Pitfalls of Accurate Protein Determination inside PLGA Nanoparticles Using the Micro BCA Assay †
Abstract
:1. Introduction
2. Experiments
2.1. Materials
2.2. Methods
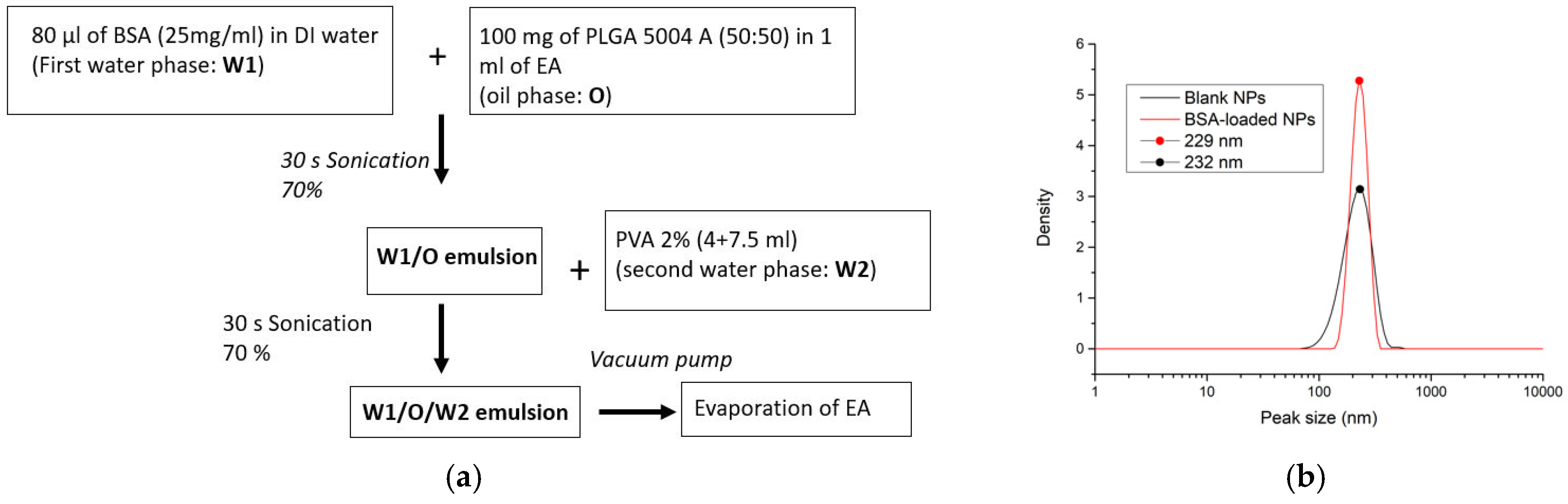
2.2.1. Preparation of PLGA NPs
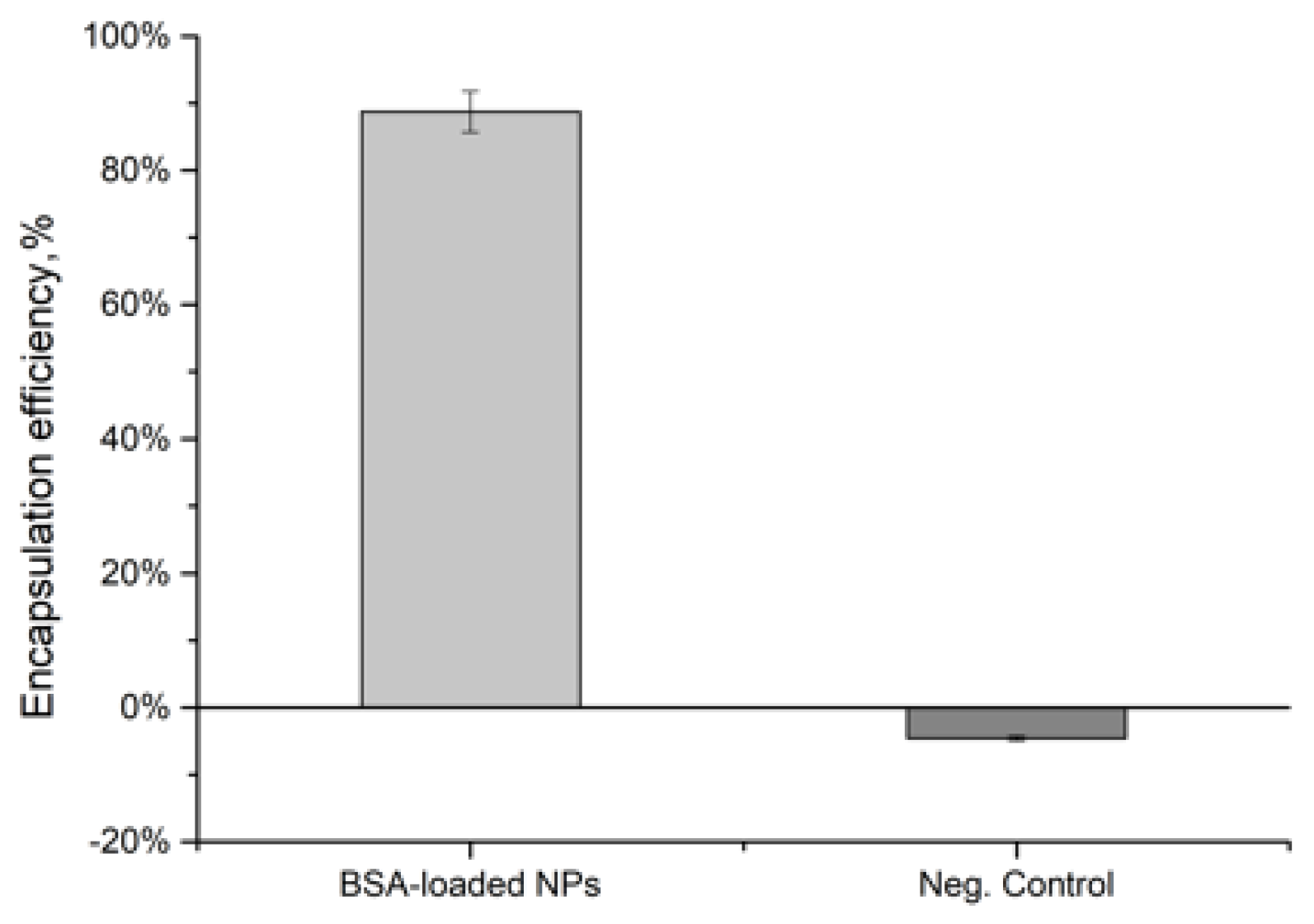
2.2.2. Indirect Encapsulation Efficiency (IEE)
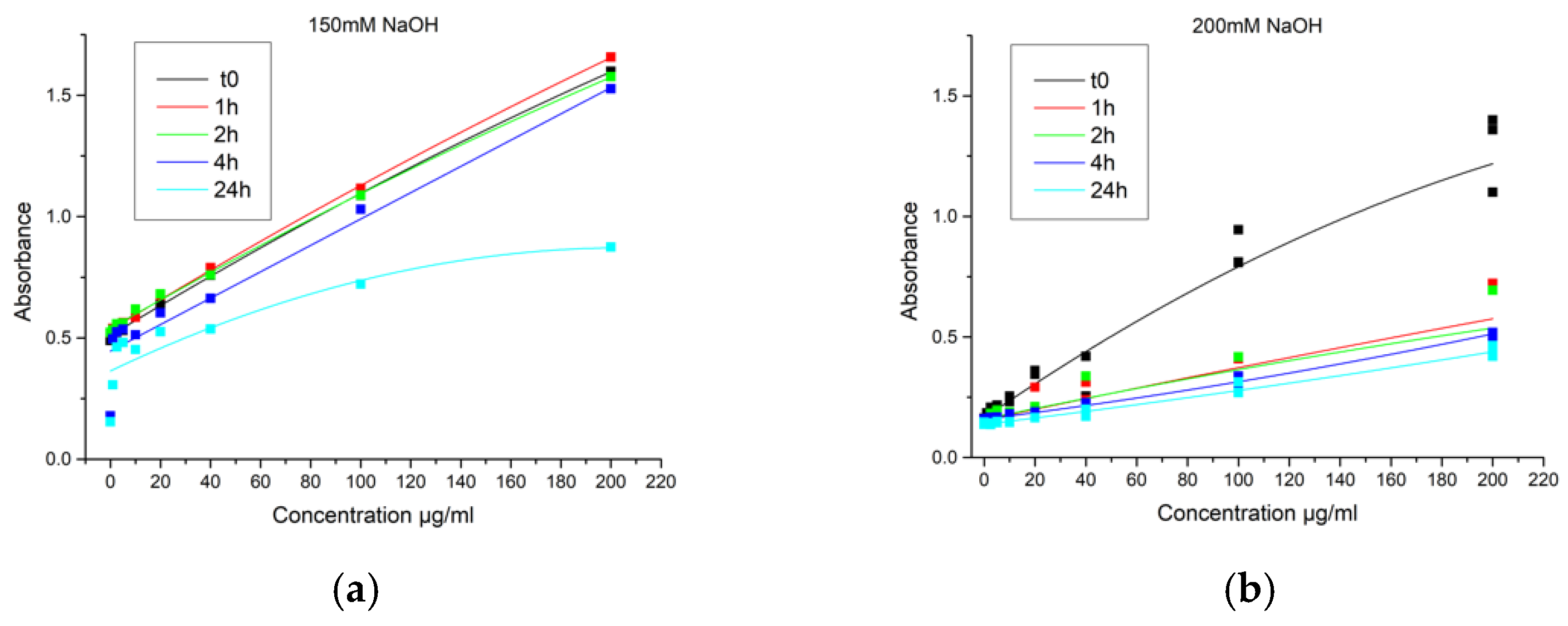
2.2.3. Direct Encapsulation Efficiency (DEE)
2.2.4. Characterization
3. Results
3.1. Indirect Encapsulation Efficiency
3.2. Direct Encapsulation Efficiency
3.2.1. DMSO Extraction
3.2.2. NaOH Extraction
4. Discussion
5. Conclusions
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
 .
.Conflicts of Interest
Abbreviations
References
- Neuse, E.W. Synthetic polymers as drug-delivery vehicles in medicine. Met.-Based. Drugs 2008, 2008, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, A.M.; Saez, V.; Cobley, A.; Graves, J.; Sukhorukov, G.B.; Mason, T.J. Controlled protein release from microcapsules with composite shells using high frequency ultrasound—Potential for in vivo medical use. Soft Matter 2011, 7, 4341–4347. [Google Scholar] [CrossRef]
- Hoshyar, N.; Gray, S.; Han, H.; Bao, G. The effect of nanoparticle size on in vivo pharmacokinetics and cellular interaction. Nanomedicine 2016, 11, 673–692. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Joshi, A.; Toor, A.P.; Verma, G. Drug delivery: Advancements and challenges. In Nanostructures for Drug Delivery; Elsevier: Amsterdam, The Netherlands, 2017; pp. 865–886. [Google Scholar] [CrossRef]
- Sah, H. A new strategy to determine the actual protein content of poly(lactide- co-glycolide) microspheres. J. Pharm. Sci. 1997, 86, 1315–1318. [Google Scholar] [CrossRef] [PubMed]
- Gornall, A.G.; Bardawill, C.J.; David, M.M. Determination of serum proteins by means of the biuret reaction. J. Biol. Chem. 1949, 177, 751–766. [Google Scholar] [CrossRef]
- Kresge, N.; Simoni, R.D.; Hill, R.L. The Most Highly Cited Paper in Publishing History: Protein Determination by Oliver H. Lowry. J. Biol. Chem. 2005, 280, e26–e28. [Google Scholar] [CrossRef]
- Smith, P.; Krohn, R.; Hermanson, G.; Mallia, A.; Gartner, F.; Provenzano, M.; Fujimoto, E.; Goeke, N.; Olson, B.; Klenk, D. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Bradford, M.M. Determinación de proteínas: Método de bradford. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- He, F. BCA (Bicinchoninic Acid) Protein Assay. Bio-Protocol 2011. [Google Scholar] [CrossRef]
- Amini, Y.; Jamehdar, S.A.; Sadri, K.; Zare, S.; Musavi, D.; Tafaghodi, M. Different methods to determine the encapsulation efficiency of protein in PLGA nanoparticles. Biomed. Mater. Eng. 2017, 28, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Rosas, J.; Bayat, A.; Gascón, A.; Igartua, M.; Guzman, F.; Patarroyo, M.A.; De Castro, M. Biodegradable PLGA microspheres as a delivery system for malaria synthetic peptide SPf66. Vaccine 2001, 19, 4445–4451. [Google Scholar] [CrossRef]
- Zheng, C.H.; Liang, W.Q.; Yu, H.Y.; Chen, H.L. Evaluation of different methods to determine the loading of proteins in PLGA microspheres. Pharmazie 2004, 59, 232–233. [Google Scholar] [PubMed]
- Gref, R.; Quellec, P.; Sanchez, A.; Calvo, P.; Dellacherie, E.; Alonso, M.J. Development and characterization of CyA-loaded poly(lactic acid)-poly(ethylene glycol)PEG micro- and nanoparticles. Comparison with conventional PLA particulate carriers. Eur. J. Pharm. Biopharm. 2001, 51, 111–118. [Google Scholar] [CrossRef]
- Fonte, P.; Soares, S.; Sousa, F.; Costa, A.; Seabra, V.; Reis, S.; Sarmento, B. Stability study perspective of the effect of freeze-drying using cryoprotectants on the structure of insulin loaded into PLGA nanoparticles. Biomacromolecules 2014, 15, 3753–3765. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Y.; Chung, T.S.; Ng, N.P. Morphology, drug distribution, and in vitro release profiles of biodegradable polymeric microspheres containing protein fabricated by double-emulsion solvent extraction/evaporation method. Biomaterials 2001, 22, 231–241. [Google Scholar] [CrossRef]
- Castellanos, I.J.; Cruz, G.; Crespo, R.; Griebenow, K. Encapsulation-induced aggregation and loss in activity of γ-chymotrypsin and their prevention. J. Control. Release 2002, 81, 307–319. [Google Scholar] [CrossRef]
- Diwan, M.; Park, T.G. Pegylation enhances protein stability during encapsulation in PLGA microspheres. J. Control. Release 2001, 73, 233–244. [Google Scholar] [CrossRef]
- Lam, X.M.; Duenas, E.T.; Daugherty, A.L.; Levin, N.; Cleland, J.L. Sustained release of recombinant human insulin-like growth factor-I for treatment of diabetes. J. Control. Release 2000, 67, 281–292. [Google Scholar] [CrossRef]
- Sousa, F.; Cruz, A.; Fonte, P.; Pinto, I.M.; Neves-Petersen, M.T.; Sarmento, B. A new paradigm for antiangiogenic therapy through controlled release of bevacizumab from PLGA nanoparticles. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clerici, M.; Nelemans, L.C.; Buzgo, M.; Simaite, A. Pitfalls of Accurate Protein Determination inside PLGA Nanoparticles Using the Micro BCA Assay. Proceedings 2021, 78, 28. https://doi.org/10.3390/IECP2020-08671
Clerici M, Nelemans LC, Buzgo M, Simaite A. Pitfalls of Accurate Protein Determination inside PLGA Nanoparticles Using the Micro BCA Assay. Proceedings. 2021; 78(1):28. https://doi.org/10.3390/IECP2020-08671
Chicago/Turabian StyleClerici, Marta, Levi Collin Nelemans, Matej Buzgo, and Aiva Simaite. 2021. "Pitfalls of Accurate Protein Determination inside PLGA Nanoparticles Using the Micro BCA Assay" Proceedings 78, no. 1: 28. https://doi.org/10.3390/IECP2020-08671
APA StyleClerici, M., Nelemans, L. C., Buzgo, M., & Simaite, A. (2021). Pitfalls of Accurate Protein Determination inside PLGA Nanoparticles Using the Micro BCA Assay. Proceedings, 78(1), 28. https://doi.org/10.3390/IECP2020-08671




