivery and on-target release. As a proof-of-concept, amylase was encapsulated inside PLGA nanoparticles using the optimized conditions discussed in the previous sections. First, amylase was precipitated with ACN to form protein nanoparticles (133.7 ± 4.2 nm). Then 10 mg/mL PLGA (final concentration) was added to the suspension, and the whole solution was precipitated in 1% F68 solution. As shown in Figure 4A, 147.7 ± 4.3 nm nanoparticles were formed. In this case, only a small (~14 nm) increase in particle size was detected, suggesting that only a small polymer shell was formed. This may be beneficial for a faster protein release. On the other hand, as seen in Figure 4B, only a small amount, 24 ± 11.2%, of amylase was encapsulated or adsorbed onto the PLGA nanoparticles.
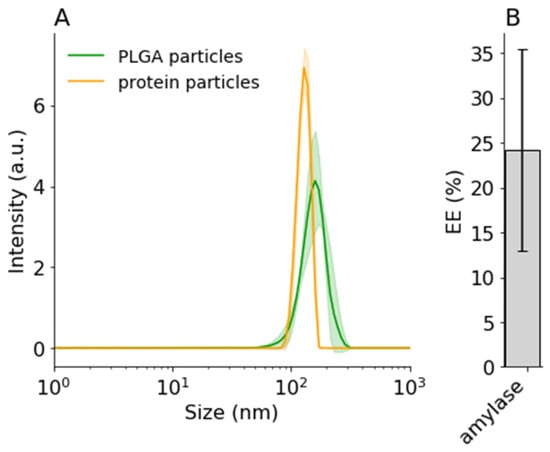
Figure 4.
DLS intensity distribution of (A) precipitated amylase (5.5:1 ACN:water) and encapsulated amylase (5002A, 1:29, 1% F68). (B) Encapsulation efficiency of amylase. Error bars and shaded areas indicate standard deviation.
Morales et al. also observed a low EE (~30%) of alpha-chemotrypsin [10]. They improved the EE by lowering the initial protein concentration, i.e., effectively decreasing the size of the protein nanoparticles. This effect of the initial protein particle size on the EE will be investigated in our further experiments. Moreover, the influence of the PLGA polymer shell on the release properties of such nanoparticles will also be studied.
4. Conclusions
In this work, we studied the influence of various process parameters on nanoprecipitation and encapsulation of proteins. We investigated the influence of several solvents and the addition speed on nanoprecipitation of proteins and showed that slow addition (0.5 mL/min) of acetonitrile was a reproducible method to obtain small (<200 nm) protein nanoparticles. We also found the optimum non-solvent:water ratio, 5.5:1, needed for an effective two-step nanoprecipitation with PLGA. Finally, we successfully precipitated amylase without losing its activity and encapsulated amylase nanoparticles in PLGA. Overall, our work validated the feasibility of larger protein precipitation, which is an important step toward nanoprecipitation of immunotherapeutics.
Author Contributions
L.C.N. conceived, designed, and performed the experiments; L.C.N. analyzed the data; L.C.N. and A.S. wrote the paper; M.B. and A.S. supervised the work and contributed insights and discussions to the theory and manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This project received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 813871.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Kintzing, J.R.; Filsinger Interrante, M.V.; Cochran, J.R. Emerging Strategies for Developing Next-Generation Protein Therapeutics for Cancer Treatment. Trends Pharmacol. Sci. 2016, 37, 993–1008. [Google Scholar] [CrossRef]
- Kontermann, R.E. Strategies for extended serum half-life of protein therapeutics. Curr. Opin. Biotechnol. 2011, 22, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Kalyane, D.; Raval, N.; Maheshwari, R.; Tambe, V.; Kalia, K.; Tekade, R.K. Employment of enhanced permeability and retention effect (EPR): Nanoparticle-based precision tools for targeting of therapeutic and diagnostic agent in cancer. Mater. Sci. Eng. C 2019, 98, 1252–1276. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Li, Y. Physicochemical characteristics of nanoparticles affect circulation, biodistribution, cellular internalization, and trafficking. Small 2013, 9, 1521–1532. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.P.M.; Chen, A.L.; Foster, A.; Drezek, R. In vivo biodistribution of nanoparticles. Nanomedicine 2011, 6, 815–835. [Google Scholar] [CrossRef] [PubMed]
- Fessi, H.; Puisieux, F.; Devissaguet, J.P.; Ammoury, N.; Benita, S. Nanocapsule formation by interfacial polymer deposition following solvent displacement. Int. J. Pharm. 1989, 55, R1–R4. [Google Scholar] [CrossRef]
- Lebouille, J.G.J.L.; Stepanyan, R.; Slot, J.J.M.; Cohen Stuart, M.A.; Tuinier, R. Nanoprecipitation of polymers in a bad solvent. Colloids Surf. A Physicochem. Eng. Asp. 2013, 460, 225–235. [Google Scholar] [CrossRef]
- Morales-Cruz, M.; Flores-Fernández, G.M.; Morales-Cruz, M.; Orellano, E.A.; Rodriguez-Martinez, J.A.; Ruiz, M.; Griebenow, K. Two-step nanoprecipitation for the production of protein-loaded PLGA nanospheres. Results Pharma Sci. 2012, 2, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Tarhini, M.; Benlyamani, I.; Hamdani, S.; Agusti, G.; Fessi, H.; Greige-Gerges, H.; Bentaher, A.; Elaissari, A. Protein-based nanoparticle preparation via nanoprecipitation method. Materials 2018, 11, 394. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.Y.R.; Nguyen, H.H.; Ryu, J.; Che, J.H.; Kang, T.S.; Lee, J.K.; Song, C.W.; Ko, S. Robust size control of bovine serum albumin (BSA) nanoparticles by intermittent addition of a desolvating agent and the particle formation mechanism. Food Chem. 2013, 141, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Von Storp, B.; Engel, A.; Boeker, A.; Ploeger, M.; Langer, K. Albumin nanoparticles with predictable size by desolvation procedure. J. Microencapsul. 2012, 29, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Rahimnejad, M.; Najafpour, G.; Bakeri, G. Investigation and modeling effective parameters influencing the size of BSA protein nanoparticles as colloidal carrier. Colloids Surf. A Physicochem. Eng. Asp. 2012, 412, 96–100. [Google Scholar] [CrossRef]
- Weber, C.; Coester, C.; Kreuter, J.; Langer, K. Desolvation process and surface characterisation of protein nanoparticles. Int. J. Pharm. 2000, 194, 91–102. [Google Scholar] [CrossRef]
- Langer, K.; Anhorn, M.G.; Steinhauser, I.; Dreis, S.; Celebi, D.; Schrickel, N.; Faust, S.; Vogel, V. Human serum albumin (HSA) nanoparticles: Reproducibility of preparation process and kinetics of enzymatic degradation. Int. J. Pharm. 2008, 347, 109–117. [Google Scholar] [CrossRef]
- Langer, K.; Balthasar, S.; Vogel, V.; Dinauer, N.; Von Briesen, H.; Schubert, D. Optimization of the preparation process for human serum albumin (HSA) nanoparticles. Int. J. Pharm. 2003, 257, 169–180. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).